The biographer Ronald Clark once described the influential evolutionary mathematician J.B.S. Haldane as “…among the last of the ‘string-and-sealing-wax' scientists, the men who could achieve results by themselves, without the aid of a committee, and who could, without the help of another committee, see how their discoveries fitted into the larger pattern of scientific knowledge” (Clark, 1984). This sentiment is also true for the equally influential evolutionary botanists Ledyard Stebbins and Edgar Anderson. Indeed, the last decade has witnessed a renaissance of studies (e.g., Arnold, 1992, 1997, 2004; Grant and Grant, 1992, 2002; Dowling and DeMarais, 1993; Rieseberg, 1997; Rieseberg et al., 2003) into the evolutionary role of natural hybridization—for example, the natural interbreeding of individuals from wild populations that can be distinguished on the basis of one or more heritable characters (Harrison, 1990; Arnold, 1997)—the process explored and described extensively by these investigators (e.g., Anderson and Stebbins, 1954). Through new insights and investigations, the general conceptual framework proposed by these authors, that natural hybridization was evolutionarily creative through the origin of new species and adaptations (or the transfer of adaptations between the hybridizing forms), has been extended greatly in the last 10 years. However, just as the work of J.B.S. Haldane remains broadly applicable to evolutionary concepts (e.g., Haldane, 1948, 1949), so also do the original experimental and conceptual studies of Anderson and Stebbins (e.g., Anderson and Hubricht, 1938; Anderson, 1948, 1949; Stebbins, 1959).
In this essay, I will consider only one of the two creative outcomes of the process of natural hybridization emphasized by these investigators, the origin/transfer of adaptations. As Anderson (1948) stated, “The commonest result of hybridization is introgression” (i.e., the transfer of genetic material between hybridizing taxa through backcrosses; Anderson and Hubricht, 1938). This is not to suggest that speciation via hybridization is less important than the transfer or origin of adaptive traits. Indeed, hybrid speciation is recognized as a major outcome when species cross in nature, particularly when one considers allopolyploid plant taxa (Stebbins, 1947; Grant, 1981; Arnold, 1997; Rieseberg, 1997). Furthermore, such hybrid speciation may result from the origin of novel adaptations (as discussed below; Rieseberg et al., 2003). Yet, I wish to emphasize the common conceptual framework of seminal work from the first half of the 20th century through to the present day that investigates the transfer and origin of adaptations through natural hybridization. I will argue that our contemporary studies are part of an historical lineage dating from at least the work of Stebbins and Anderson—that many of us, like Newton, are indeed “…standing on the shoulders of giants.”
INTROGRESSION OF ADAPTATIONS—EVIDENCE FROM PLANTS, ANIMALS, AND MICROBES
“If introgression proves to be a primary factor in evolution it will be because it so greatly enriches variation in the participating species…” (Anderson, 1949). This quote reflects the importance that Anderson, Stebbins, and other workers placed on the process of gene transfer between hybridizing forms. But the transfer of genetic material through introgression was not seen as the evolutionarily important end product. Instead, as Anderson and Stebbins (1954) hypothesized, “By introgressive hybridization elements of an entirely foreign genetic adaptive system can be carried over into a previously stabilized one, permitting the rapid reshuffling of varying adaptations and complex modifier systems. Natural selection is presented…with segregating blocks of genic material belonging to entirely different adaptive systems.”
The transfer of adaptations from one of the hybridizing forms into the other could lead to a geographic range extension by the introgressed taxon, reflecting the invasion of ecological settings available previously to only the alternate parental taxon. An examination of evolutionary literature published during and subsequent to the neo-Darwinian synthesis identifies empirical and conceptual studies that describe phenomena consistent with the transfer of adaptations through introgressive hybridization. It is a truism that a comparative approach is necessary for strong evolutionary inferences. In this regard, I will discuss seven examples, including three cases for plants (Iris: Riley, 1938; Anderson, 1949; Arnold and Bennett, 1993; Helianthus: Heiser, 1951; Kim and Rieseberg, 1999; Cowania/Purshia: Stutz and Thomas, 1964) and two examples each from animals (Dacus, now Bactrocera: Lewontin and Birch, 1966; Morrow et al., 2000; Anopheles: Wang et al., 2001; Gentile et al., 2002; Besansky et al., 2003) and microbes (Haemophilus influenzae: Kroll et al., 1998; Smoot et al., 2002; Trypanosoma cruzi: Machado and Ayala, 2001).
Iris
Anderson (1949) used the Louisiana Iris species complex and, in particular, Iris fulva and I. hexagona as his typical example for the process of introgressive hybridization. Natural hybridization between these species was seen as a type for understanding the process of introgression from its initiation to its diverse evolutionary and ecological outcomes. Because Anderson (and Stebbins) viewed the potential evolutionary outcomes of this process as pervasive and important, it is instructive to reexamine one of the examples thought by them to be so illustrative. In particular, given the current topic, it is important to ask if there is evidence for the exchange of adaptations between these species, allowing the alternate forms to invade habitats characteristic of their partner in hybridization. The evidence for the transfer of adaptations between I. fulva and I. hexagona comes from both experimental and descriptive studies.
