Abstract
RCAS avian viruses have been used to deliver oncogene expression and induce tumors in transgenic mice expressing the virus receptor TVA. Here we report the generation and characterization of a novel RCAS-Cre-IRES-PyMT (RCI-PyMT) virus designed to specifically knockout genes of interest in tumors generated in appropriate mutant mouse hosts. FGF receptor 1 (FGFR1) is a gene that is amplified in human breast cancer, but there have been no definitive studies on its function in mammary tumorigenesis, progression, and metastasis in vivo in spontaneous tumors in mice. We used the retroviral tumor knockout, or TuKO, strategy to delete fgfr1 in PyMT-induced mammary tumors in K19-tva/fgfr1loxP/loxP mice. The similarly injected control K19-tva mice developed mammary tumors exhibiting high metastasis to lung, making this an ideal model for breast cancer metastasis. The fgfr1 TuKO tumors showed significantly decreased primary tumor growth and, most importantly, greatly reduced metastasis to lung. In contrast to previous reports, FGFR1 action in this spontaneous mammary tumor model does not significantly induce epithelial-to-mesenchymal transition. Loss of FGFR1 does generate a gene signature that is reverse correlated with FGFR1 gene amplification and/or upregulation in human breast cancer. Our results suggest that FGFR1 signaling is a key pathway driving breast cancer lung metastasis and that targeting FGFR1 in breast cancer is an exciting approach to inhibit metastasis.
Abbreviations: RCI-PyMT, RCAS-Cre-IRES-PyMT virus; TuKO, tumor specific knockout
Introduction
The fibroblast growth factor receptor family consists of four members: FGFR1 to 4. FGFR1 is emerging as a therapeutic target for breast cancer (BCa). FGFR1 was initially identified as a potential druggable therapeutic target for BCa in an unbiased genomic and transcriptional aberration screening [1]. FGFR1 amplification was detected in a subset of BCa and is an independent predictor of overall survival [2]. The induced activation of an engineered FGFR1 in mammary luminal epithelial cells leads to alveolar hyperplasia and invasive lesions in vivo [3] and epithelial-to-mesenchymal transition (EMT) and invasive behavior in 3D culture in vitro [4]. Targeting FGFRs using a pan-FGFR inhibitor in mouse mammary tumor cell lines highly expressing FGFR2 inhibited xenograft growth and metastasis [5]. Finally, a recent study revealed that human breast cancers with high-level clonal amplification of FGFR2 are highly sensitive to FGFR inhibition [6]. However, it remains unknown how specifically abolishing FGFR1 signaling could affect spontaneous mammary tumorigenesis, progression, and metastasis.
The avian retroviral–based gene expression vector RCAS is designed to deliver foreign genes into transgenic mouse lines expressing the virus receptor TVA. Mammary tumors can be induced in the MMTV-tva mice by injection of RCAS-PyMT virus into mammary gland via the nipple duct [7]. These mammary tumors recapitulate many features of human breast cancer but seldom metastasize to lung [7].
Here we report the generation of the RCAS-Cre-IRES-PyMT (RCI-PyMT) viruses. When injected into transgenic mice expressing TVA, these allow the simultaneous expression of Cre to knock out the loxP flanked target gene along with the PyMT oncogene to induce tumorigenesis. Injection of RCI-PyMT viruses into the mammary gland of K19-tva mice [8] induces mammary tumor growth with metastasis to lung at high frequency. Knockout of fgfr1 in this model significantly inhibits mammary tumor growth; prolongs host mice survival; and, most importantly, strongly decreases metastasis to lung.
Materials and Methods
Plasmids
The pZ/EG plasmid was kindly provided by Dr. Lobe [9]. To generate the RCAS-Cre-IRES-PyMT (RCI-PyMT) vector, the IRES fragment was PCR amplified from the pBMN-I-eGFP vector [10] using forward primer 5′ ATAAGAATGCGGCCGCTACGTAAATTCCGCCCCTC 3′ carrying the NotI restriction site and reverse primer 5′ CCTTAATTAAGTTGTGGCCATATTATCATCG 3′ carrying the PacI restriction site, and cloned into the RCAS-Y vector between NotI and PacI restriction sites. This produced the RCAS-IRES vector. The Cre cDNA was PCR amplified from the HRMMPCreGFP vector [10] using forward primer 5′ ATAAGAATGCGGCCGCCACCATGTCCAATTTACTGACCGTACA 3′ and reverse primer 5′ ATAAGAATGCGGCCGCTAATCGCCATCTTCCAGC 3′ that both carry NotI restriction site, and cloned into the RCAS-IRES vector using NotI site. This produced the RCAS-Cre-IRES vector. Finally, the C-terminally HA tagged PyMT cDNA was PCR amplified from the RCAS-Y vector using forward primer 5′ CCTTAATTAAGCCACCATGGATAGAGTTCTGAGCAGAG 3′ and reverse primer 5′ CCTTAATTAACTAAGCGTAATCTGGAACATCGTATGGGTAGAAATGCCGGGAACGT 3′ that both carry PacI restriction sites, and cloned into the RCAS-Cre-IRES vector using PacI restriction site. This produced the RCAS-Cre-IRES-PyMT (RCI-PyMT) vector. The sequence and/or orientation of each element was verified by Sanger sequencing.
