Abstract
Background:
Previous studies have investigated the relationship between GSTA1, GSTM1, GSTP1, and GSTT1 polymorphisms and bladder cancer (BCa) susceptibility, respectively, but the results remain inconsistent. So, we conducted this meta-analysis including 79 case–control studies to explore such relationships.
Methods:
We searched PubMed, EMBASE, Cochrane library, Web of Science, and CNKI for relevant available studies. The pooled odds ratios (ORs) with 95% confidence intervals (CIs) were implemented to evaluate the intensity of associations. Publication bias was estimated using Begg funnel plots and Egger regression test. To assess the stability of the results, we used sensitivity analysis with the method of calculating the results again by omitting 1 single study each time. Between-study heterogeneity was tested using the I2 statistic.
Results:
No significant association between GSTA1 polymorphism and BCa susceptibility (OR = 1.05, 95% CI 0.83–1.33) was noted. Besides, meaningful association between individuals who carried the GSTM1 null genotype and increased BCa risk was detected (OR = 1.39, 95%CI 1.28–1.51). When stratified by ethnicity, significant difference was found in both Caucasian (OR = 1.39, 95% CI 1.23–1.58) and Asian populations (OR = 1.45, 95% CI 1.31–1.61). Moreover, in the subgroup analysis by source of controls (SOC), the results were significant in both hospital-based control groups (OR = 1.49, 95% CI 1.35–1.64) and population-based control groups (OR = 1.21, 95% CI = 1.07–1.37). Additionally, the analysis revealed no significant association between GSTP1 polymorphism and BCa risk (OR = 1.07, 95% CI 0.96–1.20). What is more, significant associations between GSTT1 polymorphism and BCa susceptibility were discovered (OR = 1.11, 95% CI 1.00–1.22). In the subgroup analysis by ethnicity, significant associations between GSTT1 null genotype and BCa risk were observed only in Caucasians (OR = 1.25, 95% CI 1.09–1.44). Furthermore, when stratified by SOC, no obvious relationship was found between the GSTT1 null genotype polymorphism with hospital-based population (OR = 1.11, 95% CI 0.97–1.28) or population-based population (OR = 1.10, 95% CI 0.96–1.27).
Conclusion:
This study suggested that GSTM1 null genotype and GSTT1 null genotype might be related to higher BCa risk, respectively. However, no associations were observed between GSTA1 or GSTP1 polymorphisms and BCa susceptibility.
Keywords: bladder cancer, glutathione S-transferases, meta-analysis, single gene polymorphism, susceptibility
1. Introduction
Bladder cancer (BCa), with an increasing incidence and mortality nowadays, has become the 9th most common cancer and the 14th leading cause of death due to cancer worldwide.[1] An estimated 429,800 new cases of BCa and 165,100 deaths took place in 2012 worldwide.[2] As a complicated and multifactorial procedure, the initiation and development of BCa are still not completely understood.[3] However, the risk factors could be mainly classified into 3 subgroups: long-term inflammation stimulation, specific chemical exposure, and genetic factors.[4] Interestingly, some people never get BCa even though exposed to specific chemicals. In contrast, many BCa patients do not have those known risk factors, suggesting that genetic factors might play a significant role in bladder carcinogenesis.[5,6]
Glutathione S-transferases (GSTs), existing in almost all living organisms, are members of a polygene family of isoenzymes.[7] GSTs are a family of multifunctional phase II enzymes that catalyze the combination of many exogenous and endogenous electrophilic compounds with glutathione, which are characterized with assisting the detoxification of various therapeutic drugs, carcinogens, products of oxidative stress, toxins, and chemical mutants.[8,9] In humans, GSTs were encoded by 8 different gene families. Among them, 4 are mainly expressed in tissues: GSTA, GSTM, GSTP, and GSTT. Accordingly, the GSTA1, GSTM1, GSTP1, and GSTT1 genes are located at chromosome 6p12.1, 1p13.3, 11q13, and 22q11.23, respectively.
Over the last 2 decades, plentiful studies have been carried out to investigate the association between GSTs and the risk of BCa, but these studies have reported conflicting results. A single study might fail to demonstrate the complicated genetic relationship due to small sample size, but meta-analysis could increase the statistical power through detecting overall effects. Previously, meta-analysis has been performed to find out the relationship between GSTM1, GSTP1, GSTT1, and BCa, respectively.[10–14] Although the results remain inconclusive or even contradictory. In addition, the relationship between GSTA1 and BCa susceptibility has not been qualitatively studied before. Some related case–control studies have been released after the previous meta-analyses, which may generate influence on the conclusions. Therefore, we conducted such meta-analysis to assess these relationships by including all eligible articles.
2. Materials and methods
2.1. Search strategy
We did a systematic search of PubMed, EMBASE, Cochrane library, Web of Science, and CNKI up to December 2015 by using the combination of the following key words: “glutathione S-transferase A1” or “GSTA1,” “glutathione Stransferase M1” or “GSTM1,” “glutathione S-transferase P1” or “GSTP1,” “glutathione S-transferase T1” or “GSTT1,” “bladder” or “urothelial,” “cancer” or “carcinoma” or “neoplasm,” and “polymorphism” or “polymorphisms” without any restriction on language. The reference lists of the selected papers were searched by hand for potentially eligible articles. We only included the study with the most recent and/or the largest sample size when several studies had partially overlapped or similar data.
2.2. Selection criteria
For this meta-analysis, the inclusion criteria were as follows: case–control studies with the original date for the evaluated associations between GSTA1, GSTM1, GSTP1 and/or GSTT1 polymorphisms, and BCa risk; the diagnosis of the patients with BCa was confirmed pathologically, and the controls were confirmed free of any cancer; and sufficient published data about the size of the sample, odds ratio (OR), and their 95% confidence interval (CI). The exclusion criteria were duplicates of previous publication; no control subjects; and patients without confirmation of BCa or mixed with other diseases.
If study populations were the same or duplicate data were published, only the study with the largest number of sample size was included. We did not need to obtain ethical approval or informed consent because our data were extracted from previous studies. Nevertheless, the included studies in our review did get patient consent, and each study was approved by an ethics committee.
2.3. Data extraction
Data were independently extracted from all eligible publications by 5 investigators (YJY, XL, CL, JYT, and ZQQ), and quality assessment was conducted by 3 authors (YJY, XL, and CL). When meeting conflicting opinions about inclusion, disagreements were resolved by discussion among team members. Relevant data were extracted from each eligible study and carefully recorded, including involved genes, 1st author name, year of publication, the ethnicity of the study population, subject source, total number of cases and controls, and different number of genotypes in cases and controls. If important unpublished information were needed, we also e-mailed the original authors. According to source of controls (SOC), studies were classified into hospital-based (HB) and population-based (PB) groups. Ethnic groups were principally defined as Caucasian, Asian, African, or Mixed.
2.4. Statistical analysis
ORs with 95% CIs were implemented. The heterogeneity was estimated using the χ2-based Q statistic, and heterogeneity was considered statistically significant when P heterogeneity ≤0.1 or I2 > 50%.[15] If the presence of heterogeneity was found, the random-effects model would be utilized. Otherwise, fixed-effects model would be performed. Then, subgroup analysis was further carried out by ethnicity and SOC properly.
To assess the stability of the results, we used sensitivity analysis with the method of calculating the results again by omitting 1 single study each time.[16] To check the publication bias between the studies, Egger linear regression test and Begg funnel plots were executed.[17] Hardy–Weinberg equilibrium was assessed by the goodness-of-fit Chi-square test, and P < 0.05 was considered as an obviously selective bias.[18] All statistical analyses tests were performed with Stata software (version 12.0; Stata Corp LP, College Station, TX). All P values below 0.05 were considered statistically significant.
3. Results
3.1. Literature search and studies characteristics
Figure 1 shows the flowchart of literature search and selection process. Finally, a total of 79 case–control studies were included according to the inclusion criteria.[19–97] Characteristics of individual study qualified for the current meta-analysis (GSTA1, GSTM1, GSTP1, and GSTT1, respectively) are presented in Tables 1–4 individually. This meta-analysis results of association between GSTs polymorphism and BCa risk are shown in Table 5.
Figure 1.