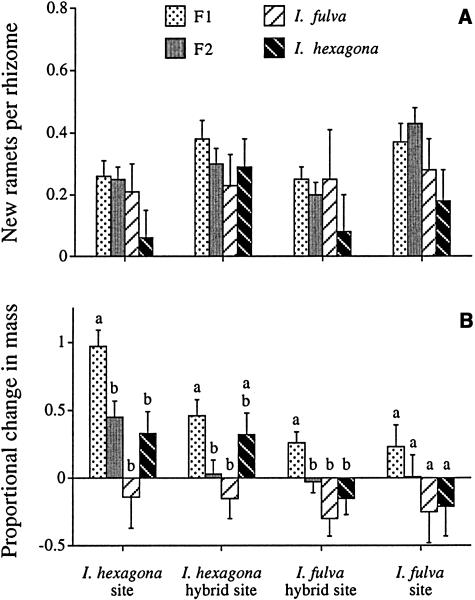
It is now well established that hybridization and introgression between I. fulva and I. hexagona (and a third species, I. brevicaulis) is common and widespread (Viosca, 1935; Riley, 1938; Arnold, 1994). This introgression has resulted in the transfer of genetic material in areas of sympatry and into allopatric populations of each species (Arnold et al., 1990; Arnold, 1993). In addition, these species and their hybrids differ in their ecological adaptations, as evidenced by their demonstrating a range of fitnesses across different environments. For example, by exposing the different genotypes to various levels of light reduction, Bennett and Grace (1990) discerned that I. fulva possessed a high level of shade tolerance compared with I. hexagona. They also found that hybrid genotypes demonstrated different fitnesses when exposed to a range of light reductions (Bennett and Grace, 1990). Finally, Emms and Arnold (1997) detected varying fitness estimates for these species and their hybrids when the genotypes were placed into natural settings in southern Louisiana (Figure 1).
Figure 1.
Clonal Reproduction of Transplanted Rhizomes of I. fulva, I. hexagona, and Their F1 and F2 Hybrids.
(A) Probability of new ramet production. Data show least-squares means + 1 se of genotype means for each plant class at each site (i.e., means are adjusted for the effect of rhizome mass).
(B) Proportional change in rhizome mass. Bars sharing a letter within each site do not differ significantly at P < 0.05 after adjustment for multiple comparisons (from Emms and Arnold, 1997).
The final piece of evidence that indicates the possible role of introgression in transferring the traits underlying these alternate adaptations comes from analyses of natural hybrid zones (i.e., a region in which natural populations of individuals that are distinguishable on the basis of one or more heritable characters overlap spatially and temporally and cross to form viable and at least partially fertile offspring; Arnold, 1997). In particular, Arnold and Bennett (1993) reported a significant association between the proportion of I. fulva genetic material in introgressed I. hexagona plants and the degree of natural light reduction they experienced. Although these plants were phenotypically I. hexagona, the individuals that possessed higher frequencies of I. fulva markers occurred in areas characterized by significantly lower light levels. This finding—I. hexagona-like plants introgressed by I. fulva growing within shade levels known to significantly reduce the fitness of I. hexagona (Bennett and Grace, 1990)—suggests strongly that the genetic architecture of the shade tolerance adaptation has been introgressed into these hybrid plants.
Helianthus
The second case of putative adaptive introgression involves the plant species Helianthus annuus and H. debilis ssp cucumerifolius. Heiser (1951) proposed that the morphological similarity of H. annuus populations in eastern Texas to H. debilis ssp cucumerifolius was attributable to introgression from the latter species into the former. Contemporary hybrid populations were identified between these two species, supporting the hypothesis of introgression. Furthermore, Heiser (1951) argued that this introgression likely involved the transfer of H. debilis ssp cucumerifolius ecological adaptations into H. annuus. He thus stated, “Helianthus annuus when it was first introduced…may have been poorly adapted and hence there might have been some selective premium placed on those hybrid forms which contained genes from H. debilis var cucumerifolius, a species already well adapted to this area.”