Animals
FVB mice were purchased from Harlan Laboratories. The fgfr1loxP/loxP mice (in ICR background) were kind gifts from Dr. Juha Partanen [11] and have been backcrossed into FVB background as previously described [12]. K19-tva mice were provided by Dr. Brian Lewis [8]. All experiments were carried out in compliance with the NIH Guide for the Care and Use of Laboratory Animals and according to Baylor College of Medicine institutional guidelines and IACUC approval.
Generation of the DF1-Z/EG Reporter Cell Line
The DF-1 chicken fibroblast cells [13] were cultured in DMEM supplemented with 10% FBS (Invitrogen). To create a reporter cell line for titration of RCAS viruses expressing Cre, including the RCI-PyMT viruses here, DF-1 cells were transfected with the pZ/EG vector and selected for resistance to G418. The resulted stable cell line was named DF1-Z/EG. As described in Figure 1, these DF1-Z/EG cells only express an enhanced GFP after infection with Cre-expressing viruses including RCI-PyMT.
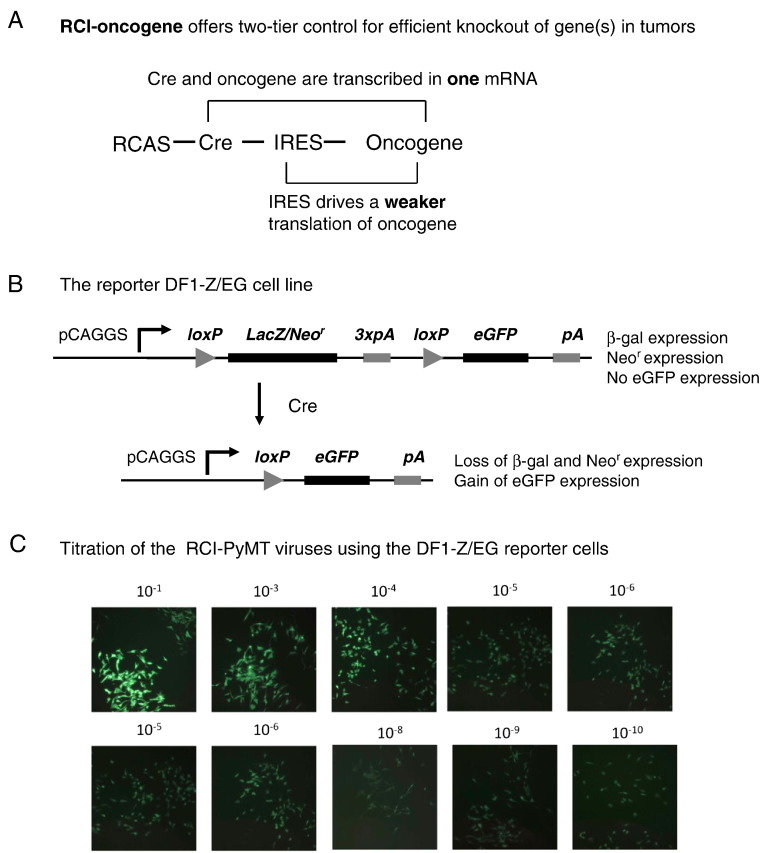
Figure 1.
Design, generation, and titration of the RCI-PyMT viruses. (A) The two-tier control for efficient and tumor specific gene manipulation using the RCI-PyMT gene delivery system. (B) Scheme of determining titers of the RCI-PyMT viruses using the DF1-Z/EG reporter cells. (C) Titration of the RCI-PyMT viruses by limiting dilution on DF1-Z/EG reporter cells. GFP+ cells indicate RCI-PyMT virus infection.
Virus Production, Titration, and Delivery to Mammary Gland
To produce viruses, DF-1 cells were transfected with the RCI-PyMT vector using Lipofectamine 2000 transfection reagent (Invitrogen). Viruses in the supernatant were concentrated 100-fold and frozen in aliquots for virus titer determination and injection of mouse as previously described [7].