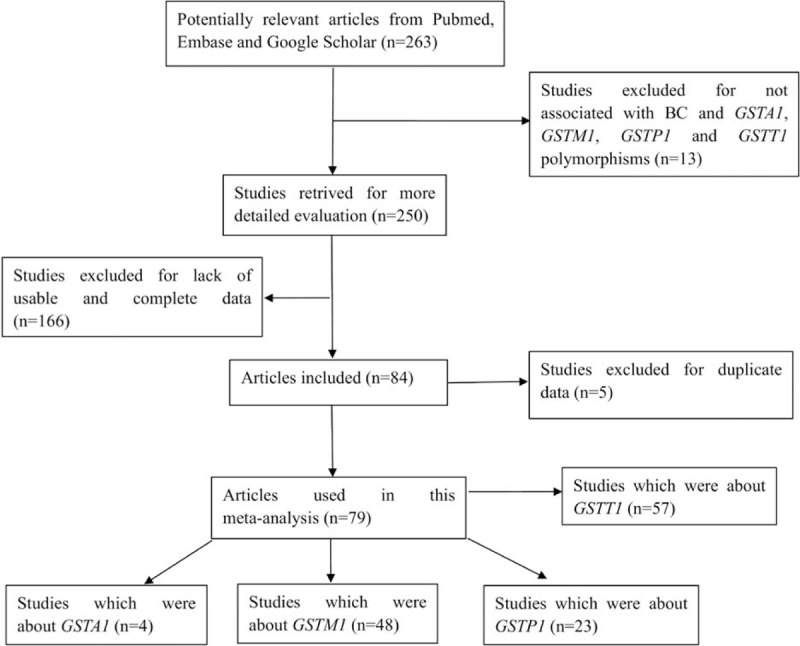
Flowchart of literature search and selection process.
Table 1.
Characteristics of individual studies included in the meta-analysis.
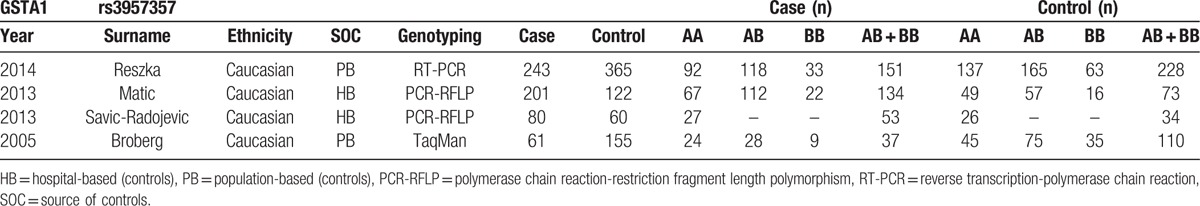
Table 4.
Characteristics of individual studies included in the meta-analysis.
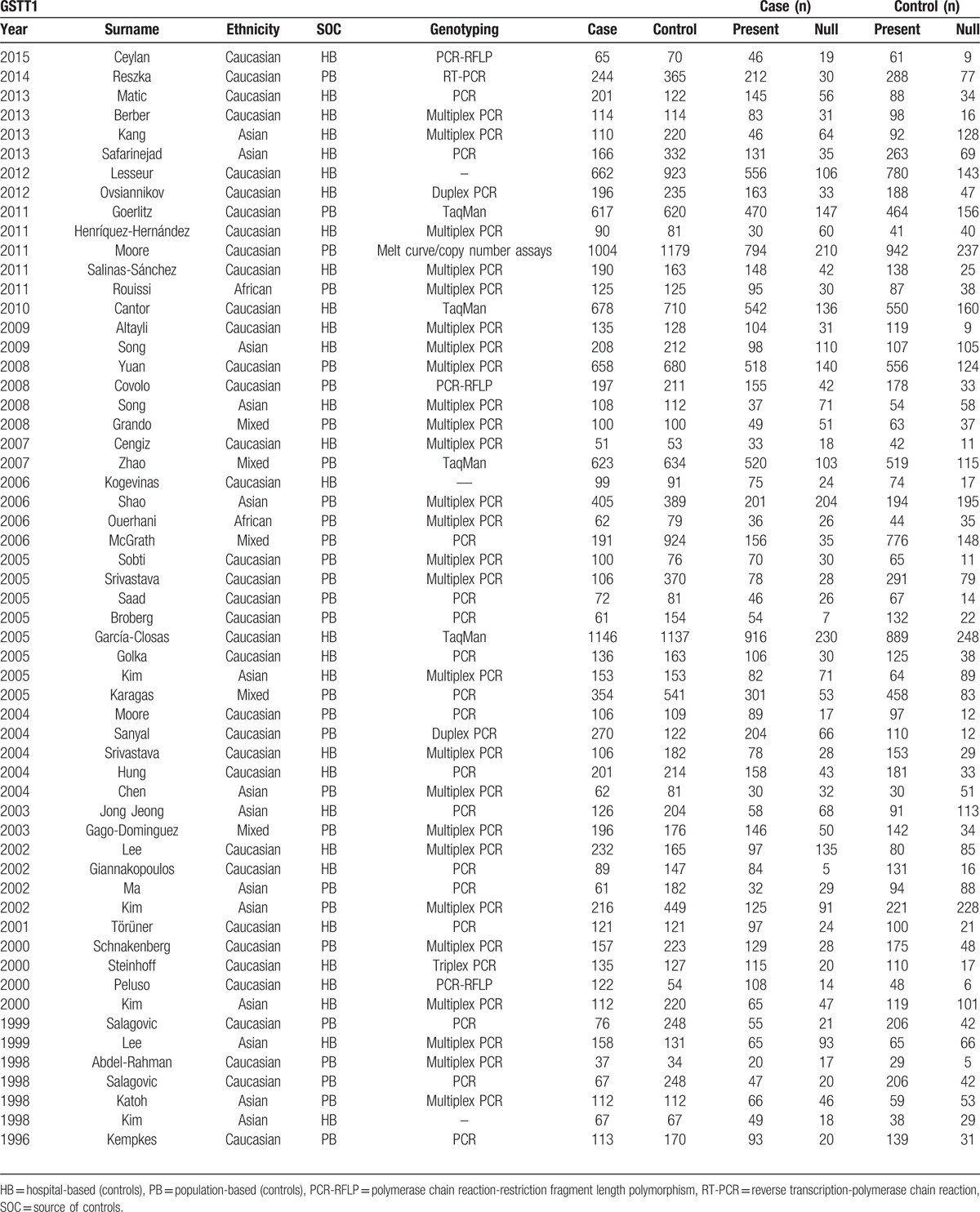
Table 5.
Meta-analysis results of association between GSTs polymorphism and bladder cancer risk.
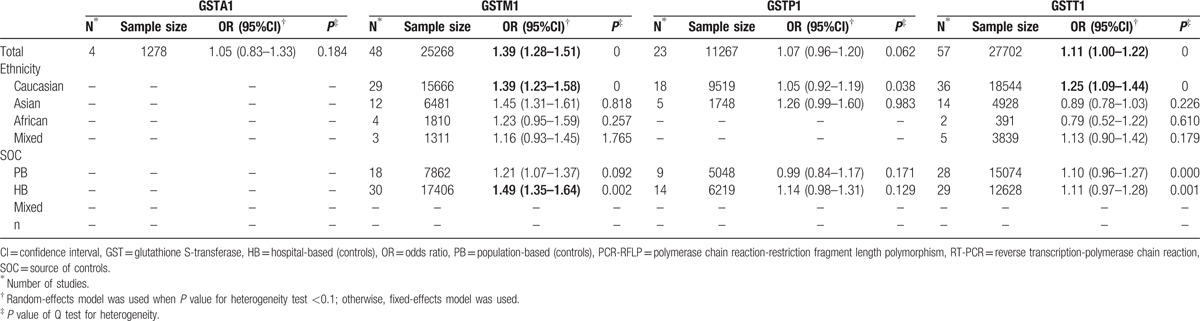
Table 2.
Characteristics of individual studies included in the meta-analysis.
