Abstract
Background:
Mounting evidence showed that microRNAs may be useful as prognostic biomarkers of cancer. Therefore, we summarize the predictive role of microRNA-218 (miR-218) for survival in patients with various cancers.
Methods:
We performed a systematic literature review and assessed the quality of included studies based on Meta-analysis of Observational Studies in Epidemiology group (MOOSE). Hazard ratios (HRs) with corresponding 95% confidence intervals (CIs) were calculated to assess the correlation between miR-218 expression and prognosis of different cancers.
Results:
We identified 10 studies for pooled analyses. For overall survival, a lower expression levels of miR-218 significantly predicted poorer survival, with the pooled HR of 2.61 (95% CI: 2.11–3.22, P < 0.001). For disease-free survival/progressive-free survival/recurrence-free survival (DFS/PFS/RFS), a lower expression level of miR-218 significantly predicted worse DFS/PFS/RFS in various carcinomas, with the pooled HR of 2.73 (95% CI: 2.08–3.58, P < 0.001). Similarly, subgroup analysis by detection method, ethnicity and cancer subtype analysis suggested that lower expression of miR-218 correlated with.
Conclusion:
Our data demonstrated that lower miR-218 expression is significantly associated with poorer overall survival (OS) and DFS/PFS/RFS and may be a novel prognostic biomarker in some cancer types.
Keywords: cancer, miR-218, prognosis, quantitative evaluation
1. Introduction
Cancer is a major public health problem in the world.[1] Although overall cancer mortality decreased by 20% between 1991 and 2010, cancer remains one of the most common causes of death worldwide.[2] The prognosis in the most cancers remains unsatisfactory, especially for advanced-stage tumors. Tumor metastasis is a complex process and a major cause of cancer deaths.[3] Therefore, it is necessary to identify valuable molecular biomarkers to promote early detection, prognostic classification, and novel therapeutic strategies for cancers.
MicroRNAs (miRNAs) are evolutionary conserved, small noncoding molecules with approximately 22 nucleotides in length, which could bind to complementary sequences in the 3′ untranslated region (3′UTR) of target mRNAs, leading to mRNA degradation or translational repression.[4] They have been shown to regulate multiple biological processes such as cell proliferation, cell differentiation, cell apoptosis, and cell cycle regulation.[5,6] Mounting evidence suggests that some miRNAs may function as oncogenes or tumor suppressors by regulating cell proliferation and other related biological behaviors.[7,8]
MicroRNA-218 (miR-218) belongs to the silt gene family, target recognition and regulatory functions as a onco-suppressor gene.[9,10] Several studies have reported that miR-218 expression was significantly downregulated in cancer tissues and played a role in cancer progression.[11,12] The role of miR-218 in the identification and characterization of tumor-initiating cells in cancers may provide new insight into understanding the relation of molecular mechanisms of tumor development.[13] Therefore, the development of new therapy options is essential.
Recent studies showed that miRNAs are associated with prognosis in various carcinomas, suggesting that they could be developed as prognostic classifiers to guide therapeutic decisions. We performed the systematic review of the data available from studies published in this field with the main aim of evaluating the role of miR-218 as a prognostic biomarker in cancer.
2. Materials and methods
Ethics committee is not applicable in this meta-analysis.
The present study was performed in accordance with the guidelines of the Meta-analysis of Observational Studies in Epidemiology group (MOOSE) issued by Stroup et al[14] and Preferred Reporting Items for Systematic Review and Meta-analyses (PRISMA) criteria.[15]
2.1. Literature search strategy
We systematically searched PubMed, Embase, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang database to identify potential studies before January 1, 2016. The search strategy employed terms related to “microRNA-218” or “miR-218” and “neoplasms” or “cancer.” The search was limited to papers published in English or Chinese language. In addition, reference lists of retrieved articles were examined manually to further identify missing relevant publications.
2.2. Inclusion and exclusion criteria
Two reviewers (FD and LD) independently assessed eligibility of the retrieved articles. Studies were included in the analysis if the following criteria were met: the study subjects were patients with any type of cancer; miR-218 expression was measured in tumor tissue or serum; investigated the survival outcome or the correlation between miR-218 expression and the clinical variables; and the full-text article was available in English or Chinese. Studies were excluded based on the following criteria: reviews, laboratory studies or letters; non-English or Chinese articles; lacked key information regarding survival outcomes, such as HRs or 95% confidence intervals (95% CIs) or unable to calculate such parameters.