The titers of the RCI-PyMT viruses were determined by limiting dilution on the DF1-Z/EG reporter cells. GFP+ signal indicates virus infection. Due to virus self-propagation after infecting the DF1-Z/EG reporter cells, the GFP signal increases over time during inoculation. Finally, viruses can also be collected from these viruses-infected DF1-Z/EG cells.
To infect mammary glands, female TVA transgenic mice at 12 to 16 weeks of age were anesthetized and intraductally injected with 10 μl of the RCI-PyMT viruses together with 0.1% bromophenol blue as tracking dye following a previously described protocol [7]. The 12- to 16-week-old animals are used here to facilitate the identification and manipulation of the nipple ducts. Three mammary glands per mouse were injected.
Genomic DNA PCR
fgfr1loxP, fgfr1wt, fgfr1KO allele, and K19-tva alleles were PCR screened and identified as previously described [8], [12]. PCR for β-casein using primers 5′ GATGTGCTCCAGGCTAAAGTT 3’and 5′ AGAAACGGAATGTTGTGGAGT 3′ was used as positive control for genomic DNA quality. PCR was done using genomic DNA from mouse tails and/or tumors.
Histology and Immunohistochemistry
Tissues were fixed in 4% paraformaldehyde and paraffin embedded. Sections (5 μm) were mounted onto slides for H&E staining and/or immunohistochemistry. Immunostaining were performed following a general protocol published previously [14]. Antigen retrieval consisted of boiling slides in 10 mM Tris, 1 mM EDTA, and 0.05% Tween 20 (pH 9.0) for 20 minutes in a microwave was used for immunofluorescence staining for E-cadherin (#3195, Cell Signaling Technology).
Western Blot
Protein samples were prepared in RIPA buffer (100 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl pH 8.0, 1% Triton X-100 with 100× protease inhibitors cocktail solution from GenDEPOT and the phosphatase inhibitors 10 mM of NaF and 20 mM of Na3VO4) and applied a brief sonication. Protein concentration was measured using the BCA protein assay kit (Piece). Equal amount (5-20 μg) of protein was used in the Western blot analysis. Antibodies used are anti-FGFR1 (#9740), anti–E-cadherin (#3195, Cell Signaling Technology), and anti–β-actin (#A5316, Sigma).
Microarray Analysis
Microarray analysis was performed using the SurePrint G3 Mouse Gene Expression v2 8 × 60 K Microarray chip (Agilent Technologies). The Feature Extraction Software v9.1.3.1 (Agilent Technologies) was used to extract and analyze the signals. The microarray data were processed using bioconductor (loess normalization). Fold changes and t tests for each gene were computed using log-transformed values. Array data have been deposited into the Gene Expression Omnibus (GSE85754). Gene signature scoring of human breast tumor expression profiles from TCGA was carried out as previously described [15], [16], with PAM50 subtype calls as previously provided [17]. The signature t score was defined as the two-sided t statistic comparing, within each tumor profile, the average of the “signature up” genes with the average of the “signature down” genes (e.g., the t score for a given tumor being high when both the “up” genes in the signature were high and the “down” genes were low). High-level gene amplification of FGFR1 in human tumors was inferred using the “thresholded” copy alteration calls of +2 as provided by Broad Firehose pipeline (http://gdac.broadinstitute.org/).
Statistics
Average tumor weight was compared between groups for statistical relevance using the unpaired Student's t test. The numbers of metastasis were compared using both the unpaired Student's t test and the Mann-Whitney test. The difference of mouse survival was analyzed using both the log-rank test and the Gehan-Breslow-Wilcoxon test. Statistical analyses were generated using Graphpad Prism 5.0 (GraphPad Software). P < .05 is considered statistically significant.
Results
Design and Generation of the RCAS-Cre-IRES-PyMT Viruses
The RCAS-PyMT viruses have been used to induce mammary tumor growth; however, unlike the MMTV-PyMT transgenic model, these virus-induced mammary tumors rarely metastasize to lung [7]. To create a new RCAS-TVA–based model that allows manipulation of the host cell genome in tumors, we designed an RCAS-Cre-IRES-PyMT (RCI-PyMT) viral construct. This expresses Cre and the PyMT oncogene in a single mRNA. Cre protein expression is driven by 5′ cap translation, whereas PyMT oncogene protein expression will be driven by the weaker IRES-mediated translation. This will drive efficient Cre expression in PyMT-induced tumor cells (Figure 1A). When these RCI-PyMT viruses are injected into the experimental K19-tva/fgfr1loxP/loxP mice and the control K19-tva mice used in this study (as described below, Figure 2A), Cre expression is expected to efficiently knock out fgfr1 in PyMT oncogene-induced mammary tumor cells in the targeted strain.