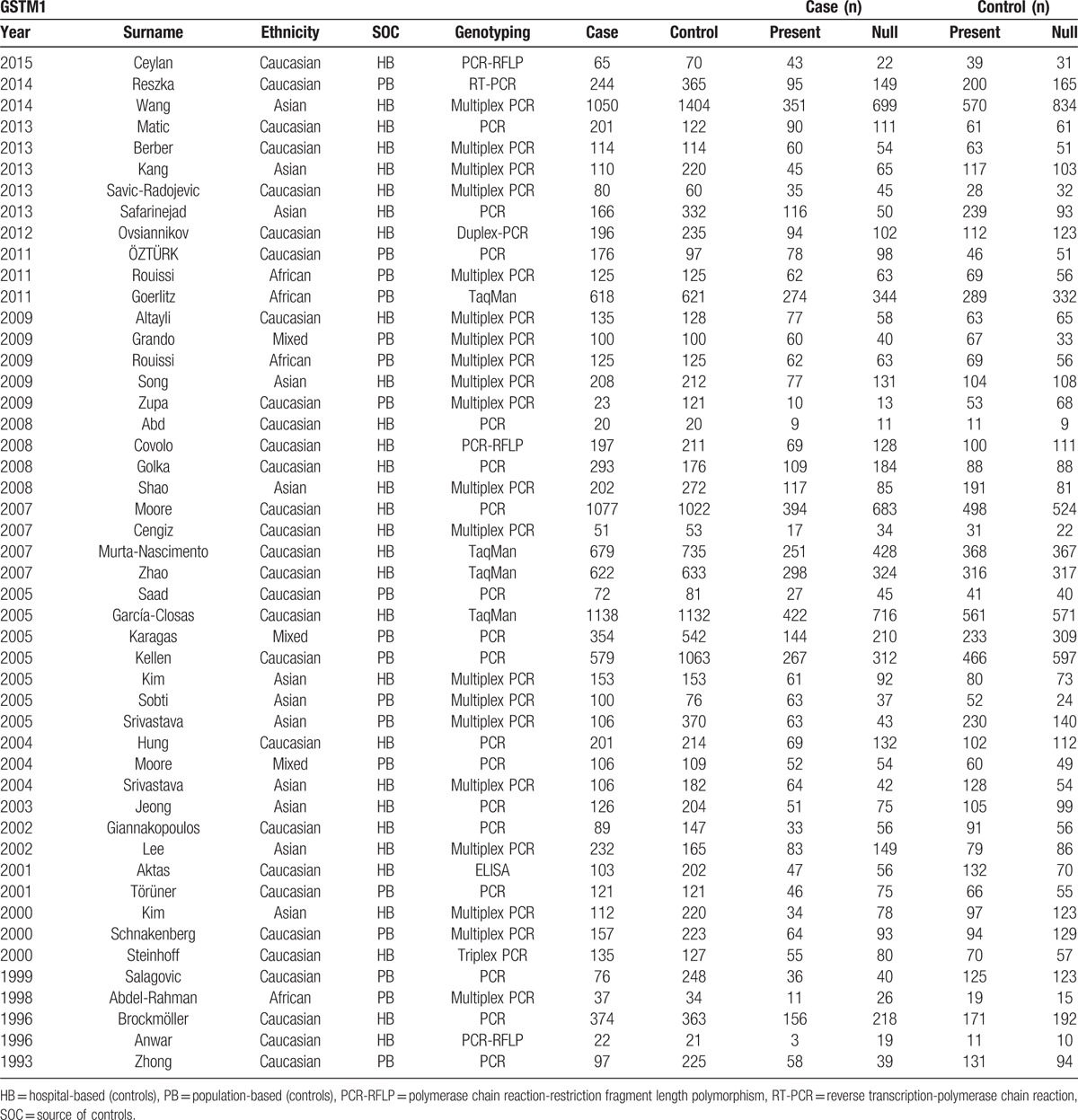
Table 3.
Characteristics of individual studies included in the meta-analysis.
3.2. GSTA1
Four studies consisting of 585 cases and 702 controls were adopted in order to evaluate the relationship between GSTA1 polymorphism and BCa risk. As shown in Fig. 2, the results indicated no significant association between GSTA1 polymorphism and BCa susceptibility (OR = 1.05, 95% CI 0.83–1.33). Subgroup analysis was not performed owing to the limited studies.
Figure 2.
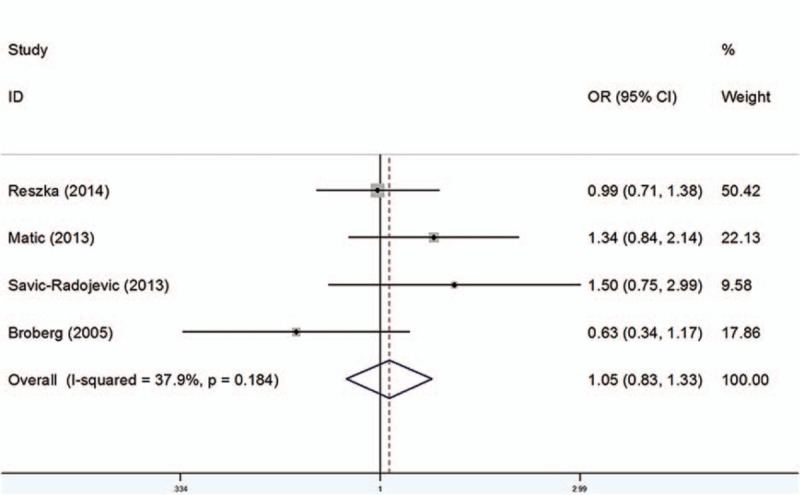
Forest plots of the association between GSTA1 polymorphism and bladder cancer susceptibility. CI = confidence interval, OR = odds ratio.
3.3. GSTM1
As shown in Table 5, 48 studies including 11,473 cases and 13,795 controls were analyzed. Overall, significant associations between individuals who carried GSTM1 null genotype and increased BCa risk were observed (OR = 1.39, 95%CI 1.28–1.51) (Fig. 3). When stratified by ethnicity, significant difference was detected in Caucasian (OR = 1.39, 95% CI 1.23–1.58) and Asian populations (OR = 1.45, 95% CI 1.31–1.61) instead of African (OR = 1.23, 95% CI 0.95–1.59) or Mixed populations (OR = 1.16, 95% CI 0.93–1.45). In addition, in the subgroup analysis by SOC, the results were significant both in HB populations (OR = 1.49, 95% CI 1.35–1.64) and PB populations (OR = 1.21, 95% CI 1.07–1.37).
Figure 3.
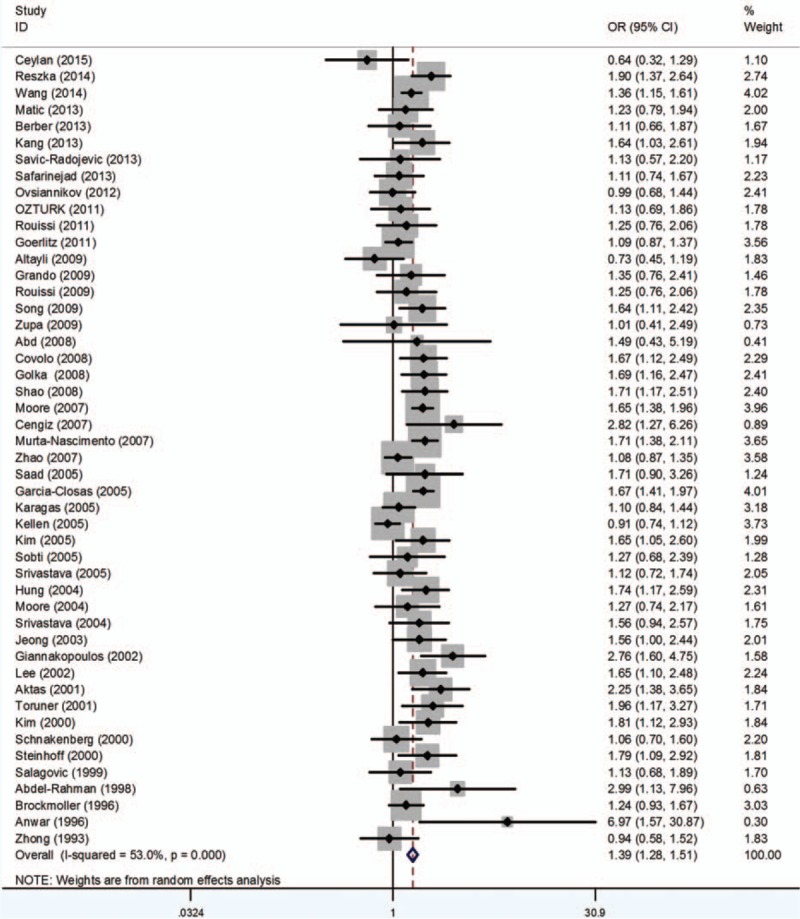
Forest plots of the association between GSTM1 polymorphism and bladder cancer susceptibility. CI = confidence interval, OR = odds ratio.