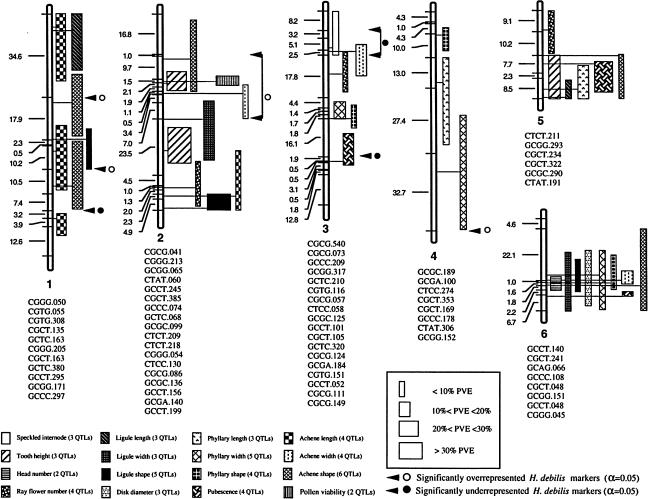
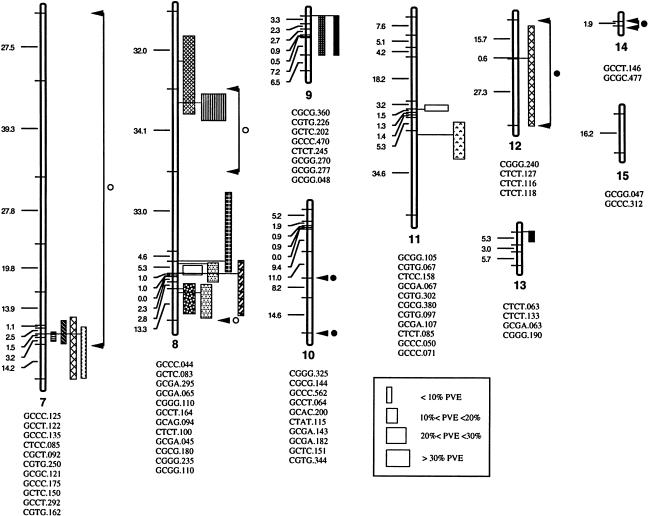
Kim and Rieseberg (1999) tested the above hypothesis using an elegant analysis of quantitative trait loci (QTL) variation associated with species-specific morphological characteristics (Figure 2). Using this approach, these authors demonstrated that the introgression of three chromosomal blocks from H. debilis ssp cucumerifolius into H. annuus was sufficient to produce the unique H. annuus phenotype found in the eastern Texas populations. Though this study was not designed to test directly the fitness effects of these QTL, the fact that these elements recreate the introgressed H. annuus phenotype suggests their involvement in adaptive trait introgression.
Figure 2.
Linkage Map and Genomic Positions of QTL Affecting Species-Specific Vegetative and Floral Characteristics Found in H. annuus and H. debilis ssp cucumerifolius.
Horizontal lines indicate genetic marker locations, with their designations given below each linkage group. Map distances between markers are indicated by numerals to the left of linkage groups. QTL positions and the magnitude of their effect are indicated to the right of the linkage groups (from Kim and Rieseberg, 1999).
Cowania/Purshia
The final example I wish to discuss, for which adaptive trait introgression among hybridizing plant species has been hypothesized, comes from the work of Stutz and Thomas (1964). These authors described extensive hybridization between the morphologically and ecologically divergent Cowania stansburyana (cliffrose) and Purshia tridentata (bitterbrush). Though the area of sympatry and hybridization included “…nearly the entire state of Utah…” (Stutz and Thomas, 1964), the two species were largely separated because of C. stansburyana and P. tridentata occurring at elevations of 5000 to 8000 feet and 3000 to 6500 feet, respectively. However, where the species overlapped, hybridization was nearly ubiquitous; thus, nonintrogressed populations in Utah were rarely detected (Stutz and Thomas, 1964).
Two findings by Stutz and Thomas (1964) suggested that introgression had facilitated the transfer of ecological adaptations. The first involved the southernmost Purshia populations. These populations (1) possessed hybrid index scores (i.e., a numeric assignment reflecting the proportion of Purshia and Cowania morphological traits) indicative of introgression from Cowania and (2) occurred in atypical (for Purshia) xeric settings. Stutz and Thomas (1964) argued that introgression from xeric-adapted Cowania populations had allowed the invasion of the southern Purshia populations into Cowania-like habitats. The second observation suggesting an adaptive role for introgression from Cowania into Purshia involved populations of the latter species that were found further north than any Cowania populations. In these northern populations, the introgression of genes from Cowania causing unpalatability was strongly inferred. Indeed, selection was hypothesized to have driven this favorable introgressed phenotype to near fixation far north of any contemporary area of overlap (Stutz and Thomas, 1964).
Bactrocera
Lewontin and Birch (1966) were the first to propose the now classic zoological example of adaptive trait introgression involving the Australian fruit fly species Bactrocera tryoni and B. neohumeralis (formerly Dacus tryoni and D. neohumeralis). According to these authors, hybridization between the Bactrocera species had resulted in (1) the introgression of a morphological marker and (2) outbreaks of B. tryoni on temperate crops, such as apples and peaches, as well as tropical fruit species, such as guavas. It was proposed that introgression from B. neohumeralis into B. tryoni allowed the latter species to expand its range into additional areas of cultivation. Although this hypothesis was subsequently discounted (Gibbs, 1968; Birch and Vogt, 1970), more recent applications of molecular genetic markers have supported the original conclusion that B. neohumeralis genes have introgressed into B. tryoni (Morrow et al., 2000). Pike et al. (2003) found that the cause of the asymmetry in gene flow between these two species—from B. neohumeralis into B. tryoni—was a difference in mating period. In conclusion, the hypothesis that introgression can greatly expand a pest species' niche (Lewontin and Birch, 1966) is supported by findings from the Bactrocera complex.