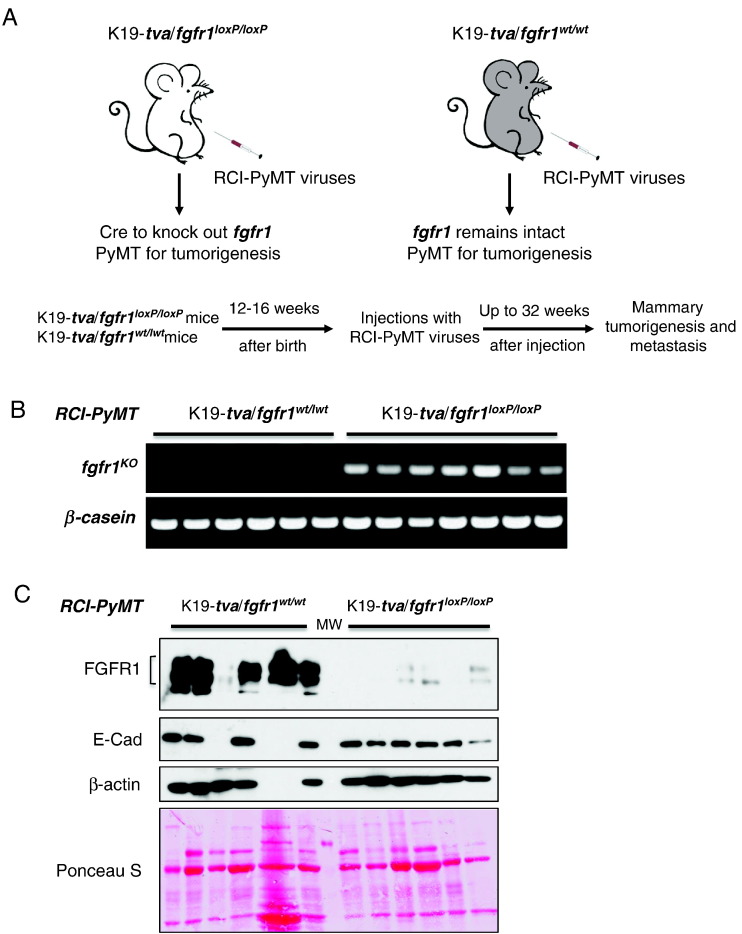
Figure 2.
TuKO of fgfr1 in the PyMT-induced mammary tumors. (A) Scheme of infecting mammary glands of K19-tva/fgfr1loxP/loxP mice and control K19-tva mice using the RCI-PyMT viruses through intraductal injections. (B) PCR for fgfr1KO-specific alleles confirms TuKO of fgfr1 in RCI-PyMT viruses–infected K19-tva/fgfr1loxP/loxP mice. (C) Western blots for the expression of FGFR1 and E-cadherin in tumors. Western blot for β-actin and Ponceau S staining of membrane are shown as loading control. One tumor expressed little β-actin. MW: molecular weight marker.
The Generation of a Reporter Cell Line and the Titration of the RCAS-Cre-IRES-PyMT Viruses
The pZ/EG vector has been used to create the Z/EG double reporter mouse line, which expresses an enhanced GFP after Cre-mediated delection of the LacZ expression cassette [9]. To titrate RCI-PyMT viruses, we created a stable DF1-Z/EG reporter cell line by transfection of the pZ/EG vector into avian DF-1 cells. As illustrated in Figure 1B, these reporter cells turn on eGFP expression upon infection with Cre-expressing viruses.
Using these cells, we confirmed the production of high-titer (>1010) RCI-PyMT viruses (Figure 1C). Due to self-propagation of the RCI-PyMT viruses from the infected DF1-Z/EG cells, these newly produced viruses will continuously infect the DF1-Z/EG cells to produce more GFP+ cells. As a result, this titration method provides extremely high sensitivity, whereas the population of GFP+ cells may not proportionally reflect virus titer/concentration.
Conditional Ablation of fgfr1 in PyMT Oncogene-Induced Mammary Tumor Model
To determine how conditional ablation of fgfr1 affects mammary tumor progression and metastasis, fgfr1loxP/loxP mice [11], [12] and the K19-tva mice [8] expressing avian virus receptor TVA in the cytokeratin 19–positive epithelial cells were crossed to produce K19-tva/fgfr1loxP/wt mice. These mice were then crossed with the fgfr1loxP/loxP mice to produce the target K19-tva/fgfr1loxP/loxP mice. K19-tva mice were used as control. We performed injections of the RCI-PyMT viruses into the nipple ducts of these mice at 12 to 16 weeks of age to infect mammary epithelial cells. As illustrated in Figure 2A, this produces tumor-specific knockout (TuKO) of fgfr1 in the K19-tva/fgfr1loxP/loxP mice, whereas FGFR1 remains intact in tumors in the control K19-tva mice. PCR amplification of the fgfr1KO-specific alleles confirmed knockout of fgfr1 in the TuKO tumors but not in the control (Figure 2B).