3.4. GSTP1
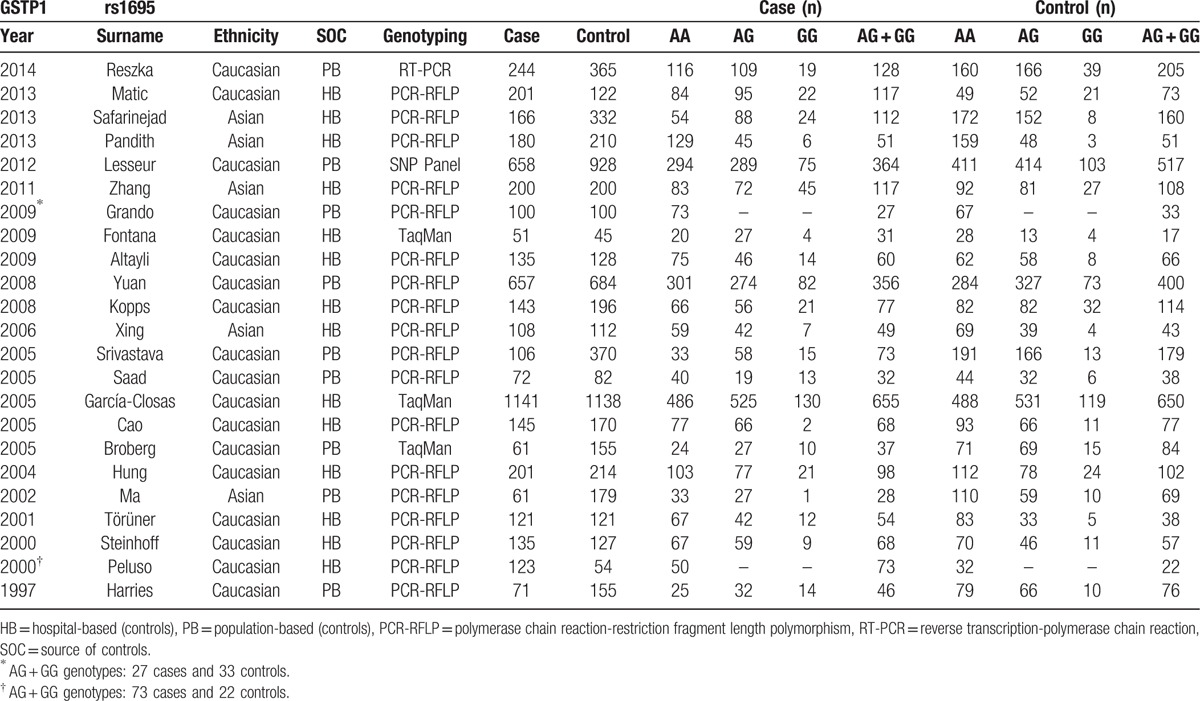
Twenty-three studies involving 5080 cases and 6187 controls were included in this study. Because a few studies provided precise data of genotypes, only dominant model could be carried out with all studies. Generally, the analysis revealed no significant association between GSTP1 Ile105Val polymorphism and BCa risk (OR = 1.07, 95% CI 0.96–1.20) (Fig. 4). No significant relationship was observed between GSTP1 polymorphism and BCa risk in patients when stratified by ethnicity. Meanwhile, there seems no relationship between GSTP1 polymorphism and the susceptibility of BCa when stratified by SOC (Table 5).
Figure 4.
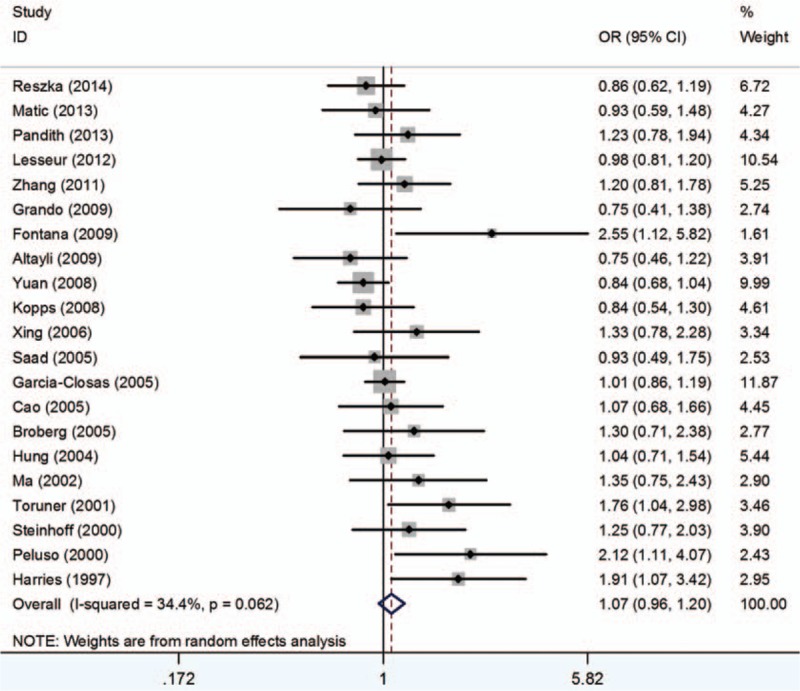
Forest plots of the association between GSTP1 polymorphism and bladder cancer susceptibility. CI = confidence interval, OR = odds ratio.
3.5. GSTT1
Fifty seven studies including 12,369 cases and 15,333 controls were analyzed. The results indicated significant association between GSTT1 polymorphism and BCa susceptibility (OR = 1.11, 95% CI 1.00–1.22) (Fig. 5). In the subgroup analysis by ethnicity, significant associations between GSTT1 null genotype and BCa risk were noted only in Caucasians (OR = 1.25, 95% CI 1.09–1.44). Additionally, when stratified by SOC, no obvious relationship was detected between the GSTT1 null genotype polymorphism with HB (OR = 1.11, 95% CI 0.97–1.28) or PB (OR = 1.10, 95% CI 0.96–1.27), respectively (Table 5).
Figure 5.
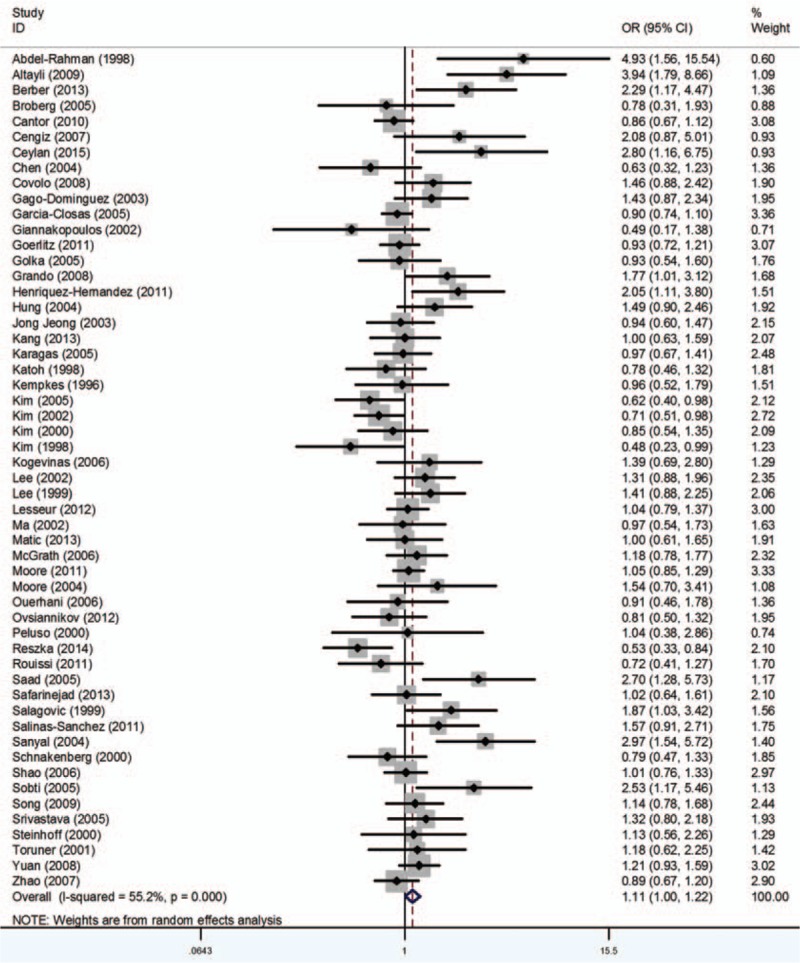
Forest plots of the association between GSTT1 polymorphism and bladder cancer susceptibility. CI = confidence interval, OR = odds ratio.
3.6. Sensitivity analysis
Sensitivity analysis was utilized to identify the influence of each study on the pooled OR by consecutively omitting 1 study each time for all subjects and subgroups. The sensitivity analysis for GSTA1, GSTM1, GSTP1, and GSTT1 polymorphism showed that no individual study affected the pooled OR significantly, which indicated that our results were reliable.
3.7. Publication bias
The publication bias of studies GSTA1, GSTM1, GSTP1, and GSTT1 were assessed, respectively, using Begg and Egger funnel plot. The overall outcomes revealed that our results were statistically dependable.
