Abstract
Trypanosomatids are an ancient family that diverged from the main eukaryotic lineage early in evolution, which display several unique features of gene organization and expression. Although genome sequencing is now complete, the nature of centromeres in these and other parasitic protozoa has not been resolved. Here, we report the functional mapping of a centromere in the American trypanosome, Trypanosoma cruzi, a parasite with an unusual mechanism of genetic exchange that involves the generation of aneuploidy by nuclear hybridization. Using a telomere-associated chromosome fragmentation approach, we show that the region required for the mitotic stability of chromosome 3 encompasses a transcriptional “strand-switch” domain constituted by a 16-kb GC-rich island. The domain contains several degenerate retrotransposon-like insertions, but atypically, lacks the arrays of satellite repeats normally associated with centromeric regions. This unusual type of organization may represent a paradigm for centromeres in T. cruzi and other primitive eukaryotes.
Centromeres are the chromosomal loci where kinetochores are assembled. This nucleoprotein complex functions as an anchor for the attachment of microtubule spindles that are necessary for segregation. Metazoan centromeres tend to be large, and in Drosophila and mammalian chromosomes, they can extend from 250 kb to >5000 kb (Sun et al. 1997; Cleveland et al. 2003). In yeast, they range from 40 to 120 kb in the case of Schizosaccharomyces pombe, but unusually in Saccharomyces cerevisiae, they are localized to 125-bp regions that are sufficient for spindle attachment (Huang 2002). Typically, centromeres are “regional” rather than “point” entities, and are made up of arrays of highly repetitive sequences interrupted by numerous retrotransposons (Sun et al. 1997; Shueler et al. 2001; Nagaki et al. 2004). Repeat sizes are similar in most organisms (150-180 bp) and approximate to the nucleosomal length, a feature that may be of functional significance (Henikoff et al. 2001). Centromeric chromatin is heterochromatic, a property that restricts access of DNA-binding factors and results in transcriptional silencing (Grewal and Moazed 2003).
The maintenance of centromeres is at least partly epigenetic. For example, following chromosome breakage in both Drosophila and humans, neocentromeres can be formed in regions adjacent to the original centromere, even though these may lack satellite arrays (du Sart et al. 1997; Williams et al. 1998). However, recent studies have also identified a role for the RNA interference (RNAi) machinery in heterochromatin formation and transcriptional silencing in S. pombe (Volpe et al. 2002; Hall et al. 2003a). Deletion of components of the RNAi machinery results in accumulation of complementary transcripts derived from centromeric repeats. This is associated with derepression of centromere-localized transgenes, disruption of centromere function, and blockage of the histone H3 lysine-9 methylation that mediates the formation of silent chromatin. These experiments suggest that low-level bidirectional transcription of centromeric repeats, in the context of an intact RNAi system, acts to maintain heterochromatin. The mechanism could involve the processing of double-stranded RNA into small RNAs that mediate the sequence-specific targeting of elements of the histone modification/chromatin silencing machinery. Although we await evidence for a role in centromere formation in other organisms, RNAi has been shown to function in other aspects of epigenetic gene silencing in Drosophila (Pal-Bhadra et al. 2004), Arabidopsis (Zilberman et al. 2003), Caenorhabditis elegans (Sijen and Plasterk 2003), and Trypanosoma brucei (Shi et al. 2004).
Over 10 million people in Latin America are infected with Trypanosoma cruzi, the insect-transmitted protozoan that causes Chagas disease (World Health Organization 2002). In common with other trypanosomatids, including Leishmania and the African trypanosome T. brucei, this parasite exhibits several unusual genetic traits. Protein-coding genes have no recognizable RNA polymerase II (pol II)-specific promoters, transcription is polycistronic, and all mRNAs are modified post-transcriptionally by the addition of a spliced leader RNA to their 5′-ends (Campbell et al. 2003). In addition, the T. cruzi genome contains an extensive range of repetitive elements (Requena et al. 1996), many genes are organized in tandem arrays, and the surface antigens are encoded by several extremely large heterogeneous gene families (Aguero et al. 2000). There is significant variation in the size of chromosomes between T. cruzi strains (Henriksson et al. 1996), and although the genome is generally diploid, the sizes of homologous chromosomes often differ considerably (Santos et al. 1997). This unusual phenomenon is also observed in T. brucei and Leishmania. Sequence elements that have a role in segregation in these organisms have not been identified. Furthermore, although many core centromeric proteins are highly conserved throughout evolution, there is a lack of obvious homologs in the trypanosomatid databases.
The T. cruzi CL Brener clone that was selected for sequencing has a haploid genome size of about 45 Mb and at least 30 chromosomes ranging from 0.45 to 3.5 Mb (Cano et al. 1995). The karyotype of this parasite remains poorly defined, and the precise relationship between chromosome homologs is unresolved. Completion of the genome project has been complicated by the finding that this clone is a hybrid of two distantly related lineages (Machado and Ayala 2001). This has had major implications. First, allelic copies of genes may vary in sequence by at least 1.5%. Second, due to recombination, chromosomes can represent a mosaic of the parental genotypes. Third, trisomy occurs in the case of some chromosomes. This latter phenomenon is probably related to the nature of genetic exchange in this organism, which unusually appears to require cell fusion within the mammalian host, followed by a return toward a diploid genotype (Gaunt et al. 2003). The mechanisms involved in chromosome loss/retention in this context are unknown.
In parallel with the T. cruzi genome project, we have developed a telomere-associated chromosome fragmentation vector as a tool to explore structural and functional aspects of chromosomes in this parasite. Here, we outline the properties of the vector and describe its use to delineate, for the first time, the sequences required for mitotic stability of a trypanosome chromosome.