2.3. Data extraction and quality assessment
Two investigators (FD and KW) evaluated and extracted the data independently from all eligible studies under the guideline of a critical review checklist. Data for analyses, including first author, year of publication, origin country, histology, sample type and size, assay, follow-up and cutoff value, HRs of miR-218 for overall survival (OS) and/or disease-free survival (DFS), progressive-free survival (PFS), recurrence-free survival (RFS), and the corresponding 95% CIs. If not available, data were calculated following Tierney et al's method.[16] If discrepancies existed, consensus would be finally reached on discussion.
The methodological quality of each study was systematically assessed according to a critical review checklist of the Dutch Cochrane Centre proposed by MOOSE to ensure their quality.[14] The key points of the basic standard are as follows: study origin of country and population, type of carcinoma, study design, outcome assessment, measurement of miR-218, cut-off of miR-218, and sufficient follow-up. The study was removed if not including the basic standard to avoid compromised quality of the meta-analysis.
2.4. Statistical analysis
We utilized RevMan 5.3 (Cochrane Collaboration, Oxford, UK) and STATA 13.1MP (StataCorp, College Station, TX) to perform all the statistical analysis.
All of the HRs and corresponding 95% CIs were used to calculate the pooled HR. Cochran Q test and Higgins I2 statistic were used to assess heterogeneity, if P-value for heterogeneity test (Pheterogeneity) < 0.05 or I2 > 50%, the sources of heterogeneity would be used for meta-regression.[17] Random or fixed-effects models were used depending on Pheterogeneity. If Pheterogeneity ≥ 0.05, we used the fixed effect model (the Mantel–Haenszel method).[18] Otherwise, random effects model (DerSimonian and Laird method) was selected.[19] The significance of merged HR was dependent on the Z test, P < 0.05 was considered statistically significant, all P values were 2-sided.
Sensitivity analysis, in which 1 study is omitted at a time, was performed to assess the quality and consistency of the results.
Publication bias was assessed by Begg test (rank correlation test)[20] and then statistically using Egger test (weighted linear regression test).[21]
3. Results
3.1. Literature search and summary of included studies
The initial literature search retrieved 1310 relevant studies and a flow diagram are shown in Fig. 1. One thousand one studies were removed because of duplication. After primary identified, 46 titles were potentially appropriate, and the corresponding abstracts were reviewed. After further identification and screening individual study, 11 eligible publications underwent full-text review, and 1 article[22] was further excluded because data were unavailable. Finally, we included 10 eligible studies[12,13,23–30] in the final evidence synthesis.
Figure 1.
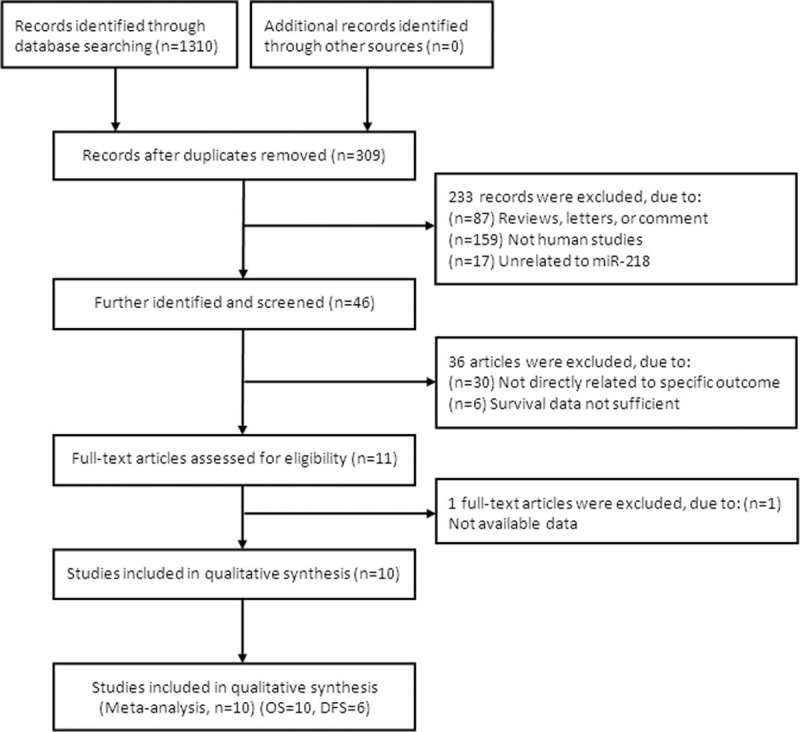
Flow chart of literature search and study selection.