Anopheles
The second zoological example I wish to highlight involves the malarial vectors, Anopheles gambiae and An. arabiensis. It is important to first emphasize that significant genetic discontinuities, indicating a level of subspecific reproductive isolation, have been detected within the widespread An. gambiae (Gentile et al., 2002). Reproductive barriers within this species are reflected by the co-occurrence, in single populations, of different genotypes at the rDNA cistron. Indeed, different genotypic variants were discovered for both the internal transcribed spacer and intergenic spacer regions (Gentile et al., 2002). The significant reproductive barriers within this species were apparent from the presence of (1) no heterozygotes for the different variants and (2) nearly total linkage disequilibrium for the pairs of internal transcribed spacer and intergenic spacer genotypes defined as S/type I and M/type II (Gentile et al., 2002). By contrast, other regions of the genome demonstrated no such genetic discontinuities (Wang et al., 2001; Gentile et al., 2002). These opposite patterns of introgression versus no introgression of different genomic regions indicate the semipermeability (Key, 1968) of the S and M genomes; some regions recombine at a high frequency, whereas others, like the rDNA cistron, recombine rarely.
The semipermeability of genomes is also apparent when we consider the genetic architectures of the sibling species An. gambiae and An. arabiensis. In particular, Lanzaro et al. (1998) and Besansky et al. (1997) reported limited and extensive introgression of nuclear and mitochondrial markers, respectively. Recent studies have supported the hypothesis of past and contemporary introgression between these two species (Wang et al., 2001; Besansky et al., 2003). With regard to the present topic, the most important conclusion was stated in the following manner: “The proposed acquisition by An. gambiae of sequences from the more arid-adapted An. arabiensis may have contributed to the spread and ecological dominance of this malaria vector” (Besansky et al., 2003). Like irises, sunflowers, and Australian fruit flies, the change in ecological tolerances and subsequent invasion of this disease vector is likely because of introgressive hybridization.
Trypanosoma
Trypanosoma cruzi is the protozoan parasite responsible for American trypanosomiasis or Chagas' disease. This protozoan species is well known for its genetic and phenotypic variability (Machado and Ayala, 2001). To test whether this variability was attributable to natural hybridization, Machado and Ayala (2001) used an analysis of DNA sequence variation at unlinked genes. They concluded that natural hybridization had played an important role in the evolutionary history of the T. cruzi lineage. Furthermore, they proposed that the variation in the adaptations associated with growth rate, pathogenicity, infectivity, and drug susceptibility was because of introgression between divergent lineages.
Haemophilus
The final example of the transfer of adaptations via introgressive hybridization comes from the bacterial species Haemophilus influenzae. This species is one of the most commonly isolated microorganisms from the human upper respiratory tract (Kroll et al., 1998). Although normally nonpathogenic, it sometimes becomes invasive, leading to serious illnesses. One of the diseases caused by H. influenzae is Brazilian purpuric fever. This disease was originally detected within a Brazilian population of children, 70% of whom died from its effects. Because Brazilian purpuric fever was characterized by meningococcal sepsis, it was originally assumed to be caused by a meningococcus species rather than by H. influenzae (Kroll et al., 1998). However, the disease was ultimately found to be caused by an infection of H. influenzae biogroup aegyptius, an organism previously known to cause only conjunctivitis (Kroll et al., 1998). H. influenzae biogroup aegyptius had thus somehow acquired the meningococcal phenotype. Kroll et al. (1998) hypothesized that the acquisition of this phenotype resulted from the transfer of virulence genes from Neisseria meningitidis into H. influenzae. This hypothesis was supported when Smoot et al. (2002) reported evidence of introgression from the pathogen into the previously nonpathogenic H. influenzae. This evidence also supported the conclusion of Kroll et al. (1998) that the transfer of the adaptations between these species facilitated the origin of a “…clone with the phenotype of rapidly fatal invasive infection….”
ORIGIN OF NOVEL ADAPTATIONS THROUGH NATURAL HYBRIDIZATION—EVIDENCE FROM PLANTS (AND MICROBES)
As with the examples of the transfer of adaptations through introgression, the de novo origin of adaptations not found in either of the hybridizing taxa would allow an ecological and geographical spread of hybrid forms. However, the origin of novel adaptations would facilitate the invasion of habitats not utilized previously by either parent (Anderson and Stebbins, 1954). The processes of exchange and origin of adaptations through natural hybridization are conceptually similar and somewhat difficult to differentiate. However, a major difference in the outcomes of the two processes is that the origin of novel adaptations may facilitate the formation of stabilized hybrid species (Grant, 1981; Arnold, 1997; Rieseberg, 1997). Anderson and Stebbins (1954) argued that this was likely because of the advent of “…new adaptive systems, adapted to new ecological niches…” (Anderson and Stebbins, 1954). I will use two examples to illustrate the process whereby ecological adaptations have arisen through hybridization between divergent lineages. The first comes again from the annual sunflowers and the second from the plant pathogen genus Phytophthora.
Helianthus
Natural hybridization between the annual sunflower species H. annuus and H. petiolaris has led to the production of at least three hybrid species (Heiser et al., 1969; Rieseberg, 1991). Yet, the fact that hybridization between these two species has led to hybrid species formation appears somewhat paradoxical. This is because early generation hybrids between H. annuus and H. petiolaris are highly infertile and inviable. For example, F1 individuals demonstrate pollen fertilities of 0 to 30%, and F2 and first backcross generation individuals produce no more than 2% viable seed (Heiser et al., 1969). Though seemingly counterintuitive, hybrids formed from strongly isolated lineages may stand a greater chance of surviving in the face of potential reproductive contact with their parents. In spite of the extremely low fitness of the initial hybrid generations, natural hybridization between H. annuus and H. petiolaris is ongoing and has produced numerous, contemporary hybrid zones (Heiser et al., 1969; Rieseberg, 1991). In regard to the present review, the most significant finding has been the confirmation of the formation of stable hybrid lineages that possess novel ecological adaptations (Lexer et al., 2003; Rieseberg et al., 2003). The evidence for these conclusions comes from (1) genomic mapping of experimental hybrid populations and the naturally occurring hybrid species and (2) QTL mapping of traits associated with the novel habitats occupied by the hybrid species (Lexer et al., 2003; Rieseberg et al., 2003). These results indicate that hybrid sunflower genotypes have formed that possess novel adaptations to habitats in which the parental genotypes are less fit (Lexer et al., 2003).