Results
The chromosome fragmentation vector pTEX-CF
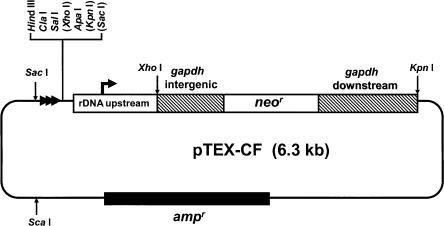
pTEX-CF (Fig. 1) was designed to facilitate telomere-associated chromosome fragmentation in T. cruzi. Our aim was to achieve this through targeted integration by single crossover, with a new telomere supplied by the vector. pTEX-CF contains the neor gene as the selectable marker with the flanking regions of the tandemly repeated T. cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase (gapdh) genes providing the necessary RNA processing elements (Kendall et al. 1990). A DNA fragment containing a ribosomal promoter, which has been shown to mediate high-level expression in transfected cells (Martinez-Calvillo et al. 1997) was inserted upstream of neor. We reasoned that inclusion of the ribosomal promoter should ensure expression of the drugselectable marker irrespective of the direction of pol II-mediated polycistronic transcription at the site of integration. Also included in the vector is a DNA fragment containing 21 copies of the telomeric repeat hexamer (5′-TTAGGG), and a multiple cloning site for insertion of the targeting sequences. Thus, following the insertion of T. cruzi genomic DNA sequences into pTEX-CF, restriction digestion with the appropriate enzyme produces a linear DNA fragment with the targeting sequences at one end and the telomeric sequences the other (see Fig. 2A).
Figure 1.
The chromosome fragmentation vector pTEX-CF. The vector was constructed by modifying the expression construct pRIBOTEX, as described in Methods. The gapdh-flanking sequences (hatched boxes) ensure correct processing of the neor transcript. Targeting fragments can be cloned in both orientations into the multiple cloning site (MCS). Bracketed restriction sites in the MCS are not unique. The location and direction of the repeated telomeric hexamers are indicated by horizontal arrowheads. The plasmid DNA (thin line) is derived from pBluescript and contains an ampr gene (filled box). Following insertion of the targeting fragment into the MCS, the construct can be used for transfection after linearization with the appropriate restriction enzyme.
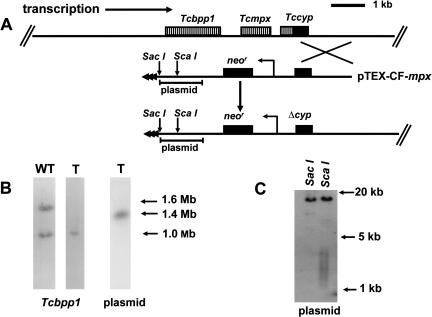
Figure 2.
(A) Map of the Tcmpx locus (top line) and integration of the pTEX-CF-mpx construct. The direction of polycistronic transcription, toward the center of the chromosome, is shown. The 1.5-kb fragment used for targeting is indicated by the crossed lines, and Δcyp identifies the disrupted cyclophilin gene formed as a result of the integration event. Expression of the neor gene is under control of the rDNA promoter. Telomeric sequences supplied by the vector are shown as horizontal arrowheads in the truncated chromosome (bottom line). The position of the plasmid sequences used as a probe in Figure 2, B and C is indicated. (B) Autoradiographs of Southern blots of T. cruzi chromosomal DNA separated by CHEFE and hybridized with a β-propeller protein gene (Tcbpp1) (Bromley et al. 2004) or plasmid DNA probes as indicated. (WT) Wild type; (T) transfected cells. (C) Southern blot of restriction-digested genomic DNA from transfected cells (see Fig. 2A for the location of the restriction sites) probed with plasmid DNA. The telomeric sequences are identifiable as a smear ranging from 1.5 to 4 kb. The top band in both lanes allows the position of the corresponding upstream restriction sites in the chromosomal DNA to be established.
Segmental monosomy in T. cruzi generated by chromosome fragmentation
To test the feasibility of generating segmental monosomy, we first targeted the mitochondrial peroxiredoxin (Tcmpx) and cytosolic iron superoxide dismutase (Tcsod) loci. In the CL Brener clone, Tcmpx is localized on chromosome homologs of 1.0 and 1.6 Mb (Wilkinson et al. 2000). Epimastigotes were transfected with a linearized pTEX-CF construct containing the targeting fragment shown in Figure 2A and drug-resistant clones isolated. When chromosomal DNA was examined after Southern blotting of CHEFE (contour clamped homogeneous electric field electrophoresis) gels, only the 1.0-Mb homolog could be detected by the Tcbpp1 probe (Fig. 2B). A plasmid DNA probe identified a single 1.4-Mb band, which we confirmed to be the truncated product of the 1.6-Mb chromosome. Southern analysis of ScaI-digested genomic DNA was also performed. This enzyme liberates telomeric repeats attached to 900 bp of plasmid DNA (Fig. 2A). The resulting fragments can be visualized as a smear ranging from 1.5 to 4 kb (Fig. 2C), compared with the 1.1 kb originally present in the integration vector. Together, these results localize the Tcmpx gene 200 kb from the end of the 1.6-Mb chromosome. It is also implicit that polycistronic transcription at this locus occurs toward the center of the chromosome. There was no detectable phenotype associated with loss of this large region of DNA.