The main characteristics of the eligible studies are summarized in Table 1. The eligible studies were published from 2010 to 2015 and included a total of 893 participants with OS data and 626 participants with DFS/PFS/RFS data from China, Taiwan, and Canada. The patients were classified as either Asian or Caucasian according to their ethnic background. The types of malignant cancers included colorectal cancer, nonsmall cell lung cancer (NSCLC), pancreatic cancer, oral cavity squamous cell carcinoma (OCSCC), nasopharyngeal carcinoma (NPC), glioma, and hepatocellular carcinoma (HCC). Frozen tissues or serum were used in eligible studies. Quantitative real-time PCR (qRT-PCR) was used in 8 studies, and immunohistochemical (IHC) was used in the remaining 2 studies.
Table 1.
Clinicopathological characteristics of eligible studies.
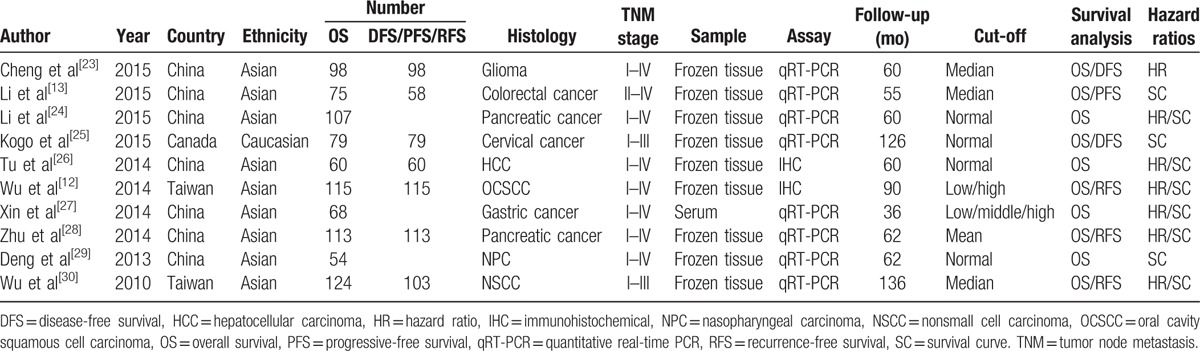
Among the eligible studies, 10 articles[12,13,23–30] evaluated both OS and DFS/PFS/RFS, 6 articles[12,13,23,25,28,30] evaluated DFS/PFS/RFS. Seven studies[12,23,24,26–28,30] directly reported HRs and 95% CIs, three studies[13,25,29] reported survival curve (SC).
3.2. Evidence synthesis and test of heterogeneity
The main results of this meta-analysis and the heterogeneity test are shown in Table 2. We firstly analyzed the association between miR-218 expression and OS, no significant heterogeneity have been found (I2 < 0.001%, P = 0.82). Therefore, the fixed effects were applied to calculate the pooled HR, a lower expression levels of miR-218 significantly predicted poorer survival, with the pooled HR of 2.61 (95% CI: 2.11–3.22, P < 0.001, Fig. 2). For evaluating the association between miR-218 expression and DFS/PFS/RFS, since the Q test of heterogeneity was not significant (I2 < 0.001%, P = 0.83), we conducted analyses using the fixed effect models. The result showed that a lower expression level of miR-218 significantly predicted worse DFS/PFS/RFS in various carcinomas, with the pooled HR of 2.73 (95% CI: 2.08–3.58, P < 0.001, Fig. 3).
Table 2.
Main results of pooled HRs in the meta-analysis.