To determine FGFR1 expression status in these RCI-PyMT–induced mammary tumors, we isolated total protein from the collected tumors and performed Western blots. FGFR1 is expressed at various levels in most of the control mammary tumors, and its expression is diminished in the experimental fgfr1 TuKO tumors (Figure 2C). Therefore, by using this newly designed RCI-PyMT/K19-tva/fgfr1loxP/loxP gene delivery system, we efficiently knocked out fgfr1 in PyMT-induced mammary tumors.
fgfr1 Ablation Inhibits Mammary Tumor Progression and Prolongs Host Survival
The mice injected with RCI-PyMT viruses were evaluated for tumor progression and time to death. We observed that a small population of mice either did not develop mammary tumors (data not shown) or survived much longer (over two times standard deviations of mean) than the rest of the mice within the same group (survival of 32 to 51 weeks in the control group and 38 to 95 weeks in the experimental fgfr1 knockout group), likely due to technical variations during virus injection. Therefore, we performed two separate sets of comparison of survival on these mice, namely, those that survived less than 32 weeks (Figure 3A) and those that survived much longer (Supplementary Figure 1).
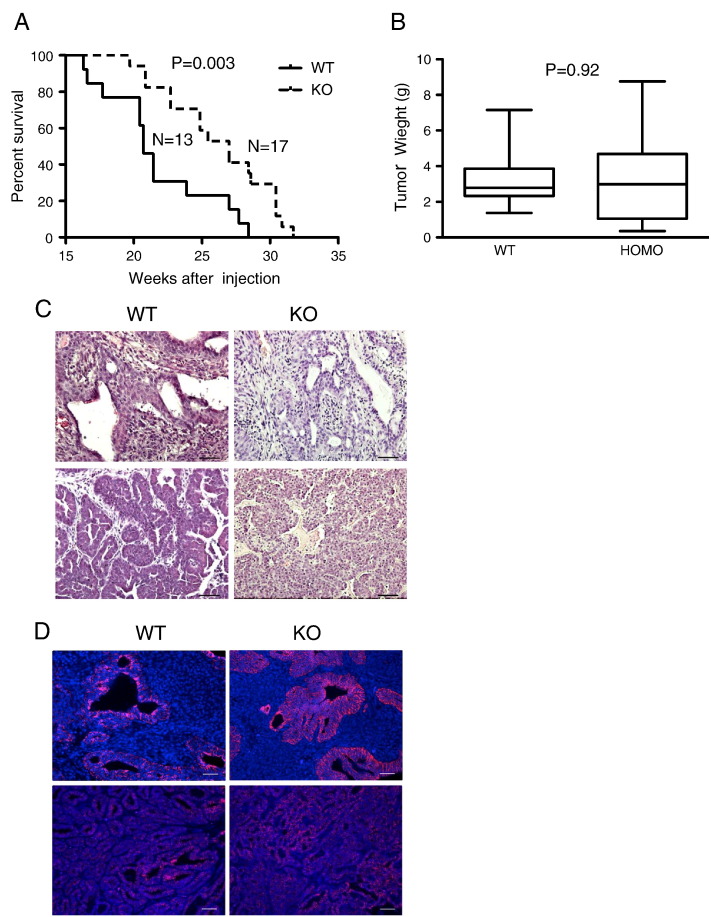
Figure 3.
TuKO of fgfr1 in the PyMT-induced mammary tumors significantly extends host mouse survival. (A) Knockout of fgfr1 in the PyMT-induced mammary tumors significantly extends hosts survival (P = .0029 for log-rank test, and P = .0033 for Gehan-Breslow-Wilcoxon test). (B) Mammary tumor weights were comparable between fgfr1 TuKO tumors and control tumors at the end of this survival study (P = .92, unpaired t test). (C) Histopathology of representative fgfr1 TuKO and control mammary tumors. (D) Immunofluorescence staining of E-cadherin on representative fgfr1 TuKO and control mammary tumors.
In both cases, the mice bearing fgfr1 TuKO tumors survived significantly longer. In the first set, the median survival of mice bearing fgfr1 ablated mammary tumors is 27.0 weeks, which is significantly longer than that of the control mice (20.7 weeks, P = .0029 for log-rank test and P = .0033 for Gehan-Breslow-Wilcoxon test, Figure 3A). The tumor weights at collection are comparable between these two groups (Figure 3B). Similarly, the fgfr1 ablated group in the second set survived a median of 56.5 weeks, which is again significantly longer than that of the control group (median survival of 34.4 weeks, P < .05 for both statistical analyses, Supplementary Figure 1).