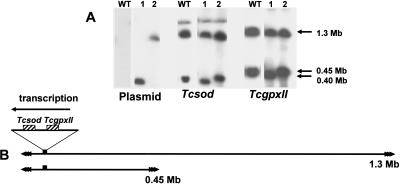
The Tcsod gene lies next to the peroxidase gene TcgpxII (Wilkinson et al. 2002) and is located on chromosome 1 (Temperton et al. 1996), which occurs as homologs of 0.45- and 1.3-Mb. When parasites were transfected with a pTEX-CF construct containing Tcsod orientated in the direction of transcription, both chromosomes could be targeted. Integration into the 0.45-Mb homolog generated a 0.4-Mb product (Fig. 3A). This is consistent with preliminary analysis of the genome sequence data, which indicates that a Tcsod gene is located 48 kb from a telomere (B. Andersson, pers. comm.). It can also be inferred that in the larger homolog, the gene is in a similar position (Fig. 3A), although we can be less precise about the distance from the telomere, due to the lack of resolution in this part of the CHEFE gel. However, it is clear that the size heterogeneity between chromosome homologs does not result from the presence of additional sequences between this Tcsod locus and the downstream telomere (Fig. 3B). Both of the truncated chromosomes were found to be stable for at least 100 generations (20 wk continuous culture), following the removal of selective drug pressure.
Figure 3.
(A) Southern analysis of T. cruzi clones representing the two types of event observed when the Tcsod locus was targeted. Chromosomal DNA from wild-type (WT) cells and transformed clones (1 and 2) was separated by CHEFE and hybridized with plasmid, Tcsod, or TcgpxII probes. The plasmid DNA identifies the truncated chromosomes that result from integration. (B) Schematic showing the location of the Tcsod locus on the 0.45- and 1.3-Mb chromosomes and the direction of polycistronic transcription (inferred from the orientation of the targeting fragment). Integration at the Tcsod locus deletes the region between the gene and the adjacent downstream telomere. The precise location of the locus on the larger homolog cannot be assessed with accuracy, because it is difficult to resolve the 1.3-Mb chromosome and its slightly smaller truncated product.
These experiments therefore demonstrate that the pTEX-CF vector can be used to generate partially monosomic cell lines. This could have several applications, including confirmation of the T. cruzi genome map, more rapid approaches to functional analysis, and delineation of sequences required for mitotic stability.
Targeted dissection of chromosome 3
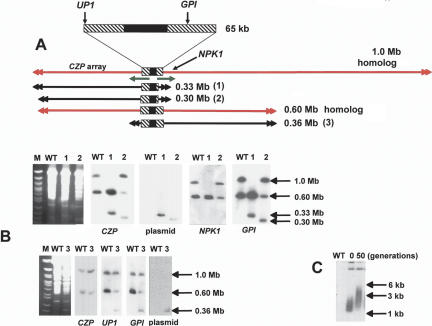
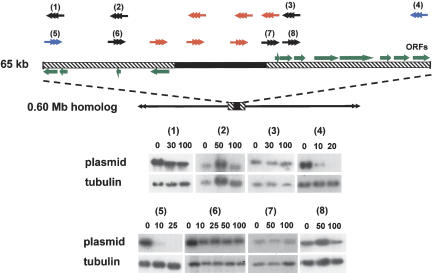
We next focused on chromosome 3, where sequencing of a 93-kb contig (Andersson et al. 1998) had identified a transcriptional “strand-switch” region, in which a 16-kb GC-rich domain, containing degenerate retroelements, separates two directional gene clusters. The corresponding region of T. brucei chromosome 1, which is syntenic with this contig, has been postulated to have centromeric properties on the basis of a low recombination frequency (Hall et al. 2003b). Chromosome 3 is present as homologs of 0.60 and 1.0 Mb in the clone CL Brener. Unusually, on the basis of hybridization intensity with several independent probes, we found that this chromosome is trisomic, with two copies of the smaller homolog and one of the larger (Fig. 4). We used single-copy targeting sequences located adjacent to the strand-switch region (Fig. 5) to generate several truncated forms of these chromosome homologs. The data presented in Figure 4 illustrate the type of analysis performed. Here, integration was targeted to sites on each side of the strand-switch region, a glucose-6-phosphate isomerase (GPI) gene (Fig. 4B) (site 8, Fig. 5), and an unidentified open reading frame (UP1) (Fig. 4B) (site 1, Fig. 5).
Figure 4.
Truncation of the 1.0- and 0.60-Mb homologs of chromosome 3. (A) A schematic showing the full-length chromosomes (drawn in red) and the truncated products (1-3) (drawn in black). Product 1 was derived from the 1.0-Mb homolog, and was generated by integration at the GPI locus. Products 2 and 3 were derived from the 0.60-Mb homolog, and were generated by integration at the GPI and UP1 loci, respectively. The 65-kb region encompassing the transcriptional strand-switch region (common to both homologs) has been expanded. The directions of polycistronic transcription are indicated by green arrows (for details, see also Fig. 5). (B) Chromosomal DNA from wild-type (WT) and transformed cells was analyzed by CHEFE (left panels, ethidium stained gels; other panels, autoradiographs of similar blots probed as indicated). (Lanes 1,2,3) The corresponding truncated chromosomes. (C) Blot of genomic DNA from cells containing the 0.36-Mb truncated product (as in lane 3, above) digested with ScaI. Comparison of telomere lengths (plasmid probe) in the initial clonal population and after 50 generations.
Figure 5.
Mitotic stability of truncated chromosomes assessed by hybridization with plasmid DNA. Expansion of the locus encompassing the strand-switch domain shows the GC-rich (filled box) and coding (hatched boxes) regions. Green arrows identify ORFs (confirmed and potential) and point in the direction of polycistronic transcription. The sites of integration-mediated chromosome fragmentation (1-8) are shown with arrowheads representing the new telomeres. This numbering system is unrelated to that in Figure 4. (Black) Mitotically stable truncated chromosomes; (blue) mitotically unstable; (red) integration not achievable. Transfected clones were grown in the absence of G418 for the generations indicated above the corresponding track. Genomic DNA was probed with plasmid DNA (autoradiographs 1-8), then rehybridized with β-tubulin to control for loading.