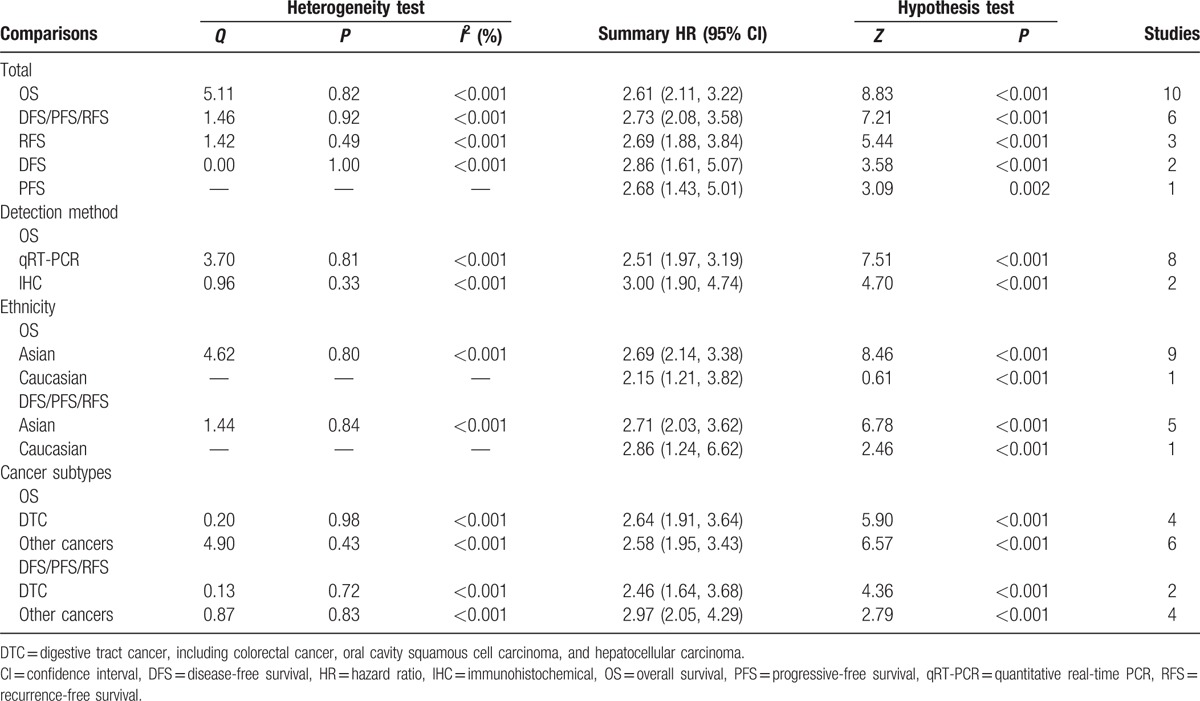
Figure 2.
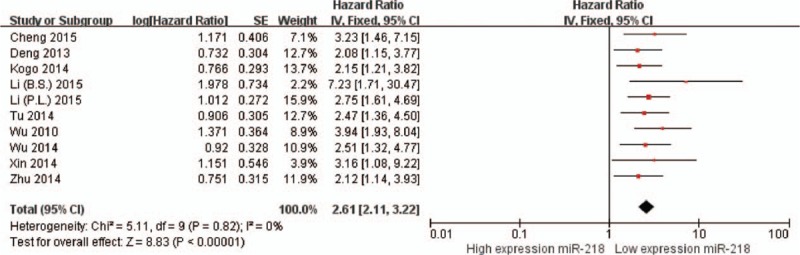
Forest plots of studies evaluating the HRs of high and low miR-218 expression with respect to OS. The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the study-specific weight. The diamond represents the pooled OR and 95% CI.
Figure 3.
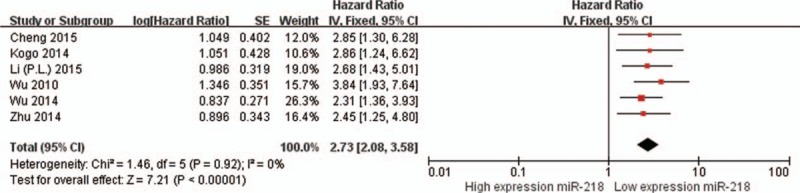
Forest plots of studies evaluating the HRs of high and low miR-218 expression with respect to DFS/PFS/RFS.