The fgfr1 TuKO and Wild-Type Tumors Exhibit Comparable Histopathology
The histopathology of the RCI-PyMT tumors is very heterogeneous among tumors and, in many cases, within the same tumor. This heterogeneity is present in both the wild-type and the fgfr1 TuKO tumors (Figure 3C). Carcinoma; papillary carcinoma; metaplastic carcinoma; carcinoma with squamous differentiation; and, in some cases, liposarcoma are present in both tumor groups, and there is no significant difference in tumor pathology between the two groups.
Because it has been previously documented that FGFR1 plays critical roles in driving EMT, we performed immunofluorescence staining for E-cadherin expression and cellular location on the primary tumors (Figure 3D). We did not observe significant changes in the E-cadherin expression pattern, indicating that loss of FGFR1 does not appear to affect EMT in these mammary tumors. Western blots further confirmed comparable protein expression levels of E-cadherin among the control and the fgfr1 TuKO tumors (Figure 2C).
fgfr1 Ablation Greatly Inhibits Mammary Tumor Metastasis to Lung
At the time of death, lung tissues were harvested and analyzed for histology. A random cross section of lung from each mouse was scored for numbers of metastatic lesions by a pathologist (Figure 4A). Mammary tumor metastases were observed in the randomly selected lung tissue section from 7 out of 13 control mice (up to 27 metastases in each lung), whereas metastases were only observed in 2 out of 16 experimental mice (maximum 2 in each lung). The incidence of metastasis to lung is strongly negatively correlated with FGFR1 knockout within the tumors (P = .0196, odds ratio = 0.114, Fisher's exact test). The average of metastases per lung of the control mice is 4.77 ± 2.23, which was greatly reduced to 0.24 ± 0.16 in the experimental mice (P = .0126, Mann-Whitney test; P = .0332, unpaired Student’s t test, Figure 4A). Together, these data strongly suggest a crucial role of FGFR1 in driving mammary tumor metastasis.
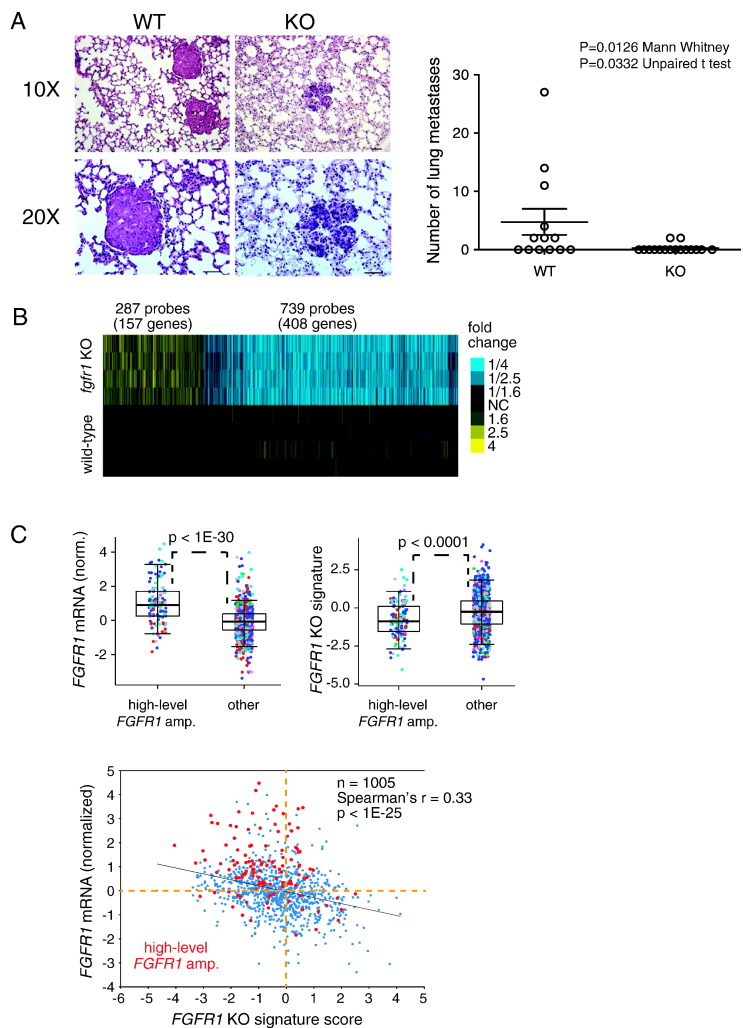
Figure 4.