The 19 clones isolated when GPI was targeted could be subdivided into two groups. The first (two clones) was characterized by the loss of the 1.0-Mb homolog and by the appearance of a 0.33-Mb product (Track 1, Fig. 4B). This was associated with an increase in the copy number of the 0.60-Mb homolog (four copies as assessed by phosphorimaging). On the basis of the hybridization pattern (Track 1), it can be concluded that the right arm of the 1.0-Mb chromosome has been lost from the cell. This deletion of almost 0.70 Mb of DNA had no effect on morphology, growth, differentiation into the metacyclic (infectious) form, or invasion of cultured macrophages (data not shown). Both clones also produced a lethal infection in the SCID mouse model (five mice per group) 19-21 d post-inoculation (Methods), the same period that was observed with nontransformed parasites. Several conclusions can be drawn from these data. First, the right arm of the 1.0-Mb chromosome contains most of the additional 400 kb of sequence that is responsible for the size heterogeneity between the two homologs. Second, although we do not yet know the nature of these additional sequences, their deletion from the larger homolog has no apparent phenotypic consequences. Finally, the observed increase in chromosome copy number suggests the presence of dose-dependent gene(s) in the deleted right arm of the 1.0-Mb homolog.
In the second group of clones (17), we identified a new chromosome band of 0.30 Mb (Track 2, Fig. 4B). Hybridization intensities were consistent with one copy each of the 0.30- and 0.60-Mb chromosomes and two copies of the 1.0-Mb homolog in these cells. It can be inferred that the 0.30-Mb band is a truncated product of the 0.60-Mb homolog, and that the left arm of this chromosome is 30 kb shorter than the corresponding region of the larger homolog (cf. Tracks 1 and 2, Fig. 4B).
When UP1 was targeted, all clones were found to contain a new chromosome band of 0.36 Mb derived, by inference, following deletion of the left arm of the 0.60-Mb homolog (Fig. 4B). Again, this was associated with duplication of the 1.0-Mb chromosome. This may reflect the presence of pseudogenes in the remaining 0.60-Mb homolog. We also investigated telomere elongation on the 0.36-Mb truncated chromosome by examining ScaI-digested genomic DNA. When a newly isolated clone was analyzed (Fig. 4C), the resulting fragments were visualized as a smear ranging from 1.2 to 3.0 kb, representing telomere lengths from 0.1 to 1.9 kb. After 50 generations, the smear extended up to 6 kb. This is a more rapid rate of de novo telomere elongation than has been observed in T. brucei, where newly formed telomeres were found to initially increase in length by ∼8 bp per generation (Horn et al. 2000).
In total, parasites were transfected with 14 different fragmentation constructs targeted at sites proximal to the chromosome 3 strand-switch region. With six of these (marked red, Fig. 5), we were unable to generate transformants in which integration had occurred at the targeted locus, despite multiple rounds of transfection. In each of these cases, the targeting fragment had been derived from the 16-kb GC-rich domain, or from sequences immediately adjacent to it. This refractoriness to integration may reflect properties associated with local chromatin, perhaps similar to those of centromeric heterochromatin, which restricts access of DNA-binding factors and confers transcriptional silencing (Grewal and Moazed 2003). In all other instances, truncated chromosomes of the predicted size were generated. These were verified as outlined in Figure 4 and Methods. The large majority of truncated chromosomes were derived from the smaller homolog, and each of these deletion events was associated with an increase in the copy number of the larger chromosome.
Mitotic stability of truncated forms of chromosome 3
To assess the effects on mitotic stability, transformed clones containing shortened forms of the 0.60-Mb homolog were cultured in the absence of G418. In these circumstances, the increased copy number of the 1.0-Mb homolog should negate any effect that dose-dependent genes might have on the selective retention of chromosomes where fragmentation had led to the deletion of a functional centromere. Genomic DNA was prepared and hybridized with a plasmid probe to determine the level of each truncated chromosome in the population (Fig. 5). All but two were found to be stable for at least 100 generations, even though in some cases (integration at sites 3 and 6; Fig. 5) the shortened chromosomes did not contain overlapping regions of sequence. This therefore excludes the possibility that the centromere in this T. cruzi chromosome is a point entity, as in S. cerevisiae (Cheeseman et al. 2002). In contrast to the above, truncated chromosomes generated by integration at the UP1 (site 5) and UP3 (site 4) loci were mitotically unstable and had largely disappeared from the population 10 generations after the removal of selective pressure. Loss of these chromosomes was confirmed by CHEFE analysis. We propose that these fragmentations delineate the functional centromere of chromosome 3 to elements within the 65-kb sequence shown in Figure 5. Alternative explanations, which implicate loss of a replication origin, are unlikely. Replication of eukaryotic chromosomes can be initiated at multiple sites (Gilbert 2001), and in T. cruzi, there are no apparent restrictions on the replication of plasmid-based constructs (Kelly et al. 1992).