To explain the heterogeneity in OS, subgroup analysis was performed by detection method, significant relevance was observed both in qRT-PCR subgroup (HR = 2.51, 95% CI: 1.79–3.13, P < 0.001) and IHC subgroup (HR = 3.00, 95% CI: 1.90–4.74, P < 0.001). Considering the large proportion of Chinese patients in the studies, we carried out a stratified analysis by classifying studies into subgroups of ethnicity (Asian and Caucasian). The expression of miR-218 was significantly correlated with OS in Asians (HR = 2.69, 95% CI: 2.14–3.38, P < 0.001) and Caucasians (HR = 2.15, 95% CI: 1.21–3.82; P < 0.001) (Table 2) and expression of miR-218 significantly associated with DFS/PFS/RFS in Asians (HR = 2.71, 95% CI: 2.03–3.62, P < 0.001) and Caucasians (HR = 2.86, 95% CI: 1.24–6.62, P = 0.01) (Table 2). When grouped by the cancer types, we found that miR-218 expression was significantly correlated with digestive tract cancer (DTC) (HR = 2.64, 95% CI: 1.91–3.64, P < 0.001 for OS; HR = 2.46, 95% CI: 1.64–3.68, P < 0.001 for DFS/PFS/RFS) and other cancers groups (HR = 2.58, 95% CI: 1.95–3.43, P < 0.001 for OS; HR = 2.97, 95% CI: 2.05–4.29, P < 0.001 for DFS/PFS/RFS) (Table 2).
3.3. Sensitivity analysis
Sensitivity analysis was performed through systematic omitting 1 study each time and calculating the pooled HRs again. As shown in Figs. 4 and 5, the stability of the entire study was not influenced by 1 individual study.
Figure 4.
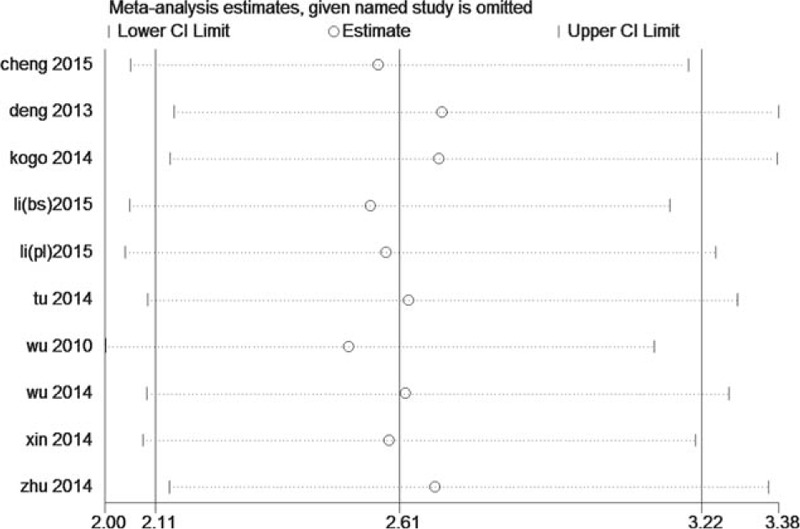
Sensitivity analysis for OS of miR-218.
Figure 5.
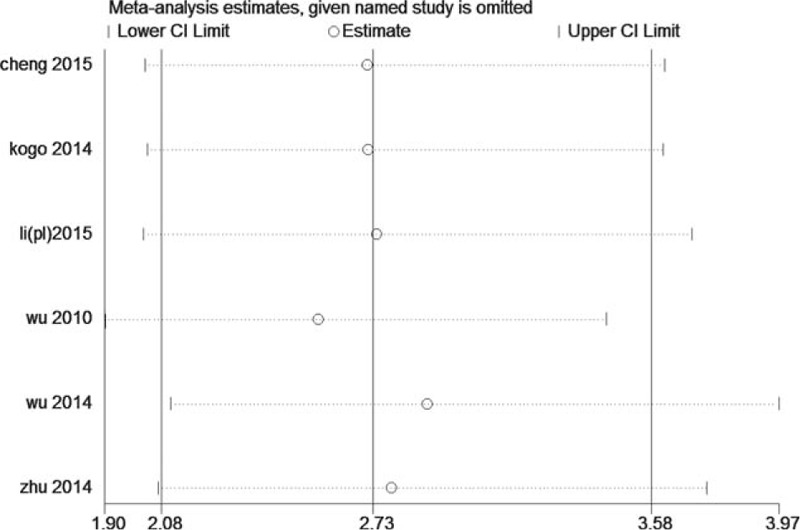
Sensitivity analysis for DFS/PFS/RFS of miR-218.
3.4. Evaluation of publication bias
Begg funnel plot and Egger linear regression test were performed to assess the publication biases of OS and DFS/PFS/RFS among included studies. The shape of the funnel plot did not reveal any evidence of obvious asymmetry (Table 3, Fig. 6A and B). Egger regression was used to provide statistical evidence of funnel plot symmetry, indicating that there was no significant publication bias (Table 3).