4. Discussion
BCa is one of the most common cancers of the urinary tract. However, the exact mechanisms of bladder carcinogenesis remain unclear. There is a growing realization that the development of BCa is caused by a complex interaction of both genetic and environmental factors.[98] Although genetic factors are considered to be a crucial part of the pathogenic process of BCa, especially the polymorphisms in metabolic pathways.[99] As one of the most important parts of phase II super family of metabolism enzymes, GSTs are composed of 7 classes (α, μ, ω, π, σ, θ, ξ).[100] Among them, GSTA1, GSTM1, GSTP1, and GSTT1 are considered to be the most important. Almost all members of the GST family show genetic polymorphism, which leads to a complete absence or lowering of enzyme activity.
GSTA1 has 3 single nucleotide polymorphisms (SNPs): −567TOG, −69COT, and −52GOA.[101] Differential expression with lower transcriptional activation of variant GSTA1∗B (−567G, −69T, and −52A) than common GSTA1∗A allele (−567T, −69C, and −52G) are resulted from these replacements.[102] GSTM1 plays an important role in preventing the development of cancers. The inherited homozygous absence of the GSTM1 gene results in the deficiency of the enzyme activity.[103] GSTP1 is an important part of GST families, and the most commonly studied GSTP1 variant is exon 5 Ile105Val, encoding an Ile/Val exchange at codon 105 (Ile105Val; A105G) (rs947894), which has been shown to be linked to lower expression of metabolic activity.[104] People with the GSTT1 null genotype was reported to have decreased enzyme activity and decreased ability to detoxify the environmental and dietary agents, especially 1,3-butadiene and ethylene oxide, which could induce chromosomal damage and make people more susceptible to cancer.[105] By catalyzing the detoxification of electrophilic compounds through conjugation with glutathione, these enzymes can prevent cells from damage.[106] Besides, GSTs are able to regulate the induction of other proteins and enzymes which is important for cellular functions. The polymorphisms affect the enzyme activity, leading to increased genotoxic damage and affect the transportation of steroid hormones, causing the development of cancer eventually.[107,108] GSTs are essential for maintaining genomic integrity because electrophilic compounds could damage the DNA.[109] Therefore, GSTA1, GSTM1, GSTP1, and GSTT1 may play an important role in the development of BCa.
Recently, there were increasing case–control studies concerned with the associations between GSTs polymorphisms and BCa susceptibility.[19–97] Nevertheless, the inconsistent results of them might owe to limited sample size, various methodologies, and race and dissimilar source of controls. Although several meta-analyses have explored the relationship between GSTM1, GSTP1, and GSTT1 polymorphisms and BCa susceptibility, respectively,[10–14] the results remain unclear. Besides, because of relatively small number of studies, no meta-analysis on GSTA1 has been performed before. What is more, additional studies have been published since the last meta-analysis. So, all these might have generated great influence to the previous conclusions. Thus, we did this meta-analysis.
For the first time, we performed meta-analysis on the relationship between GSTA1 polymorphism and BCa susceptibility. We included 4 case–control studies in this meta-analysis, and the results suggested that there was no association. According to the published papers, the conclusion on the relationship between GSTA1 polymorphism and BCa susceptibility is inconsistent. The exact mechanism of the influence of GSTA1 polymorphism on BCa is still unclear. However, in association with smoking, low activity GSTA1 seems to increase individual susceptibility to BCa.[20] The limited amount of involved studies may become a major factor which could influence the evaluation of the real association between GSTA1 polymorphism and BCa risk.
The analysis of the present studies indicated that the null genotype of GSTM1 polymorphism significantly increases BCa susceptibility. Jiang et al[10] performed a meta-analysis indicating the similar results with ours in 2011, which included 33 studies. Nevertheless, 48 studies were involved in our meta-analysis, which could provide more comprehensive and reliable results.
Meanwhile, similar to the outcome of the meta-analysis conducted by Gong et al in 2012,[14] significant associations between GSTT1 polymorphism and BCa susceptibility were discovered. However, we included 7 more studies, which could be more credible.
In the aspect of GSTP1, contrary to the previous meta-analysis (Wang et al[13] and Kellen et al[111]), this analysis revealed no significant relationship between GSTP1 polymorphism and BCa risk. Wang et al internalized 25 studies, but there were 2 duplicates of previous publication, so we excluded them. Although the result changes, we think it is more believable. And Kellen et al included 16 studies (4273 cases and 5081 controls), whereas we selected 23 studies involving 5080 cases and 6187 controls. Additional studies have been published since the last meta-analysis, which would might change the previous results. So, we think the result of our study is more reliable. More studies are required to validate our results.
Alternatively, subgroup analysis was performed according to ethnicity in GSTM1, GSTP1, or GSTT1 genetic variants. For GSTM1, when stratified by ethnicity, significant difference was detected in both Caucasian and Asian populations instead of African populations and Mixed populations. No significant relationship was observed between GSTP1 polymorphism and BCa risk in patients when stratified by ethnicity. For GSTT1, significant associations were observed only in Caucasians. As a complicated multigenetic disease, cancer has diversity among different ethnic populations, and the existence of the discrepancy might owing to different genetic background.[110] As a result of ethnic differences, the incidence of gene polymorphisms may vary notably among different phyletic populations. Although the possible reasons of the conflicting results were unknown, there might be several explanations for it. First, among different ethnic groups, various environmental factors and genetic backgrounds might not be exposed sheerly, which might also be affected by unidentified genes. Second, the selection bias and limitation of sample size should also be taken into consideration.
In the present meta-analysis, the cases and controls were from dissimilar sources. The results suggest that there is association between GSTM1 null genotype and BCa susceptibility both in the subgroup analysis of studies with HB and PB controls. Our meta-analysis also revealed there is no relationship between other GSTs polymorphisms and BCa risk in their respective SOC groups. Subgroup analysis was not performed owing to the limited studies for GSTA1. However, more prospective studies should be performed to evaluate if there has indeed an association between the other GSTs polymorphisms and BCa risk exists in the subgroup analysis of SOC.
Despite the certain conclusions generated in this study, there still exist several limitations. First, the sample sizes of GSTM1, GSTP1, and GSTT1 were large enough, nevertheless which caused possible false positive conclusions. Second, the number of some subgroups was relatively small, and it is hard to search for the reliable association with limited statistical power. Third, when it comes to GSTA1 polymorphism, the sample size was too small. Additional studies with higher quality and larger sample size should be included in the future to verify our result. Fourth, BCa results from complex interactions between a variety of genetic and environmental factors, thereby suggesting that BCa susceptibility would not be influenced by any single gene. BCa is a multifactorial disease, so the complex interactions like gene-environment factors could not be ignored. Last, the total outcomes were based on unadjusted effect estimates without enough data for the adjustment by other covariates, such as smoking status, age, gender, and so on. The influence of confounding factors should be payed more attention. Hence, a more precise meta-analysis could be conducted if detailed data of some individual studies can be accessed.
The results indicated that the GSTM1 null genotype might elevate BCa susceptibility, and the GSTT1 polymorphism might enhance BCa risk. No significant associations were observed between GSTA1 or GSTP1 polymorphism and BCa risk. For the 1st time, we performed this meta-analysis to evaluate the association between GSTA1 polymorphism and BCa risk. However, taking the restriction of sample size into consideration, analysis with larger and more well-designed studies is required to validate our results. In the future, the analysis of different combinations of polymorphisms of the 4 isoforms could be performed if the data is available.
Footnotes
Abbreviations: BCa = bladder cancer, CI = confidence interval, GST = glutathione S-transferase, HB = hospital-based, OR = odds ratio, PB = population-based, SOC = source of controls.
YY, XL, and CL contributed equally to this work.
The authors have no funding and conflicts of interest to disclose.