Phytophthora
Brasier et al. (1999) have concluded that some plant disease epidemics may be because of “…accelerated pathogen evolution…as a consequence of genetic exchange between introduced, or introduced and resident, fungal pathogens….” Brasier et al. (1999) emphasized the importance of testing for the effects of natural hybridization on the evolution of plant pathogens because they might include “…the acquisition of new host specificities…” and the “…emergence of entirely new pathogen taxa.” As discussed above, both outcomes have been postulated for nonpathogenic species as well. The acquisition of greater ecological diversity and the origin of new species via hybridization and introgression have been inferred for numerous species complexes (Arnold, 1997; Rieseberg, 1997; Schweitzer et al., 2002; Rieseberg et al., 2003).
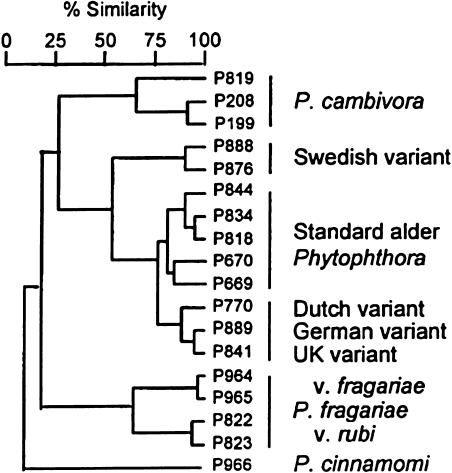
The evolution of hybrid fungal pathogens is exemplified well by the genus Phytophthora. This fungal taxon includes species that infest cultivated (e.g., potato blight) and native species (e.g., forest dieback; Brasier et al., 1999). Significantly, pathogenic isolates (1) possess hybrid genotypes and (2) are genetically very similar to the nonpathogenic species, Phytophthora cambivora (Brasier et al., 1999). It was thus concluded that the pathogenic isolates had formed from hybridization between nonpathogenic strains (Figure 3). Though their parents were not adapted to a pathogenic life history, within 3 years of their being detected, the hybrids had caused the deaths of >10,000 trees (Brasier et al., 1999).
Figure 3.
Genetic Relationships among the Hybrid Pathogenic (Swedish, Standard Alder, Dutch, German, and UK Variants) Phytophthora and Their Possible Parents, P. cambivora and P. fragariae (from Brasier et al., 1999).
The origin of the pathogenic Phytophthora from natural hybridization between nonpathogens is instructive for understanding both evolutionary processes and disease control. On the one hand, this example demonstrates the same process whereby natural hybridization produces new evolutionary lineages that possess novel phenotypes and ecological amplitudes (Anderson and Stebbins, 1954). On the other hand, though the origin of some fungal lineages via hybridization may lead to beneficial interactions (e.g., plant endosymbionts; Tsai et al., 1994), the origin of plant pathogens obviously poses a threat to artificial and natural ecosystems (Brasier et al., 1999). However, the positive and negative outcomes (as judged by humans) both reflect the common hypothesis emphasized in this review; hybridization and/or introgression can produce hybrid genotypes that possess novel adaptations that allow the invasion of additional niches (Anderson, 1948, 1949; Anderson and Stebbins, 1954).
FUTURE DIRECTIONS
In a 1992 review, I suggested that “At the present time, the greatest lack in analyses of natural hybridization, as with most areas of evolutionary biology, involves experimentation. Reciprocal transplants, life history tables, experimental hybridization and gene flow analyses in natural and experimental hybrid populations are necessary to measure interactions between the environment and genotype. Such studies will allow an assessment of the relative roles of deterministic and stochastic processes in the evolution of hybrid populations and, thus lead to predictions concerning the evolutionary consequences of natural hybridization” (Arnold, 1992). I believe that several research groups, including our own, have made great strides in addressing the need for experimentation (e.g., Grant and Grant, 2002; Johnston et al., 2003; Rieseberg et al., 2003). For example, it is now recognized that hybrid genotypes demonstrate a range of fitnesses when they are exposed to a variety of habitats (e.g., Arnold, 1992, 1997; Arnold et al., 1999; Fritz et al., 2003). This reflects a major shift from the conceptual framework established in the neo-Darwinian synthesis that assumed uniform hybrid unfitness (see Arnold, 1997 for a discussion). There are also numerous data sets concerning the extent of introgression (e.g., Dowling and Secor, 1997; Williams and Arnold, 2001) and in a few instances, the fitness effects of such gene flow (e.g., Parsons et al., 1993; Cruzan and Arnold, 1994; Rieseberg et al., 1999) in natural hybrid zones.