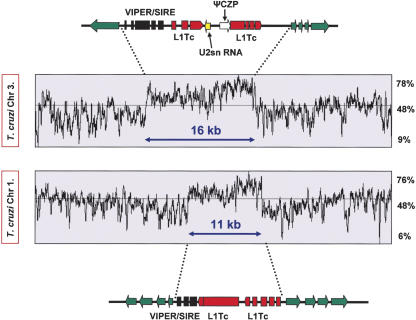
The region deleted in these experiments has been fully sequenced (Andersson et al. 1998). It encompasses the 16-kb GC-rich central core, which separates two gene clusters transcribed on opposite strands (Fig. 6). This domain is composed predominantly of degenerate retroelements, including a composite VIPER (vestigial interposed retroelement)/SIRE (short interspersed repetitive element) and two degenerate non-LTR retrotransposon sequences related to the L1Tc autonomous retroelement (Vazquez et al. 2000; Bringaud et al. 2002). These flank both sides of a U2snRNA gene and a truncated cruzipain pseudogene. Unusually, this region lacks any satellite repeats similar to those present in the centromeres of other eukaryotes. This represents a major structural difference that may be a feature of centromeres in T. cruzi. The retention of mitotic stability in the absence of this GC-rich core (integration at sites 3 and 6, Fig. 5) could be indicative of neocentromere formation. Neocentromeres can arise after chromosome breakage in higher eukaryotes (du Sart et al. 1997; Williams et al. 1998) as a result of the spread of heterochromatin into adjacent euchromatic regions, despite the fact that such regions may lack characteristic centromere sequences. This phenomenon limits the potential for more accurately defining the boundary of functional centromeres using the fragmentation approach.
Figure 6.
Organization of the GC-rich strand-switch regions on T. cruzi chromosomes 1 (lower)and3(upper). Green arrows identify the first ORFs of the polycistronic gene clusters and indicate the direction of transcription. The degenerate retrotransposon-like VIPER/SIRE element (black) and the L1Tc autonomous retroelements (red) are indicated. This region of chromosome 3 also contains a truncated cruzipain pseudogene (czp) and a U2snRNA gene. In chromosome 1, the strand-switch domain is located ∼120 kb from the telomere, 70-kb upstream of the Tcsod locus (Fig. 2), between ORFs identified by temporary systematic-ids Tc00.1047053508707.234 and 270.
Discussion
There are no experimental data on the structure or organization of centromeres in trypanosomatids, or other parasitic protozoa. To address this issue in T. cruzi, we have developed a fragmentation vector able to delete defined regions of chromosomal DNA. This approach has been used previously to investigate chromosome structure in other organisms (Farr et al. 1992; Itzhaki et al. 1992; Tamar and Papadopoulou 2001). Initially, we generated partially monosomic cell lines by targeting the Tcmpx and Tcsod loci, and deleted 200 kb and 50 kb of DNA, respectively. With the completion of the T. cruzi genome project, it should be feasible to undertake a systematic functional screen of all of the genes in these deleted regions using a “singleknockout” strategy. This could be extended to other areas of the genome as the appropriate monosomic lines are generated. New, more high-throughput procedures such as these are urgently required if the genome sequence is to be fully dissected at the functional level, since targeted gene deletion is very time consuming in T. cruzi (each round of gene disruption taking at least 3 mo), and the parasite lacks the enzymatic machinery necessary for RNAi-based aproaches (DaRocha et al. 2004; our own observations).
When targeting chromosome 3, we were able to delete up to 700 kb in the case of the larger homolog. With the smaller homolog, both arms could be deleted in separate experiments. However, in all of these cases, we observed that fragmentation was associated with an increase in copy number, even though this chromosome is already trisomic. To our knowledge, this is the first report of apparent “diploid insufficiency” in any organism. One explanation could be that the trisomy of chromosome 3 has allowed deleterious mutations in some dosedependent genes to be maintained in the CL Brener clone. Subsequent loss of nonmutant allele(s) due to fragmentation could then be lethal or detrimental, resulting in a selective advantage to those cells containing additional copies of the chromosome. The plasticity of trypanosomatid genomes has been well documented (Myler 1993; Lanzer et al. 1995; Gueiros-Filho and Beverley 1996).
It has proved difficult to accurately map the boundaries of centromeres in most organisms (Choo 2000; Henikoff et al. 2001), and experimental verification is rarely straightforward. Furthermore, in higher eukaryotes, their large size and highly repetitive structure has restricted complete sequence analysis to a single instance (Nagaki et al. 2004). In parasitic protozoa, the nature of centromeres is a major gap in our understanding of chromosome structure and propagation. A putative centromere has been delineated to a 400-kb fragment of chromosome 5 of the trypanosomatid Leishmania (Tamar and Papadopoulou 2001), but the sequence of this region is not yet available. With Leishmania chromosome 1, large targeted deletions have led to the tentative suggestion that clusters of subtelomeric repeats may have a role in mitotic stability (Dubessay et al. 2001, 2002). In T. brucei, numerous minichromosomes, which act as a store of divergent surface antigen genes, are constructed around palindromes based on 177-bp repeats. It has been postulated that this type of organization may have a role in centromeric function (Wickstead et al. 2004). In the malaria parasite Plasmodium falciparum, an 8-12-kb noncoding domain present on each chromosome has been proposed as a candidate centromere (Bowman et al. 1999). This region contains an extremely AT-rich core that includes several repeat elements. However, there are no experimental data to confirm that these domains have a role in conferring mitotic stability.
In T. cruzi, our data imply that the locus required for the mitotic stability of chromosome 3 centers on a 16-kb GC-rich strand-switch region that is composed predominantly of degenerate retrotransposon-like elements, but lacks the arrays of satellite repeats characteristic of centromeric DNA in higher eukaryotes. These areas between polycistronic transcription units are thought to be one of the few transcriptionally quiescent parts of the T. cruzi genome. However, this property cannot be a sole indicator of centromeric function, since chromosome 3, like most trypanosomatid chromosomes contains more that one strand-switch region (B. Andersson, pers. comm.). When a fully contiguous T. cruzi sequence is available, it should be possible to test experimentally regions of the genome with a similar type of organization to the GC-rich strand-switch domain of chromosome 3 and to identify the elements that define a functional centromere in this organism. Currently, there is a 300-kb contig that contains more than half the sequence of the small homolog of chromosome 1 (Fig. 6). This extends from a telomere toward the center of the chromosome and encompasses the Tcsod locus (Fig. 3). Examination of this contig reveals the presence of a single 11-kb GC-rich strand-switch domain that has remarkable organizational similarity with the element that we characterized on chromosome 3 (Fig. 6), including the presence of sequences related to the VIPER, SIRE, and L1Tc retroelement families. Importantly, this GC-rich region is retained on the stable truncated chromosome generated following integration at the Tcsod locus.