Table 3.
Publication bias of miR-218 for Begg test and Egger test.

Figure 6.
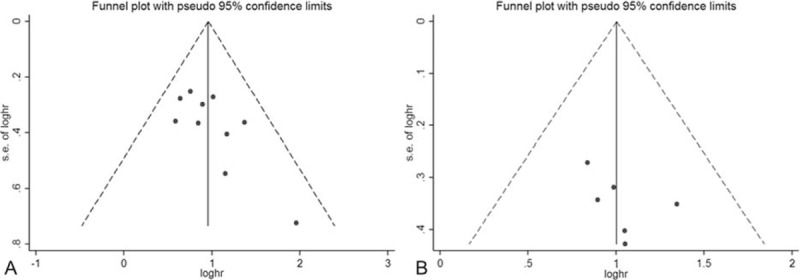
(A) Funnel plot of publication bias on the relationship between miR-218 expression and OS. The vertical line in the funnel plot indicates the fixed-effects summary estimate, whereas the sloping lines indicate the expected 95% CI for a given SE. (B) Funnel plot of publication bias on the relationship between miR-218 expression and DFS/PFS/RFS.
4. Discussion
Recently, mounting evidence shows that miRNAs in cancer research has substantially changed the understanding of gene regulation, as an important cellular molecules involved in the normal and pathological states,[31] miRNAs are important regulators of gene expression in tumor development by target genes and tumor suppressors or via directly exerting corresponding functions as oncogenes or tumor suppressors.[32,33] In recent years, numerous studies have investigated that aberrantly expressed miRNAs in different types of cancer, they can be used as novel prognostic biomarkers of tumor.[34–36]
MiR-218 is a vertebrate-specific miRNA that has been predicted and experimentally confirmed to play a crucial role in tumorigenesis and tumor progression by regulating the expression of potential targets.[37,38] MiR-218 have found to serve as a candidate tumor suppressor in targeting multiple cancer by regulation of relative gene expression.[39,40] Mathew et al[41] identified a miR-218-RTK-HIF2α signaling axis which promotes tumor angiogenesis and glioblastoma multiforme (GBM) cell survival, especially for necrotic mesenchymal tumors. Meanwhile, it was demonstrated that silencing of miRNA-21 promotes migration and invasion of breast cancer through Slit2-Robo1 pathway.[42] Importantly, these results suggested that miR-218 acts as a potential tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma, including RICTOR, CDK6, and cathepsin B (CTSB).[43,44] However, significance of miR-218 expression with clinicopathological factors and/or prognosis of cancers are unclear.
In the present study, we conducted this analysis of the published literature to identify a group of miR-218 for which the data support validation as prognostic biomarkers of cancer outcomes. Due to the included studies used a variety of indices to evaluate tumor progression, such as DFS, PFS, and RFS, we combined these indices to evaluate the prognostic value of miR-218.
To the best of our knowledge, our meta-analysis is the first to critically examine available literature and identify the prognostic role of miR-218 in various cancers. The results demonstrated that expression of miR-218 was significantly correlated with OS (HR = 2.61, 95% CI: 2.11–3.22, P < 0.001) and DFS/PFS/RFS (HR = 2.73, 95% CI: 2.08–3.58, P < 0.001) in cancer, further demonstrating the predictive value of miR-218. Our stratified analysis suggested a closer relationship between rising miR-218 levels and poor survival in Asians and Caucasians. Among 10 studies reporting, four were related to DTC. Therefore, we performed a subgroup analysis of DTC. The result also revealed that reduced miR-218 yielded worse OS and DFS/PFS/RFS in DTC. Due to the lack of eligible studies reporting for each cancer type, further studies are required to determine whether pathological cancer types impact the prognostic role of miR-218.
Studies show that the main reason for the high mortality of cancer is the invasion and metastasis.[44] Elevated expression of miR-218 inhibited the invasion and migration of cancer cells,[45–47] it is currently believed that several types of deregulated miR-218 and its downregulation is associated with a poor prognosis.[48] These results show that miR-218 play a tumor suppressor and decreased miR-218 expression in the tissue or serum was associated with OS and DFS/PFS/RFS. However, numerous published studies have been reported that miR-218 can regulate tumor invasion,[49,50] the exact clinicopathologic significance and prognostic of miRNA-218 in cancers remain inconclusive.