In spite of the empirical and conceptual advances made in the last decade, there remains a great need for experimental work that addresses the effects of environmental selection on specific genotypes. For example, several studies have addressed the effect of different hybrid plant phenotypes on pollinator behavior (Campbell et al., 1997; Emms and Arnold, 2000; Wesselingh and Arnold, 2000; Hodges et al., 2002; Bradshaw and Schemske, 2003). However, each of these studies either lacked the genotypic resolution necessary to finely dissect the effect of genotype on the phenotype and, thus, on behavior or in the one case where the genetic basis of a floral color polymorphism is well understood (Bradshaw and Schemske, 2003), deals with a potentially unique case of genes with large effects. It is necessary that a diverse set of genetically well-defined floral traits be presented to natural pollinator assemblages to test their effects on gene flow and fitness. Furthermore, though it can now be assumed that the environment plays a role in the evolution of plant, animal, and microbe hybrid zones (Anderson, 1948; Endler, 1977; Moore, 1977; Howard, 1986; Harrison, 1986; Arnold, 1997), the genetic basis of adaptations to microhabitats present in these zones is virtually unknown. Without these data, strong inferences concerning the pattern of introgression of these and other adaptations across areas of overlap are impossible to construct.
From the above, we see that the measurement of “…interactions between the environment and genotype” (Arnold, 1992) remains the great prize for students of natural hybridization. As comparative data sets are assembled that allow tests for common patterns, it should become possible to construct models that take into account genotypic and environmental variation and thus predict hybrid zone evolution. As this is accomplished, we will be better able to discern the evolutionary significance of the transfer and origin of adaptations via natural hybridization and introgression.
I think it is important to conclude with what may be obvious to some but transparent to the rest of us. We are unable to confidently predict the important concepts and questions that will arise from future work on natural hybridization. Just as Anderson and Stebbins, for example, could not foresee the relative importance of additive versus epistatic interactions, we cannot, with any measure of certainty, outline the relative effect of the environment on the fitness of hybrid genotypes. However, I would suggest that the results from future research “…must be regarded more as we regard those cathedrals where work of many different periods is mixed and produces a total effect, admirable indeed but never foreseen nor intended by any one of the successive builders” (Lewis, 1964).
Acknowledgments
The author wishes to thank A. Bouck, S. Cornman, D. Rosenthal, and two anonymous reviewers for comments on the manuscript. During the preparation of this essay, National Science Foundation Grant DEB-0074159 supported the author.
References
- Anderson, E. (1948). Hybridization of the habitat. Evolution 2, 1–9. [Google Scholar]
- Anderson, E. (1949). Introgressive Hybridization. (New York: John Wiley and Sons).
- Anderson, E., and Hubricht, L. (1938). Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am. J. Bot. 25, 396–402. [Google Scholar]
- Anderson, E., and Stebbins, G.L., Jr. (1954). Hybridization as an evolutionary stimulus. Evolution 8, 378–388. [Google Scholar]
- Arnold, M.L. (1992). Natural hybridization as an evolutionary process. Annu. Rev. Ecol. Syst. 23, 237–261. [Google Scholar]
- Arnold, M.L. (1993). Iris nelsonii: Origin and genetic composition of a homoploid hybrid species. Am. J. Bot. 80, 577–583. [DOI] [PubMed] [Google Scholar]
- Arnold, M.L. (1994). Natural hybridization and Louisiana irises. BioSci. 44, 141–147. [Google Scholar]
- Arnold, M.L. (1997). Natural Hybridization and Evolution. (Oxford: Oxford University Press).
- Arnold, M.L. (2004). Natural hybridization and the evolution of domesticated, pest, and disease organisms. Mol. Ecol., in press. [DOI] [PubMed]
- Arnold, M.L., and Bennett, B.D. (1993). Natural hybridization in Louisiana irises: Genetic variation and ecological determinants. In Hybrid Zones and the Evolutionary Process, R.G. Harrison, ed (Oxford: Oxford University Press), pp. 115–139.
- Arnold, M.L., Bennett, B.D., and Zimmer, E.A. (1990). Natural hybridization between Iris fulva and I. hexagona: Pattern of ribosomal DNA variation. Evolution 44, 1512–1521. [DOI] [PubMed] [Google Scholar]
- Arnold, M.L., Bulger, M.R., Burke, J.M., Hempel, A.L., and Williams, J.H. (1999). Natural Hybridization – How Low Can You Go? (And Still Be Important). Ecology 80, 371–381. [Google Scholar]
- Bennett, B.D., and Grace, J.B. (1990). Shade tolerance and its effect on the segregation of two species of Louisiana iris and their hybrids. Am. J. Bot. 77, 100–107. [Google Scholar]
- Besansky, N.J., Krzywinski, J., Lehmann, T., Simard, F., Kern, M., Mukabayire, O., Fontenille, D., Touré, Y., and Sagnon, N.F. (2003). Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: Evidence from multilocus DNA sequence variation. Proc. Natl. Acad. Sci. USA 100, 10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky, N.J., Lehmann, T., Fahey, G.T., Fontenille, D., Braack, L.E.O., Hawley, W.A., and Collins, F.H. (1997). Patterns of mitochondrial variation within and between African malaria vectors, Anopheles gambiae and An. arabiensis, suggest extensive gene flow. Genetics 147, 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch, L.C., and Vogt, W.G. (1970). Plasticity of taxonomic characters of the Queensland fruit flies Dacus tryoni and Dacus neohumeralis (Tephritidae). Evolution 24, 320–343. [DOI] [PubMed] [Google Scholar]
- Bradshaw, H.D., and Schemske, D.W. (2003). Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426, 176–178. [DOI] [PubMed] [Google Scholar]
- Brasier, C.M., Cooke, D.E.L., and Duncan, J.M. (1999). Origin of a new Phytophthora pathogen through interspecific hybridization. Proc. Natl. Acad. Sci. USA 96, 5878–5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, D.R., Waser, N.M., and Melendez-Ackerman, E.J. (1997). Analyzing pollinator-mediated selection in a plant hybrid zone: Hummingbird visitation patterns on three spatial scales. Am. Nat. 149, 295–315. [Google Scholar]
- Clark, R. (1984). JBS – The Life and Work of J.B.S. Haldane. (Oxford: Oxford University Press).