T. brucei chromosome 1 and Leishmania major chromosome 12 are syntenic with chromosome 3 of T. cruzi at the strand-switch locus (Ghedin et al. 2004). The corresponding T. brucei strand-switch region (∼40 kb) contains two full-length Ingi retrotransposon-like elements, closely linked to a DIRE (degenerated Ingi/L1Tc-related element), and shows evidence of low recombination frequency, a feature of centromeric DNA (Hall et al. 2003a). In addition, this domain includes a 5.5-kb AT-rich stretch composed predominantly of 58-bp degenerate repeats. A DIRE is also present in the 10-kb strand-switch domain of L. major chromosome 12. Previously, retroelements had not been identified in L. major (Ghedin et al. 2004). It will be interesting to determine whether the conserved organization of these strand-switch regions is of functional significance.
There is a large epigenetic component involved in centromere maintenance and recognition, although recent evidence also implicates RNA-mediated mechanisms. In fission yeast, the RNAi system is necessary for transcriptional silencing at centromeres (Hall et al. 2003b) and for heterochromatin formation (Volpe et al. 2002). Disruption of RNAi in T. brucei has been reported variously to be associated with increased abundance of retroposon transcripts (Shi et al. 2004) and with defects in segregation and assembly of the mitotic spindle (Durand-Dubief and Bastin 2003). In T. cruzi, however, the lack of functional RNAi excludes a role for this system and highlights a possible major difference in the mechanisms of centromere maintenance between trypanosome species.
In higher eukaryotes, a role for sequence-specific binding in centromere function has been inferred from the adaptive evolution of the satellite repeat sequences and the kinetochoreassociated histone CenH3. This dynamic process has been proposed as an important mechanism in speciation and seems to be driven by events within the developing oocyte, where four copies of each chromosome are formed, only one of which is retained in the haploid nucleus (Henikoff et al. 2001). Thus, centromeres that more efficiently bind kinetochore proteins gain a slight selective advantage over the other homologs. In turn, mutations in centromeric proteins, such as CenH3, that act to restore parity of centromere binding are positively selected. This process of “meiotic drive,” allied with the potential of centromeric DNA to undergo rearrangement by various mechanisms, is thought to lead to the extensive divergence of centromeric sequences, even between closely related species (Henikoff and Malik 2002).
Although reproduction in T. cruzi is predominantly asexual, genetic exchange does occur and involves cell fusion within the mammalian host, followed by a return toward diploidy (Gaunt et al. 2003). This explains the wide range of DNA content in naturally occurring T. cruzi isolates (McDaniel and Dvorak 1993), including the occurrence of trisomy in the CL Brener clone. Factors that influence chromosome loss/retention following nuclear hybridization are unknown, but must reflect looser restraints on ploidy and tension-mediated separation of sister kinetochores (Dewar et al. 2004). We propose that a process analogous to meiotic drive, involving competition between homologous centromeres, may also occur as hybrid progeny move from tetraploidy to diploidy. The nature of the chromosome 3 centromere, which is made up predominantly of degenerate retroelements, offers considerable scope for sequence divergence. This could provide a mechanism for the reproductive isolation of parasite lineages.
Methods
Vector construction
The chromosome fragmentation vector pTEX-CF (Fig. 1) was constructed by modifying the T. cruzi expression plasmid pRIBOTEX (Martinez-Calvillo et al. 1997) (a gift from Dr. Roberto Hernandez, U.N.A.M., Mexico City). Briefly, the multiple cloning site (MCS) in this vector was first replaced with the MCS from pBluescript (Stratagene), into which was inserted a BamHI/HindIII fragment containing 21 copies of the telomere hexamer repeat 5′-TTAGGG. Targeting fragments to be inserted into the MCS were amplified by PCR, verified by sequencing, and confirmed as single copy by Southern analysis of genomic DNA. Vector DNA to be used for transfection was linearized with HindIII and purified by gel electrophoresis.
Parasite growth and analysis of transformants
T. cruzi epimastigotes were cultured at 28°C in RPMI 1640 medium (Sigma) containing the supplements described (Kendall et al. 1990). For transfection experiments, linear DNA (∼1-2 μg) was introduced into epimastigotes by electroporation (Kelly et al. 1992). Transformed cells were selected over 6-8 wk in the presence of 100 μg mL-1 G418, and then cloned by limiting dilution. Clones were analyzed by Southern blotting of CHEFE-separated chromosomal DNA and restriction-digested genomic DNA, and by sequence determination of transformant-specific PCR fragments generated with primers that straddled the expected integration site. Intact T. cruzi chromosomes for CHEFE analysis were extracted using an agarose-embedding technique (Gibson and Miles 1986) and separated with a Bio-Rad CHEF Mapper system using an auto-algorithm set to the appropriate molecular mass range. Gels were blotted onto nylon membranes using standard techniques, and where required, signal intensity following hybridization was quantified by PhosphorImager analysis (Molecular Dynamics).
Assessing mitotic stability
Transformed clones containing various truncated forms of the 0.60-Mb homolog of chromosome 3 were generated (Fig. 5). These were continuously cultured (five generations per week) in the absence/presence of G418, and DNA isolated at various time points. Mitotic stability was assessed by Southern hybridization using plasmid DNA as a probe for truncated chromosomes. Blots were reprobed with the β-tubulin gene to control for loading. Results were confirmed by CHEFE analysis.