Knockout of fgfr1 in the PyMT-induced mammary tumors significantly inhibits metastasis to lung. (A) Representative micrographs of lung tissue from mice carrying fgfr1 TuKO tumors and the controls. Knockout of fgfr1 significantly inhibits metastasis to lung (P = .033, unpaired Student’s t test; P = .013, Mann-Whitney test). (B) Heat map of top genes differentially expressed in KO vs wt (P < .01, fold change> 1.4). (C) (top) Box plots of FGFR1 mRNA expression (left, expression normalized to standard deviations from the median) and FGFR1 KO gene signature scoring (right) in human breast tumor expression profiles from TCGA comparing tumors with high-level gene amplification of FGFR1 versus other tumors. P values by t test. Box plots represent 5%, 25%, 50%, 75%, and 95%. Data points colored according to PAM50 subtype (red, basal-like; blue, LumA; cyan, LumB; pink, HER2-e; green, normal-like). (Bottom) Scatterplot of normalized FGFR1 mRNA expression versus FGFR1 KO signature scoring in TCGA human breast tumors. P value by Spearman's rank correlation.
FGFR1 Gene Signature Derived from RCI-PyMT Mammary Tumors Correlates with FGFR1 Gene Amplification and Expression in Human Breast Cancer
To reveal the underlying mechanisms for FGFR1-dependent mammary tumor metastasis, we performed expression microarray analysis on four representative primary mammary tumors from each group (four from control group and four from fgfr1 TuKO group). One thousand twenty-six transcript probes were differentially expressed, including 739 (408 genes) downregulated and 287 (157 genes) upregulated in the fgfr1 TuKO tumors (fold change >1.4, P < .01, Figure 4B).
We defined this list of differentially regulated genes between the fgfr1 TuKO group and the wild-type group as the FGFR1 KO mammary tumor gene signature and investigated whether this signature is reverse correlated with FGFR1 gene amplification in human breast cancers in TCGA (n = 1005 cases). This analysis assessed the direction of expression in the human cancers relative to their induction or repression in the mouse tumors. We found that human breast cancers with FGFR1 gene amplification have significantly lower expression of this gene signature, as expected. Furthermore, this gene signature was inversely correlated with FGFR1 expression across breast cancers (Figure 4C), which supports the notion that this mouse mammary tumor–derived gene transcriptional program is similarly regulated by FGFR1 in human breast cancer.
Discussion
Based on the previously established RCAS-TVA gene delivery system, we designed the RCI-oncogene (RCI-PyMT used in this study)/TVA model for accurate and efficient genetic manipulation in oncogene-induced tumors in vivo [18]. By injecting the RCI-PyMT viruses into the nipple duct of the experimental K19-tva/fgfr1loxP/loxP mice and the control K19-tva mice, we demonstrated that our new model allows accurate and efficient Cre-mediated knockout of fgfr1 within the PyMT oncogene-induced mammary tumors.
The original RCAS-PyMT/TVA mammary tumor model exhibits little metastasis to lung and is not suitable for studying mammary tumor metastasis [7]. In contrast, our RCI-PyMT/TVA model exhibits high lung metastasis, making it an ideal model for breast cancer metastasis. The basis for this difference is not clear, but the significantly prolonged mammary tumor progression, potentially due to weaker PyMT oncogene expression in our model, might allow more time for tumor metastasis.
The RCI vector is designed to allow incorporation of different oncogenes (or mini-oncogenes). Along with the use of K19-tva mice carrying different floxed genes of interest, our model system provides a rapid method to examine the biological functions of gene(s) of interest in mammary tumors driven by different oncogenes. When infecting transgenic mice expressing TVA in different tissues/organs, this model can be easily adapted to study different types of tumors.
Our current study strongly suggests a crucial role of FGFR1 signaling in driving breast tumor metastasis in addition to promoting primary tumor growth. This phenocopies our previous study on prostate-specific knockout of fgfr1 in a spontaneous prostate tumor model where we demonstrated an essential role of FGFR1 in driving prostate cancer metastasis [12]. Together, these studies indicate that targeting FGFR1 may provide a rare therapeutic opportunity to inhibit cancer metastasis. Currently, many small molecule inhibitors targeting FGFR1 are being developed and/or tested in clinical trials for various types of human cancers. There are also FDA-approved drugs including ponatinib, nintedanib, pazopanib, and regorafenib, which are all multikinase inhibitors targeting FGFR1 with low specificity [19]. It will be important to investigate whether inhibiting FGFR1 in clinical setting also inhibits the metastasis of breast cancer, prostate cancer, and/or other types of human cancers.
It has been previously reported that FGFR1 gene amplification predicts poor prognosis of BCa. However, as a receptor tyrosine kinase, neither gene amplification nor FGFR1 expression directly confirms FGFR1 signaling activation. The FGFR1 gene signature generated in this study may help to more accurately evaluate FGFR1 activation status in BCa and identify patients who may be prioritized for FGFR1-targeted therapy.