- Cruzan, M.B., and Arnold, M.L. (1994). Assortative mating and natural selection in an Iris hybrid zone. Evolution 48, 1946–1958. [DOI] [PubMed] [Google Scholar]
- Dowling, T.E., and DeMarais, B.D. (1993). Evolutionary significance of introgressive hybridization in cyprinid fishes. Nature 362, 444–446. [Google Scholar]
- Dowling, T.E., and Secor, C.L. (1997). The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 28, 593–619. [Google Scholar]
- Emms, S.K., and Arnold, M.L. (1997). The effect of habitat on parental and hybrid fitness: Reciprocal transplant experiments with Louisiana Irises. Evolution 51, 1112–1119. [DOI] [PubMed] [Google Scholar]
- Emms, S.K., and Arnold, M.L. (2000). Site-to-site differences in pollinator visitation patterns in a Louisiana Iris hybrid zone. Oikos 91, 568–578. [Google Scholar]
- Endler, J.A. (1977). Geographic Variation, Speciation, and Clines. (Princeton, NJ: Princeton University Press). [PubMed]
- Fritz, R.S., Hochwender, C.G., Brunsfeld, S.J., and Roche, B.M. (2003). Genetic architecture of susceptibility to herbivores in hybrid willows. J. Evol. Biol. 16, 1115–1126. [DOI] [PubMed] [Google Scholar]
- Gentile, G., della Torre, A., Maegga, B., Powell, J.R., and Caccone, A. (2002). Genetic differentiation in the African Malaria vector, Anopheles gambiae s.s., and the problem of taxonomic status. Genetics 161, 1561–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, G.W. (1968). The frequency of interbreeding between two sibling species of Dacus (Diptera) in wild populations. Evolution 22, 667–683. [DOI] [PubMed] [Google Scholar]
- Grant, P.R., and Grant, B.R. (1992). Hybridization of bird species. Science 256, 193–197. [DOI] [PubMed] [Google Scholar]
- Grant, P.R., and Grant, B.R. (2002). Unpredictable evolution in a 30-year study of Darwin's finches. Science 296, 707–711. [DOI] [PubMed] [Google Scholar]
- Grant, V. (1981). Plant Speciation. (New York: Columbia University Press).
- Haldane, J.B.S. (1948). The theory of a cline. J. Genet. 48, 277–284. [DOI] [PubMed] [Google Scholar]
- Haldane, J.B.S. (1949). Suggestions as to the quantitative measurement of rates of evolution. Evolution 3, 51–56. [DOI] [PubMed] [Google Scholar]
- Harrison, R.G. (1986). Pattern and process in a narrow hybrid zone. Heredity 56, 337–349. [Google Scholar]
- Harrison, R.G. (1990). Hybrid zones: Windows on evolutionary process. Oxford Surv. Evol. Biol. 7, 69–128. [Google Scholar]
- Heiser, C.B., Jr. (1951). Hybridization in the annual sunflowers: Helianthus annuus X H. debilis var. cucumerifolius. Evolution 5, 42–51. [Google Scholar]
- Heiser, C.B., Jr., Smith, D.M., Clevenger, S.B., and Martin, W.C., Jr. (1969). The North American sunflowers (Helianthus). Mem. Torr. Bot. Club 22, 1–213. [Google Scholar]
- Hodges, S.A., Whittall, J.B., Fulton, M., and Yang, J.Y. (2002). Genetics of floral traits influencing reproductive isolation between Aquilegia formosa and Aquilegia pubescens. Am. Nat. 159, S51–S60. [DOI] [PubMed] [Google Scholar]
- Howard, D.J. (1986). A zone of overlap and hybridization between two ground cricket species. Evolution 40, 34–43. [DOI] [PubMed] [Google Scholar]
- Johnston, J.A., Arnold, M.L., and Donovan, L.A. (2003). High hybrid fitness at seed and seedling life history stages in Louisiana Irises. J. Ecol. 91, 438–446. [Google Scholar]
- Key, K.H.L. (1968). The concept of stasipatric speciation. Syst. Zool. 17, 14–22. [Google Scholar]
- Kim, S.-C., and Rieseberg, L.H. (1999). Genetic architecture of species differences in annual sunflowers: Implications for adaptive trait introgression. Genetics 153, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, J.S., Wilks, K.E., Farrant, J.L., and Langford, P.R. (1998). Natural genetic exchange between Haemophilus and Neisseria: Intergenic transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA 95, 12381–12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzaro, G.C., Touré, Y.T., Carnahan, J., Zheng, L., Dolo, G., Traoré, S., Petrarca, V., Vernick, K.D., and Taylor, C. (1998). Complexities in the genetic structure of Anopheles gambiae populations in west Africa as revealed by microsatellite DNA analysis. Proc. Natl. Acad. Sci. USA 95, 14260–14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, C.S. (1964). The Discarded Image – An Introduction to Medieval and Renaissance Literature. (Cambridge, UK: Cambridge University Press).