References
- 1.Mahdavifar N, Ghoncheh M, Pakzad R, et al. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac J Cancer Prev 2015; 17:381–386. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65:87–108. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Carbayo M. Hypermethylation in bladder cancer: biological pathways and translational applications. Tumour Biol 2012; 33:347–361. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman DS, Shipley WU, Feldman AS, et al. Bladder cancer. Lancet 2009; 374:239–249. [DOI] [PubMed] [Google Scholar]
- 5.Pandith AA, Siddiqi MA. Burden of cancers in the valley of Kashmir: 5 year epidemiological study reveals a different scenario. Tumour Biol 2012; 33:1629–1637. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Hildebrandt MA, Chang DW. Genome-wide association studies of bladder cancer risk: a field synopsis of progress and potential applications. Cancer Metastasis Rev 2009; 28:269–280. [DOI] [PubMed] [Google Scholar]
- 7.Simic T, Savic-Radojevic A, Pljesa-Ercegovac M, et al. Glutathione S-transferases in kidney and urinary bladder tumors. Nat Rev Urol 2009; 6:281–289. [DOI] [PubMed] [Google Scholar]
- 8.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev 1997; 6:733–743. [PubMed] [Google Scholar]
- 9.Tang JJ, Wang MW, Jia EZ, et al. The common variant in the GSTM1 and GSTT1 genes is related to markers of oxidative stress and inflammation in patients with coronary artery disease: a case-only study. Mol Biol Rep 2010; 37:405–410. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Z, Li C, Wang X, et al. Glutathione S-transferase M1 polymorphism and bladder cancer risk: a meta-analysis involving 33 studies. Exp Biol Med 2011; 236:723–728. [DOI] [PubMed] [Google Scholar]
- 11.Zhang RG, Xu GY, Chen WJ, et al. Genetic polymorphisms of glutathione S-transferase M1 and bladder cancer risk: a meta-analysis of 26 studies. Mol Biol Rep 2011; 38:2491–2497. [DOI] [PubMed] [Google Scholar]
- 12.Wu K, Wang X, Xie Z, et al. Glutathione S-transferase P1 gene polymorphism and bladder cancer susceptibility: an updated analysis. Mol Biol Rep 2013; 40:687–695. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Xue L, Chong T, et al. Quantitative assessment of the association between glutathione S-transferase P1 Ile105Val polymorphism and bladder cancer risk. Tumor Biol 2013; 34:1651–1657. [DOI] [PubMed] [Google Scholar]
- 14.Gong M, Dong W, An R, et al. Glutathione S-transferase T1 polymorphism contributes to bladder cancer risk: a meta-analysis involving 50 studies. DNA Cell Biol 2012; 31:1187–1197. [DOI] [PubMed] [Google Scholar]
- 15.Higgins JP, Thompson SG. Quantifying heterogeneity in a metaanalysis. Stat Med 2002; 21:1539–1558. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Guo W, Shi X, et al. Association between alcohol consumption and the risk of Barrett's esophagus: a meta-analysis of observational studies. Medicine (Baltimore) 2015; 94:e1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egger M, Davey SG, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 1992; 48:361–372. [PubMed] [Google Scholar]
- 19.Reszka E, Jablonowski Z, Wieczorek E, et al. Polymorphisms of NRF2 and NRF2 target genes in urinary bladder cancer patients. J Cancer Res Clin Oncol 2014; 140:1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matic M, Pekmezovic T, Djukic T, et al. GSTA1, GSTM1, GSTP1, and GSTT1 polymorphisms and susceptibility to smoking-related bladder cancer: a case-control study. Urol Oncol 2013; 31:1184–1192. [DOI] [PubMed] [Google Scholar]
- 21.Savic-Radojevic A, Djukic T, Simic T, et al. GSTM1-null and GSTA1-low activity genotypes are associated with enhanced oxidative damage in bladder cancer. Redox Rep 2013; 18:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broberg K, Björk J, Paulsson K, et al. Constitutional short telomeres are strong genetic susceptibility markers for bladder cancer. Carcinogenesis 2005; 26:1263–1271. [DOI] [PubMed] [Google Scholar]
- 23.Ceylan GG, Ceylan C, Taşdemir S, et al. The effect of glutathione-S-transferases in the susceptibility to bladder cancer. Ir J Med Sci 2015; 184:851–854. [DOI] [PubMed] [Google Scholar]
- 24.Wang M, Chu H, Lv Q, et al. Cumulative effect of genome- wide association study-identified genetic variants for bladder cancer. Int J Cancer 2014; 135:2653–2660. [DOI] [PubMed] [Google Scholar]
- 25.Berber U, Yilmaz I, Yilmaz O, et al. CYP1A1 (Ile 462 Val), CYP1B1 (Ala 119 Ser and Val 432 Leu), GSTM1 (null), and GSTT1 (null) polymorphisms and bladder cancer risk in a turkish population. Asian Pac J Cancer Prev 2013; 14:3925–3929. [DOI] [PubMed] [Google Scholar]
- 26.won Kang H, Song PH, Ha YS, et al. Glutathione S-transferase M1 and T1 polymorphisms: susceptibility and outcomes in muscle invasive bladder cancer patients. Eur J Cancer 2013; 49:3010–3019. [DOI] [PubMed] [Google Scholar]
- 27.Safarinejad MR, Safarinejad S, Shafiei N, et al. Association of genetic polymorphism of glutathione S-transferase (GSTM1, GSTT1, GSTP1) with bladder cancer susceptibility. Urol Oncol 2013; 31:1193–1203. [DOI] [PubMed] [Google Scholar]
- 28.Ovsiannikov D, Selinski S, Lehmann ML, et al. Polymorphic enzymes, urinary bladder cancer risk, and structural change in the local industry. J Toxicol Environ Health Part A 2012; 75:557–565. [DOI] [PubMed] [Google Scholar]
- 29.Öztürk T, Kahraman ÖT, Toptaş B, et al. The effect of CYP1A1 and GSTM1 gene polymorphisms in bladder cancer development in a Turkish population. In Vivo 2011; 25:663–668. [PubMed] [Google Scholar]
- 30.Rouissi K, Ouerhani S, Hamrita B, et al. Smoking and polymorphisms in xenobiotic metabolism and DNA repair genes are additive risk factors affecting bladder cancer in Northern Tunisia. Pathol Oncol Res 2011; 17:879–886. [DOI] [PubMed] [Google Scholar]
- 31.Goerlitz D, El Daly M, Abdel-Hamid M, et al. GSTM1, GSTT1 null variants, and GPX1 single nucleotide polymorphism are not associated with bladder cancer risk in Egypt. Cancer Epidemiol Biomarkers Prev 2011; 20:1552–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altayli E, Gunes S, Yilmaz AF, et al. CYP1A2, CYP2D6, GSTM1, GSTP1, and GSTT1 gene polymorphisms in patients with bladder cancer in a Turkish population. Int Urol Nephrol 2009; 41:259–266. [DOI] [PubMed] [Google Scholar]
- 33.Grando JPS, Kuasne H, Losi-Guembarovski R, et al. Association between polymorphisms in the biometabolism genes CYP1A1, GSTM1, GSTT1 and GSTP1 in bladder cancer. Clin Exp Med 2009; 9:21–28. [DOI] [PubMed] [Google Scholar]
- 34.Rouissi K, Ouerhani S, Marrakchi R, et al. Combined effect of smoking and inherited polymorphisms in arylamine N-acetyltransferase 2, glutathione S-transferases M1 and T1 on bladder cancer in a Tunisian population. Cancer Genet Cytogenet 2009; 190:101–107. [DOI] [PubMed] [Google Scholar]
- 35.Song DK, Xing DL, Zhang LR, et al. Association of NAT2, GSTM1, GSTT1, CYP2A6, and CYP2A13 gene polymorphisms with susceptibility and clinicopathologic characteristics of bladder cancer in Central China. Cancer Detect Prev 2009; 32:416–423. [DOI] [PubMed] [Google Scholar]
- 36.Zupa A, Sgambato A, Bianchino G, et al. GSTM1 and NAT2 polymorphisms and colon, lung and bladder cancer risk: a case-control study. Anticancer Res 2009; 29:1709–1714. [PubMed] [Google Scholar]
- 37.Abd El Hameed AH, Negm OE, El-Gamal OM, et al. Genetic polymorphism of glutathione S-transferases M1 and T1 in Egyptian patients with bilharzial bladder cancer. Urol Oncol 2008; 1009.1015. [DOI] [PubMed] [Google Scholar]
- 38.Covolo L, Placidi D, Gelatti U, et al. Bladder cancer, GSTs, NAT1, NAT2, SULT1A1, XRCC1, XRCC3, XPD genetic polymorphisms and coffee consumption: a case–control study. Eur J Epidemiol 2008; 23:355–362. [DOI] [PubMed] [Google Scholar]