Acknowledgments
We thank our colleagues Bjorn Andersson, David Baker, David Horn, and Michael Miles for advice and for critical reading of this manuscript, and Roberto Hernandez for providing the pRIBOTEX construct. We acknowledge Andersson et al. (1998), who produced the chromosome 3 DNA data utilized in this work (Accession nos. AF052831-AF052833), and thank Bjorn Andersson and Daniel Nilsson (Karolinska Institute) for permission to use the chromosome 1 sequence, which was generated as part of the T. cruzi Genome Project. This investigation was funded by the Wellcome Trust and the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR).
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2895105.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: B. Andersson and R. Hernandez.]
References
- Aguero, F., Verdun, R.E., Frasch, A.C., and Sanchez, D.O. 2000. A random sequencing approach for the analysis of the Trypanosoma cruzi genome: General structure, large gene and repetitive DNA families, and gene discovery. Genome Res. 10: 1996-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, B., Aslund, L., Tammi, M., Tran, A.N., Hoheisel, J.D., and Pettersson, U. 1998. Complete sequence of a 93.4-kb contig from chromosome 3 of Trypanosoma cruzi containing a strand-switch region. Genome Res. 8: 809-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, S., Lawson, D., Basham, D., Brown, D., Chillingworth, T., Churcher, C.M., Craig, A., Davies, R.M., Devlin, K., Feltwell, T., et al. 1999. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature 400: 532-538. [DOI] [PubMed] [Google Scholar]
- Bringaud, F., Garcia-Perez, J.L., Heras, S.R., Ghedin, E., El-Sayed, N.M., Andersson, B., Baltz, T., and Lopez, M.C. 2002. Identification of non-autonomous non-LTR-retrotransposons in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 124: 73-78. [DOI] [PubMed] [Google Scholar]
- Bromley, E.V., Taylor, M.C., Wilkinson, S.R., and Kelly, J.M. 2004. The amino terminal domain of a novel WD repeat protein from Trypanosoma cruzi contains a non-canonical mitochondrial targeting signal. Int. J. Parasit. 34: 63-71. [DOI] [PubMed] [Google Scholar]
- Campbell, D.A., Thomas, S., and Sturm, N.R. 2003. Transcription in kinetoplastid protozoa: Why be normal? Microbes Infect. 5: 1231-1240. [DOI] [PubMed] [Google Scholar]
- Cano, M.I., Gruber, A., Vazquez, M., Cortes, A., Levin, M.J., Gonzalez, A., Degrave, W., Rondinelli, E., Zingales, B., Ramirez, J.L., et al. 1995. Molecular karyotype of clone CL Brener chosen for the Trypanosoma cruzi genome project. Mol. Biochem. Parasitol. 71: 273-278. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Drubin, D.G., and Barnes, G. 2002. Simple centromere, complex kinetochore: Linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157: 199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H.A. 2000. Centromerization. Trends Cell Biol. 10: 182-188. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y., and Sullivan, K.F. 2003. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signalling. Cell 112: 407-421. [DOI] [PubMed] [Google Scholar]
- DaRocha, W.D., Otsu, K., Teixeira, S.M., and Donelson, J.E. 2004. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol. Biochem. Parasitol. 133: 175-186. [DOI] [PubMed] [Google Scholar]
- Dewar, H., Tanaka, K., Nasmyth, K., and Tanaka, T.U. 2004. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428: 32-33. [DOI] [PubMed] [Google Scholar]
- Dubessay, P., Ravel, C., Bastien, P., Lignon, M.F., Ullman, B., Pages, M., and Blaineau, C. 2001. Effect of large targeted deletions on the mitotic stability of an extra chromosome mediating drug resistance in Leishmania. Nucleic Acid Res. 29: 3231-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubessay, P., Ravel, C., Bastien, P., Stuart, K., Dedet, J.P., Blaineau, C., and Pages, M. 2002. Mitotic stability of a coding DNA sequence-free version of Leishmania major chromosome 1 generated by targeted chromosome fragmentation. Gene 289: 151-159. [DOI] [PubMed] [Google Scholar]
- Durand-Dubief, M. and Bastin, P. 2003. TbAGO1, an Argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart, D., Cancilla, M.R., Earle, E., Mao, J.I., Saffery, R., Tainton, K.M., Kalitsis, P., Martyn, J., Barry, A.E., and Choo, K.H. 1997. A functional neocentromere formed through activation of a latent human centromere and consisting of non-α satellite DNA. Nat. Genet. 16: 144-153. [DOI] [PubMed] [Google Scholar]
- Farr, C.J., Stevanovic, M., Thomson, E.J., Goodfellow, P.N., and Cooke, H.J. 1992. Telomere-associated chromosome fragmentation: Applications in genome manipulation and analysis. Nat. Genet. 2: 275-282. [DOI] [PubMed] [Google Scholar]
- Gaunt, M.W., Yeo, M., Frame, I.A., Stothard, J.R., Carrosco, H.J., Taylor, M.C., Meno, S.S., Veazey, P., Miles, G.A., Acosta, N., et al. 2003. Mechanism of genetic exchange in American trypanosomes. Nature 421: 936-939. [DOI] [PubMed] [Google Scholar]
- Ghedin, E., Bringuad, F., Peterson, J., Myler, P., Berriman, M., Ivens, A., Andersson, B., Bontempi, E., Eisea, J., Angiuoli, S., et al. 2004. Gene synteny and evolution of genomic architecture in trypanosomatids. Mol. Biochem. Parasitol. 134: 183-191. [DOI] [PubMed] [Google Scholar]