- Lewontin, R.C., and Birch, L.C. (1966). Hybridization as a source of variation for adaptation to new environments. Evolution 20, 315–336. [DOI] [PubMed] [Google Scholar]
- Lexer, C., Welch, M.E., Durphy, J.L., and Rieseberg, L.H. (2003). Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: Implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol. Ecol. 12, 1225–1235. [DOI] [PubMed] [Google Scholar]
- Machado, C.A., and Ayala, F.J. (2001). Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 98, 7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, W.S. (1977). An evaluation of narrow hybrid zones in vertebrates. Q. Rev. Biol. 52, 263–277. [Google Scholar]
- Morrow, J., Scott, L., Congdon, B., Yeates, D., Frommer, M., and Sved, J. (2000). Close genetic similarity between two sympatric species of Tephritid fruit fly reproductively isolated by mating time. Evolution 54, 899–910. [DOI] [PubMed] [Google Scholar]
- Parsons, T.J., Olson, S.L., and Braun, M.J. (1993). Unidirectional spread of secondary sexual plumage traits across an avian hybrid zone. Science 260, 1643–1646. [DOI] [PubMed] [Google Scholar]
- Pike, N., Wang, W., and Meats, A. (2003). The likely fate of hybrids of Bactrocera tryoni and Bactrocera neohumeralis. Heredity 90, 365–370. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H. (1991). Homoploid reticulate evolution in Helianthus (Asteraceae): Evidence from ribosomal genes. Am. J. Bot. 78, 1218–1237. [Google Scholar]
- Rieseberg, L.H. (1997). Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 28, 359–389. [Google Scholar]
- Rieseberg, L.H., Raymond, O., Rosenthal, D.M., Lai, Z., Livingstone, K., Nakazato, T., Durphy, J.L., Schwarzbach, A.E., Donovan, L.A., and Lexer, C. (2003). Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H., Whitton, J., and Gardner, K. (1999). Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152, 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, H.P. (1938). A character analysis of colonies of Iris fulva, Iris hexagona var. giganticaerulea and natural hybrids. Am. J. Bot. 25, 727–738. [Google Scholar]
- Schweitzer, J.A., Martinsen, G.D., and Whitham, T.G. (2002). Cottonwood hybrids gain fitness traits of both parents: A mechanism for their long-term persistence? Am. J. Bot. 89, 981–990. [DOI] [PubMed] [Google Scholar]
- Smoot, L.M., Franke, D.D., McGillivary, G., and Actis, L.A. (2002). Genomic analysis of the F3031 Brazilian purpuric Fever clone of Haemophilus influenzae biogroup aegyptius by PCR-based subtractive hybridization. Infect. Immun. 70, 2694–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, G.L., Jr. (1947). Types of polyploids: Their classification and significance. Adv. Gen. 1, 403–429. [DOI] [PubMed] [Google Scholar]
- Stebbins, G.L., Jr. (1959). The role of hybridization in evolution. Proc. Am. Phil. Soc. 103, 231–251. [Google Scholar]
- Stutz, H.C., and Thomas, L.K. (1964). Hybridization and introgression in Cowania and Purshia. Evolution 18, 183–195. [Google Scholar]
- Tsai, H.-F., Liu, J.-S., Staben, C., Christensen, M.J., Latch, G.C.M., Siegel, M.R., and Schardl, C.L. (1994). Evolutionary diversification of fungal endophytes of tall fescue grass by hybridization with Epichloë species. Proc. Natl. Acad. Sci. USA 91, 2542–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viosca, P., Jr. (1935). The irises of southeastern Louisiana A taxonomic and ecological interpretation. Bull. Am. Iris Soc. 57, 3–56. [Google Scholar]
- Wang, R., Zheng, L., Touré, Y.T., Dandekar, T., and Kafatos, F.C. (2001). When genetic distance matters: Measuring genetic differentiation at microsatellite loci in whole-genome scans of recent and incipient mosquito species. Proc. Natl. Acad. Sci. USA 98, 10769–10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselingh, R.A., and Arnold, M.L. (2000). Pollinator behaviour and the evolution of Louisiana Iris hybrid zones. J. Evol. Biol. 13, 171–180. [Google Scholar]
- Williams, J.H., Jr., and Arnold, M.L. (2001). Environmental, historical, and introgressive sources of genetic structure in the woody perennial, Betula occidentalis. Int. J. Plant Sci. 162, 1097–1109. [Google Scholar]