- 39.Golka K, Schmidt T, Seidel T, et al. The influence of polymorphisms of glutathione S-transferases M1 and M3 on the development of human urothelial cancer. J Toxicol Environ Health 2008; 71:881–886. [DOI] [PubMed] [Google Scholar]
- 40.Shao J, Gu M, Zhang Z, et al. Genetic variants of the cytochrome P450 and glutathione S-transferase associated with risk of bladder cancer in a south-eastern Chinese population. Int J Urol 2008; 15:216–221. [DOI] [PubMed] [Google Scholar]
- 41.Moore LE, Malats N, Rothman N, et al. Polymorphisms in one-carbon metabolism and trans-sulfuration pathway genes and susceptibility to bladder cancer. Int J Cancer 2007; 120:2452–2458. [DOI] [PubMed] [Google Scholar]
- 42.Cengiz M, Ozaydin A, Ozkilic AC, et al. The investıgatıon of GSTT1, GSTM1 and SOD polymorphism in bladder cancer patıents. Int Urol Nephrol 2007; 39:1043–1048. [DOI] [PubMed] [Google Scholar]
- 43.Murta-Nascimento C, Silverman DT, Kogevinas M, et al. Risk of bladder cancer associated with family history of cancer: do low-penetrance polymorphisms account for the increase in risk. Cancer Epidemiol Biomarkers Prev 2007; 16:1595–1600. [DOI] [PubMed] [Google Scholar]
- 44.Zhao H, Lin J, Grossman HB, et al. Dietary isothiocyanates, GSTM1, GSTT1, NAT2 polymorphisms and bladder cancer risk. Int J Cancer 2007; 120:2208–2213. [DOI] [PubMed] [Google Scholar]
- 45.Saad AA, O’Connor PJ, Mostafa MH, et al. Glutathione S-transferase M1, T1 and P1 polymorphisms and bladder cancer risk in Egyptians. Int J Biol Markers 2004; 20:69–72. [DOI] [PubMed] [Google Scholar]
- 46.García-Closas M, Malats N, Silverman D, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet 2005; 366:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karagas MR, Park S, Warren A, et al. Gender, smoking, glutathione-S-transferase variants and bladder cancer incidence: a population-based study. Cancer Lett 2005; 219:63–69. [DOI] [PubMed] [Google Scholar]
- 48.Kellen E, Zeegers M, Paulussen A, et al. Does occupational exposure to PAHs, diesel and aromatic amines interact with smoking and metabolic genetic polymorphisms to increase the risk on bladder cancer? The Belgian case control study on bladder cancer risk. Cancer Lett 2007; 245:51–60. [DOI] [PubMed] [Google Scholar]
- 49.Kim EJ, Jeong P, Quan C, et al. Genotypes of TNF-α, VEGF, hOGG1, GSTM1, and GSTT1: useful determinants for clinical outcome of bladder cancer. Urology 2005; 65:70–75. [DOI] [PubMed] [Google Scholar]
- 50.Sobti RC, Al-Badran AI, Sharma S, et al. Genetic polymorphisms of CYP2D6, GSTM1, and GSTT1 genes and bladder cancer risk in North India. Cancer Genet Cytogenet 2005; 156:68–73. [DOI] [PubMed] [Google Scholar]
- 51.Srivastava DSL, Mishra DK, Mandhani A, et al. Association of genetic polymorphism of glutathione S-transferase M1, T1, P1 and susceptibility to bladder cancer. Eur Urol 2005; 48:339–344. [DOI] [PubMed] [Google Scholar]
- 52.Hung RJ, Boffetta P, Brennan P, et al. GST, NAT, SULT1A1, CYP1B1 genetic polymorphisms, interactions with environmental exposures and bladder cancer risk in a high-risk population. Int J Cancer 2004; 110:598–604. [DOI] [PubMed] [Google Scholar]
- 53.Moore LE, Wiencke JK, Bates MN, et al. Investigation of genetic polymorphisms and smoking in a bladder cancer case–control study in Argentina. Cancer Lett 2004; 211:199–207. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava DSL, Kumar A, Mittal B, et al. Polymorphism of GSTM1 and GSTT1 genes in bladder cancer: a study from North India. Arch Toxicol 2004; 78:430–434. [DOI] [PubMed] [Google Scholar]
- 55.Jeong HJ, Kim HJ, Seo IY, et al. Association between glutathione S-transferase M1 and T1 polymorphisms and increased risk for bladder cancer in Korean smokers. Cancer Lett 2003; 202:193–199. [DOI] [PubMed] [Google Scholar]
- 56.Giannakopoulos X, Charalabopoulos K, Baltogiannis D, et al. The role of N-acetyltransferase-2 and glutathione S-transferase on the risk and aggressiveness of bladder cancer. Anticancer Res 2001; 22:3801–3804. [PubMed] [Google Scholar]
- 57.Lee SJ, Cho SH, Park SK, et al. Combined effect of glutathione S-transferase M1 and T1 genotypes on bladder cancer risk. Cancer Lett 2002; 177:173–179. [DOI] [PubMed] [Google Scholar]
- 58.Aktas D, Ozen H, Atsu N, et al. Glutathione S-transferase M1 gene polymorphism in bladder cancer patients: a marker for invasive bladder cancer. Cancer Genet Cytogenet 2001; 125:1–4. [DOI] [PubMed] [Google Scholar]
- 59.Törüner GA, Akyerli C, Uçar A, et al. Polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and bladder cancer susceptibility in the Turkish population. Arch Toxicol 2001; 75:459–464. [DOI] [PubMed] [Google Scholar]
- 60.KIM WUNJAE, LEE HLAE, LEE SC, et al. Polymorphisms of N-acetyltransferase 2, glutathione S-transferase mu and theta genes as risk factors of bladder cancer in relation to asthma and tuberculosis. J Urol 2000; 164:209–213. [PubMed] [Google Scholar]
- 61.Schnakenberg E, Lustig M, Breuer R, et al. Gender-specific effects of NAT2 and GSTM1 in bladder cancer. Clin Genet 2000; 57:270–277. [DOI] [PubMed] [Google Scholar]
- 62.Steinhoff C, Franke KH, Golka K, et al. Glutathione transferase isozyme genotypes in patients with prostate and bladder carcinoma. Arch Toxicol 2000; 74:521–526. [DOI] [PubMed] [Google Scholar]
- 63.Salagovic J, Kalina H, Hrivnak M. The role of human glutathione S-transferases M1 and T1 in individual susceptibility to bladder cancer. Physiol Res 1999; 48:465–471. [PubMed] [Google Scholar]
- 64.Abdel-Rahman SZ, Anwar WA, Abdel-Aal WE, et al. GSTM1 and GSTT1 genes are potential risk modifiers for bladder cancer. Cancer Detect Prev 1997; 22:129–138. [DOI] [PubMed] [Google Scholar]
- 65.Brockmöller J, Cascorbi I, Kerb R, et al. Combined analysis of inherited polymorphisms in arylamine N-acetyltransferase 2, glutathione S-transferases M1 and T1, microsomal epoxide hydrolase, and cytochrome P450 enzymes as modulators of bladder cancer risk. Cancer Res 1996; 56:3915–3925. [PubMed] [Google Scholar]
- 66.Anwar WA, Abdel-Rahman SZ, El-Zein RA, et al. Genetic polymorphism of GSTM1, CYP2E1 and CYP2D6 in Egyptian bladder cancer patients. Carcinogenesis 1996; 17:1923–1929. [DOI] [PubMed] [Google Scholar]
- 67.Zhong S, Wyllie AH, Barnes D, et al. Relationship between the GSTM1 genetic polymorphism and susceptibility to bladder, breast and colon cancer. Carcinogenesis 1993; 14:1821–1824. [DOI] [PubMed] [Google Scholar]
- 68.Pandith AA, Lateef A, Shahnawaz S, et al. GSTP1 gene Ile105Val polymorphism causes an elevated risk for bladder carcinogenesis in smokers. Asian Pac J Cancer Prev 2013; 14:6375–6378. [DOI] [PubMed] [Google Scholar]
- 69.Lesseur C, Gilbert-Diamond D, Andrew AS, et al. A case-control study of polymorphisms in xenobiotic and arsenic metabolism genes and arsenic-related bladder cancer in New Hampshire. Toxicol Lett 2012; 210:100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang RG, Xu GY, Chen WJ, et al. Genetic polymorphisms of glutathione S-transferase P1 and bladder cancer susceptibility in a Chinese population. Genet Test Mol Biomarkers 2011; 15:85–88. [DOI] [PubMed] [Google Scholar]
- 71.Fontana L, Delort L, Joumard L, et al. Genetic polymorphisms in CYP1A1, CYP1B1, COMT, GSTP1 and NAT2 genes and association with bladder cancer risk in a French cohort. Anticancer Res 2009; 29:1631–1635. [PubMed] [Google Scholar]
- 72.Yuan JM, Chan KK, Coetzee GA, et al. Genetic determinants in the metabolism of bladder carcinogens in relation to risk of bladder cancer. Carcinogenesis 2008; 29:1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kopps S, Angeli-Greaves M, Blaszkewicz M, et al. Glutathione S-transferase P1 ILE105Val polymorphism in occupationally exposed bladder cancer cases. J Toxicol Environ Health 2008; 71:898–901. [DOI] [PubMed] [Google Scholar]