Although meta-analysis is robust, our study also has several limitations that should be acknowledged. Firstly, the reliability of our results is questionable in light of the number of eligible studies for OS and DFS/RFS/RFS. Additionally, the patient populations were limited to Asia, and North America, lacking data from other regions, which might impact the statistical power of analysis, and ethnic bias might be possible, even though the statistical test did not show it. Secondly, the number of individual prognostic studies dealing with certain tumor type was not sufficient, which might impact the statistical power of analysis. Therefore, well-designed clinical studies with larger sample sizes should be carried out in the future. Thirdly, a clear definition should be made about the cutoff value of miR-218 level for outcomes. To date, most investigators use median or mean value in their studies as the cutoff value and the accurate value were different. Fourthly, due to not all survival data of the eligible studies were given directly, some data were extracted from survival curves. These calculated HRs with corresponding 95% CIs might be brought several tiny errors. Finally, although there was no significant evidence of publication bias in this analysis, cautions should be taken, and the tendency for journals to publish positive results could also make certain bias.
In summary, our data demonstrated that lower miR-218 expression is significantly associated with poorer OS and DFS/PFS/RFS and may be a novel prognostic biomarker in some cancer types, further multicenter prospective clinical studies are needed to determine the association between miR-218 and cancer prognosis.
Footnotes
Abbreviations: CIs = confidence intervals, DFS = disease-free survival, HRs = hazard ratios, miR-218 = microRNA-218, miRNAs = microRNAs, PFS = progressive-free survival, RFS = recurrence-free survival.
Funding: This research was supported by People's Republic of China National Natural Science Foundation of China (no. 81202278) and Excellent Youth Foundation of He’nan Scientific Committee (no. 124100510007).
Author contributions: All authors contributed significantly to this work. FD and KW designed and drafted the manuscript. LD, XZ, and SC collected studies, summarized data, and copyedited this article. YF and CS collected articles, summarized data, and did statistical work. All authors reviewed this manuscript and approved the final draft.
The authors have no conflicts of interest to disclose.
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010; 60:277–300. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014; 64:9–29. [DOI] [PubMed] [Google Scholar]
- 3.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006; 127:679–695. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012; 13:271–282. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 6.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009; 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nohata N, Hanazawa T, Kinoshita T, et al. MicroRNAs function as tumor suppressors or oncogenes: aberrant expression of microRNAs in head and neck squamous cell carcinoma. Auris Nasus Larynx 2013; 40:143–149. [DOI] [PubMed] [Google Scholar]
- 8.Babashah S, Soleimani M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer 2011; 47:1127–1137. [DOI] [PubMed] [Google Scholar]
- 9.He X, Dong Y, Wu CW, et al. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Mol Med 2013; 8:1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu Y, Gao X, Li G, et al. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res 2013; 73:6046–6055. [DOI] [PubMed] [Google Scholar]
- 11.Gao X, Jin W. The emerging role of tumor-suppressive microRNA-218 in targeting glioblastoma stemness. Cancer Lett 2014; 353:25–31. [DOI] [PubMed] [Google Scholar]
- 12.Wu D-W, Chuang C-Y, Lin W-L, et al. Paxillin promotes tumor progression and predicts survival and relapse in oral cavity squamous cell carcinoma by microRNA-218 targeting. Carcinogenesis 2014; 35:1823–1829. [DOI] [PubMed] [Google Scholar]
- 13.Li P-L, Zhang X, Wang L-L, et al. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis 2015; 36:1484–1493. [DOI] [PubMed] [Google Scholar]
- 14.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. JAMA 2000; 283:2008–2012. [DOI] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 2009; 151:264–269. [DOI] [PubMed] [Google Scholar]
- 16.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007; 8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson SG, Higgins JP. How should meta-regression analyses be undertaken and interpreted? Stat Med 2002; 21:1559–1573. [DOI] [PubMed] [Google Scholar]
- 18.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies. J Natl Cancer Inst 1959; 22:719–748. [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177–188. [DOI] [PubMed] [Google Scholar]
- 20.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994; 50:1088–1101. [PubMed] [Google Scholar]