Among the genes that are altered by loss of FGFR1, we have begun to identify potential candidates that might contribute to the tumor-promoting and/or metastasis-promoting activity of FGFR1 in BCa. Heparanase, which was greatly downregulated in the FGFR1 TuKO tumors (Gene Expression Omnibus GSE85754), is especially interesting. Heparanase degrades and remodels the extracellular matrix and promotes metastasis through its endoglycosidase activity. It also induces upregulation/release of growth factors such as VEGF, FGF-2, and HB-EGF for tumor-promoting activity [20]. Therefore, FGFR1 activation of heparanase may provide a direct avenue for FGFR1 promotion of cancer progression and/or metastasis. Further functional studies are needed to evaluate the significance of heparanase or other candidate genes in mediating the tumor- and/or metastasis-promoting activity of FGFR1 in BCa.
The following is the supplementary data related to this article.
TuKO of fgfr1 in the PyMT-induced mammary tumors significantly extends host mouse survival. (A) Knockout of fgfr1 in the PyMT-induced mammary tumors significantly extends hosts survival (P < .05 for both log-rank test and Gehan-Breslow-Wilcoxon test). All mice survived more than 32 weeks. (B) Mammary tumor weights were comparable between fgfr1 TuKO tumors and control tumors at the end of this survival study (P = .29, unpaired t test).
Acknowledgements
We thank Dr. Juha Partanen for providing the fgfr1loxP/loxP mice, Dr. Brian Lewis for providing the K19-tva mice, Dr. Corrinne Lobe for providing the pZ/EG vector, Dr. Garry Nolan for providing the pBMN-I-eGFP vector, and Drs. Daniel Silver and David Livingston for providing the HRMMPCreGFP vector. This study was supported by Department of Defense grants (W81XWH-09-1-0618, W81XWH-09-1-0651, W81XWH-13-1-0162, W81XWH-13-1-0163), Cancer Prevention Research Institute of Texas grant (RP130651) to F. Y., and National Institutes of Health grant (P30 CA125123) to C. J. C.
References
- 1.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welm BE, Freeman KW, Chen M, Contreras A, Spencer DM, Rosen JM. Inducible dimerization of FGFR1: development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol. 2002;157:703–714. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol. 2005;171:663–673. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dey JH, Bianchi F, Voshol J, Bonenfant D, Oakeley EJ, Hynes NE. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 6.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, Kilgour E, Smith NR, Geh C, Rooney C. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016;6:838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Z, Podsypanina K, Huang S, McGrath A, Toneff MJ, Bogoslovskaia E, Zhang X, Moraes RC, Fluck M, Allred DC. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci U S A. 2006;103:17396–17401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morton JP, Mongeau ME, Klimstra DS, Morris JP, Lee YC, Kawaguchi Y, Wright CV, Hebrok M, Lewis BC. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc Natl Acad Sci U S A. 2007;104:5103–5108. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 10.Yang F, Strand DW, Rowley DR. Fibroblast growth factor-2 mediates transforming growth factor-beta action in prostate cancer reactive stroma. Oncogene. 2008;27:450–459. doi: 10.1038/sj.onc.1210663. [DOI] [PubMed] [Google Scholar]
- 11.Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 12.Yang F, Zhang Y, Ressler SJ, Ittmann MM, Ayala GE, Dang TD, Wang F, Rowley DR. FGFR1 is essential for prostate cancer progression and metastasis. Cancer Res. 2013;73:3716–3724. doi: 10.1158/0008-5472.CAN-12-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 14.Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65:8887–8895. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- 15.N. Cancer Genome Atlas Research Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F, Zhang Y, Senbabaoglu Y, Ciriello G, Yang L, Reznik E, Shuch B, Micevic G, De Velasco G, Shinbrot E. Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 2016;14:2476–2489. doi: 10.1016/j.celrep.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Dong B, Ittmann MM, Yang F. A versatile gene delivery system for efficient and tumor specific gene manipulation in vivo. Discoveries. 2016;4:e58. doi: 10.15190/d.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helsten T, Schwaederle M, Kurzrock R. Fibroblast growth factor receptor signaling in hereditary and neoplastic disease: biologic and clinical implications. Cancer Metastasis Rev. 2015;34:479–496. doi: 10.1007/s10555-015-9579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem Sci. 2009;34:511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TuKO of fgfr1 in the PyMT-induced mammary tumors significantly extends host mouse survival. (A) Knockout of fgfr1 in the PyMT-induced mammary tumors significantly extends hosts survival (P < .05 for both log-rank test and Gehan-Breslow-Wilcoxon test). All mice survived more than 32 weeks. (B) Mammary tumor weights were comparable between fgfr1 TuKO tumors and control tumors at the end of this survival study (P = .29, unpaired t test).