- 74.Xing DL, et al. Association study of polymorphisms in the human drug metabolism enzyme gene and bladder cancer risk. Zhengzhou Daxue 2006; 12:1–61. [Google Scholar]
- 75.Cao W, Cai L, Rao JY, et al. Tobacco smoking, GSTP1 polymorphism, and bladder carcinoma. Cancer 2005; 104:2400–2408. [DOI] [PubMed] [Google Scholar]
- 76.Qing-wen MA, Guo-fang LIN, JI-GANG C, et al. Polymorphism of glutathione S-transferase T1, M1 and P1 genes in a Shanghai population: patients with occupational or non-occupational bladder cancer. Biomed Environ Sci 2002; 15:253–260. [PubMed] [Google Scholar]
- 77.Peluso M, Airoldi L, Magagnotti C, et al. White blood cell DNA adducts and fruit and vegetable consumption in bladder cancer. Carcinogenesis 2000; 21:183–187. [DOI] [PubMed] [Google Scholar]
- 78.Harries LW, Stubbins MJ, Forman D, et al. Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis 1997; 18:641–644. [DOI] [PubMed] [Google Scholar]
- 79.Henríquez-Hernández LA, Navarro P, Luzardo OP, et al. Polymorphisms of glutathione S-transferase μ and θ, MDR1 and VEGF genes as risk factors of bladder cancer: a case-control study. Urol Oncol 2012; 30:660–665. [DOI] [PubMed] [Google Scholar]
- 80.Moore LE, Baris DR, Figueroa JD, et al. GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis 2011; 32:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salinas-Sánchez AS, Sánchez-Sánchez F, Donate-Moreno MJ, et al. Polymorphic deletions of the GSTT1 and GSTM1 genes and susceptibility to bladder cancer. BJU Int 2011; 107:1825–1832. [DOI] [PubMed] [Google Scholar]
- 82.Cantor KP, Villanueva CM, Silverman DT, et al. Polymorphisms in GSTT1, GSTZ1, and CYP2E1, disinfection by-products, and risk of bladder cancer in Spain. Environ Health Perspect 2010; 118:1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song DK, Xing DL, Zhang LR, et al. Relationship between polymorphism of glutathione stransferase and genetic susceptibility to bladder cancer. Chin J Urol 2008; 29:80–83. [Google Scholar]
- 84.Kogevinas M, Fernandez F, Garcia-Closas M, et al. Hair dye use is not associated with risk for bladder cancer: evidence from a case-control study in Spain. Eur J Cancer 2006; 42:1448–1454. [DOI] [PubMed] [Google Scholar]
- 85.Shao CX, Xiang YB, Zhang W, et al. Polymorphisms of GSTM1 and GSTT1 with smoking and bladder cancer risk: a population-based case control study. Tumor 2006; 26:346–351. [Google Scholar]
- 86.Ouerhani S, Tebourski F, Slama MRB, et al. The role of glutathione transferases M1 and T1 in individual susceptibility to bladder cancer in a Tunisian population. Ann Hum Biol 2006; 33:529–535. [DOI] [PubMed] [Google Scholar]
- 87.McGrath M, Michaud D, Vivo I. Polymorphisms in GSTT1, GSTM1, NAT1 and NAT2 genes and bladder cancer risk in men and women. BMC Cancer 2006; 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Golka K, Seidel T, Dietrich H, et al. [Occupational and non-occupational risk factors in bladder cancer patients in an industrialized area located in former East-Germany]. Aktuelle Urologie 2005; 36:417–422. [DOI] [PubMed] [Google Scholar]
- 89.Sanyal S, Festa F, Sakano S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis 2004; 25:729–734. [DOI] [PubMed] [Google Scholar]
- 90.Chen Y, Xu L, Guo YL, et al. Polymorphisms in GSTT1 and p53 and urinary transitional cell carcinoma in south-western Taiwan: a preliminary study. Biomarkers 2004; 9:386–394. [DOI] [PubMed] [Google Scholar]
- 91.Gago-Dominguez M, Bell DA, Watson MA, et al. Permanent hair dyes and bladder cancer: risk modification by cytochrome P4501A2 and N-acetyltransferases 1 and 2. Carcinogenesis 2003; 24:483–489. [DOI] [PubMed] [Google Scholar]
- 92.Kim WJ, Kim H, Kim CH, et al. GSTT1-null genotype is a protective factor against bladder cancer. Urology 2002; 60:913–918. [DOI] [PubMed] [Google Scholar]
- 93.Lee SJ, Kang D, Cho SH, et al. Association of genetic polymorphism of glutathione s-transferase M1 and T1 and bladder cancer. J Korean Cancer Assoc 1999; 31:548–555. [Google Scholar]
- 94.Salagovic J, Kalina I, Stubna J, et al. Genetic polymorphism of glutathione S-transferases M1 and T1 as a risk factor in lung and bladder cancers. Neoplasma 1997; 45:312–317. [PubMed] [Google Scholar]
- 95.Katoh T, Inatomi H, Kim H, et al. Effects of glutathione S-transferase (GST) M1 and GSTT1 genotypes on urothelial cancer risk. Cancer Lett 1998; 132:147–152. [DOI] [PubMed] [Google Scholar]
- 96.Kim H, Kim WJ, Lee HL, et al. A case-control study on the effects of the genetic polymorphisms of N-acetyltransferase 2 and glutathione S-transferase mu and theta on the risk of bladder cancer. Korean J Prevent Med 1998; 31:275–284. [Google Scholar]
- 97.Kempkes M, Golka K, Reich S, et al. Glutathione S-transferase GSTM1 and GSTT1 null genotypes as potential risk factors for urothelial cancer of the bladder. Arch Toxicol 1996; 71:123–126. [DOI] [PubMed] [Google Scholar]
- 98.Malats N, Real FX. Epidemiology of bladder cancer. Hematol Oncol Clin North Am 2015; 29:177–189. [DOI] [PubMed] [Google Scholar]
- 99.Rodrigues IS, Kuasne H, Losi-Guembarovski R, et al. Evaluation of the influence of polymorphic variants CYP1A1 2B, CYP1B1 2, CYP3A4 1B, GSTM1 0, and GSTT1 0 in PCa. Urol Oncol 2011; 29:654–663. [DOI] [PubMed] [Google Scholar]
- 100.McIlwain CC, Townsend DM, Tew KD, et al. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene 2006; 25:1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology 2000; 61:154–166. [DOI] [PubMed] [Google Scholar]
- 102.Coles BF, Morel F, Rauch C, et al. Effect of polymorphism in the human glutathione S-transferase A1 (hGSTA1) promoter on hepatic GSTA1 and GSTA2 expression. Pharmacogenetics 2001; 11:663–669. [DOI] [PubMed] [Google Scholar]
- 103.Coles FB, Kadlubar FF. Human class glutathione S-transferases: genetic polymorphism, expression, and susceptibility to disease. Methods Enzymol 2005; 401:9–42. [DOI] [PubMed] [Google Scholar]
- 104.Zhong S, Wyllie AH, Barnes D, et al. Relationship between the GSTM1 genetic polymorphism and susceptibility to bladder, breast and colon cancer. Carcinogenesis 1993; 14:1821–1824. [DOI] [PubMed] [Google Scholar]
- 105.Harris MJ, Coggan M, Langton L, et al. Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients. Pharmacogenetics 1998; 8:27–31. [DOI] [PubMed] [Google Scholar]
- 106.Wiencke JK, Pemble S, Ketterer B, et al. Gene deletion of glutathione S-transferase theta: correlation with induced genetic damage and potential role in endogenous mutagenesis. Cancer Epidemiol Biomarkers Prev 1995; 4:253–259. [PubMed] [Google Scholar]
- 107.McIlwain CC, Townsend DM, Tew KD, et al. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene 2006; 25:1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Listowsky I, Abramovitz M, Homma H, et al. Intracellular binding and transport of hormones and xenobiotics by glutathione-S-transferases. Drug Metab Rev 1988; 19:305–318. [DOI] [PubMed] [Google Scholar]
- 109.Huang W, Shi H, Hou Q, et al. GSTM1 and GSTT1 polymorphisms contribute to renal cell carcinoma risk: evidence from an updated meta-analysis. Sci Rep 2015; 5:17971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hirschhorn JN, Lohmueller K, Byrne E, et al. A comprehensive review of genetic association studies. Genet Med 2002; 4:45–61. [DOI] [PubMed] [Google Scholar]
- 111.Kellen E, Hemelt M, Broberg K, et al. Pooled analysis and meta-analysis of the glutathione S-transferase P1 Ile 105Val polymorphism and bladder cancer: a HuGE-GSEC review. Am J Epidemiol 2007; 165:1221–1230. [DOI] [PubMed] [Google Scholar]