- 21.Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alajez NM, Lenarduzzi M, Ito E, et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res 2011; 71:2381–2391. [DOI] [PubMed] [Google Scholar]
- 23.Cheng MW, Wang LL, Hu GY. Expression of microRNA-218 and its clinicopathological and prognostic significance in human glioma cases. Asian Pac J Cancer Prev 2015; 16:1839–1843. [DOI] [PubMed] [Google Scholar]
- 24.Li BS, Liu H, Yang WL. Reduced miRNA-218 expression in pancreatic cancer patients as a predictor of poor prognosis. Genet Mol Res 2015; 14:16372–16378. [DOI] [PubMed] [Google Scholar]
- 25.Kogo R, How C, Chaudary N, et al. The microRNA-218∼Survivin axis regulates migration, invasion, and lymph node metastasis in cervical cancer. Oncotarget 2015; 6:1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu K, Li C, Zheng X, et al. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep 2014; 32:1571–1577. [DOI] [PubMed] [Google Scholar]
- 27.Xin S-Y, Feng X-S, Zhou L-Q, et al. Reduced expression of circulating microRNA-218 in gastric cancer and correlation with tumor invasion and prognosis. World J Gastroenterol 2014; 14:6906–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Xu Y, Du J, et al. Expression of microRNA-218 in human pancreatic ductal adenocarcinoma and its correlation with tumor progression and patient survival. J Surg Oncol 2014; 109:89–94. [DOI] [PubMed] [Google Scholar]
- 29.Deng M, He Z-m, Liu J-f, et al. Expression of microRNA-218 in nasopharyngeal carcinoma and its clinical significance. TUMOR 2013; 33:177–180.189. [Google Scholar]
- 30.Wu D-W, Cheng Y-W, Wang J, et al. Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res 2010; 70:10392–10401. [DOI] [PubMed] [Google Scholar]
- 31.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014; 20:460–469. [DOI] [PubMed] [Google Scholar]
- 32.Cho WC. MicroRNAs in cancer—from research to therapy. Biochim Biophys Acta 2010; 1805:209–217. [DOI] [PubMed] [Google Scholar]
- 33.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol 2010; 42:1273–1281. [DOI] [PubMed] [Google Scholar]
- 34.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010; 101:2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem 2009; 55:623–631. [DOI] [PubMed] [Google Scholar]
- 36.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012; 4:143–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassan MQ, Maeda Y, Taipaleenmaki H, et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem 2012; 287:42084–42092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Zhang E, Lin C. MicroRNAs in tumor angiogenesis. Life Sci 2015; 136:28–35. [DOI] [PubMed] [Google Scholar]
- 39.Zhang BG, Li JF, Yu BQ, et al. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep 2012; 27:1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatarano S, Chiyomaru T, Enokida H, et al. 505 miR-218 on the genomic loss region of chromosome 4p15. 31 Functions as a tumor suppressor in bladder cancer. J Urol 2011; 185:e205. [DOI] [PubMed] [Google Scholar]
- 41.Mathew LK, Skuli N, Mucaj V, et al. miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. Proc Natl Acad Sci 2014; 111:291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, Li Q, Wang Q, et al. Silencing of miRNA-218 promotes migration and invasion of breast cancer via Slit2-Robo1 pathway. Biomed Pharmacother 2012; 66:535–540. [DOI] [PubMed] [Google Scholar]
- 43.Venkataraman S, Birks DK, Balakrishnan I, et al. MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J Biol Chem 2013; 288:1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang BB, Liu SY, Zhan Y, et al. microRNA-218 expression and its association with the clinicopathological characteristics of patients with cervical cancer. Exp Ther Med 2015; 10:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin J, Cai L, Liu Z-M, et al. miRNA-218 inhibits osteosarcoma cell migration and invasion by down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev 2013; 14:3681–3684. [DOI] [PubMed] [Google Scholar]
- 46.He H, Hao S-j, Yao L, et al. MicroRNA-218 inhibits cell invasion and migration of pancreatic cancer via regulating ROBO1. Cancer Biol Ther 2014; 15:1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishikawa R, Goto Y, Sakamoto S, et al. Tumor-suppressive microRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci 2014; 105:802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Yu X, Shen J, et al. MicroRNA expression and its implications for diagnosis and therapy of gallbladder cancer. Oncotarget 2015; 6:13914–13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uesugi A, Kozaki K-i, Tsuruta T, et al. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res 2011; 71:5765–5778. [DOI] [PubMed] [Google Scholar]
- 50.Song L, Huang Q, Chen K, et al. miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-β. Biochem Biophys Res Commun 2010; 402:135–140. [DOI] [PubMed] [Google Scholar]