- Gibson, W.C. and Miles, M.A. 1986. The karyotype and ploidy of Trypanosoma cruzi. EMBO J. 5: 1299-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, D.M. 2001. Making sense of eukaryotic DNA replication origins. Science 294: 96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, S.I. and Moazed, D. 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798-802. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho, F.J. and Beverley, S.M. 1996. Selection against the dihydrofolate reductase-thymidylate synthase (DHFR-TS) locus as a probe of genetic alterations in Leishmania major. Mol. Cell Biol. 16: 5655-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I.M.K., Noma, K., and Grewal, S.I. 2003a. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. 100: 193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, N., Berriman, M., Lennard, N.J., Harris, B.R., Hertz-Fowler, C., Bart-Delabesse, E.N., Gerrard, C.S., Atkin, R.J., Barron, A.J., Bowman, S., et al. 2003b. The DNA sequence of chromosome I of an African trypanosome: Gene content, chromosome organization, recombination and polymorphism. Nucleic Acid Res. 31: 4864-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. and Malik, H.S. 2002. Centromeres: Selfish drivers. Nature 417: 227. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098-1102. [DOI] [PubMed] [Google Scholar]
- Henriksson, J., Aslund, L., and Pettersson, U. 1996. Karyotype variability in Trypanosoma cruzi. Parasitol. Today 12: 108-114. [DOI] [PubMed] [Google Scholar]
- Horn, D., Spence, C., and Ingram, A.K. 2000. Telomere maintenance and length regulation in Trypanosoma brucei. EMBO J. 19: 2332-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acid Res. 30: 1465-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki, J.E., Barnett, M.A., MacCarthy, A.B., Buckle, V.J., Brown, W.R., and Porter, A.C. 1992. Targeted breakage of a human chromosome mediated by cloned human telomeric DNA. Nat. Genet. 2: 283-287. [DOI] [PubMed] [Google Scholar]
- Kelly, J.M., Ward, H.W., Miles, M.A., and Kendall, G. 1992. A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucleic Acid Res. 20: 3963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, G., Wilderspin, A.F., Ashall, F., Miles, M.A., and Kelly, J.M. 1990. Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase does not conform to the “hotspot” topogenic signal model. EMBO J. 9: 2751-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzer, M., Fischer, K., and Le Blancq, S.M. 1995. Parasitism and chromosome dynamics in protozoan parasites: Is there a connection? Mol. Biochem. Parasitol. 70: 1-8. [DOI] [PubMed] [Google Scholar]
- Machado, C.A. and Ayala, F.J. 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. 98: 7396-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Calvillo, S., Lopez, I., and Hernandez, R. 1997. pRIBOTEX expression vector: A pTEX derivative for rapid selection of Trypanosoma cruzi transfectants. Gene 199: 71-76. [DOI] [PubMed] [Google Scholar]
- McDaniel, J.P. and Dvorak, J.A. 1993. Identification, isolation, and characterization of naturally-occurring Trypanosoma cruzi variants. Mol. Biochem. Parasitol. 57: 213-222. [DOI] [PubMed] [Google Scholar]
- Myler, P.J. 1993. Molecular variation in trypanosomes. Acta Trop. 53: 205-225. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138-145. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Leibovitch, B.A., Gandhi, S.G., Rao, M., Bhadra, U., Birchler, J.A., and Elgin, S.C. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669-672. [DOI] [PubMed] [Google Scholar]
- Requena, J.M., Lopez, M.C., and Alonso, C. 1996. Genomic repetitive DNA elements in Trypanosoma cruzi. Parasitol. Today 12: 297-283. [DOI] [PubMed] [Google Scholar]
- Santos, M.R., Cano, M.I., Schijman, A., Lorenzi, H., Vazquez, M., Levin, M.J., Ramirez, J.L., Brandao, A., Degrave, W.M., and da Silveira, J.F. 1997. The Trypanosoma cruzi genome project: Nuclear karyotype and gene mapping of clone CL Brener. Mem. Inst. Oswaldo Cruz. 92: 821-828. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109-115. [DOI] [PubMed] [Google Scholar]
- Shi, H., Djikeng, A., Tschudi, C., and Ullu, E. 2004. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: Control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell Biol. 24: 420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T. and Plasterk, R.H. 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426: 310-314. [DOI] [PubMed] [Google Scholar]
- Sun, X., Wahlstrom, J., and Karpen, G. 1997. Molecular structure of a functional Drosophila centromere. Cell 91: 1007-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamar, S. and Papadopoulou, B. 2001. A telomere-mediated chromosome fragmentation approach to assess mitotic stability and ploidy alterations of Leishmania chromosomes. J. Biol. Chem. 276: 11662-11673. [DOI] [PubMed] [Google Scholar]
- Temperton, N.J., Wilkinson, S.R., and Kelly, J.M. 1996. Cloning of an Fe-superoxide dismutase gene homologue from Trypanosoma cruzi. Mol. Biochem. Parasitol. 76: 339-343. [DOI] [PubMed] [Google Scholar]
- Vazquez, M., Ben-Dov, C., Lorenzi, H., Moore, T., Schijman, A., and Levin, M.J. 2000. The short interspersed repetitive element of Trypanosoma cruzi, SIRE, is part of VIPER, an unusual retroelement related to long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. 97: 2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Wickstead, B., Ersfeld, K., and Gull, K. 2004. Small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 14: 1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, S.R., Temperton, N.J., Mondragon, A., and Kelly, J.M. 2000. Distinct mitochondrial and cytosolic enzymes mediate trypanothione-dependent peroxide metabolism in Trypanosoma cruzi. J. Biol. Chem. 275: 8220-8225. [DOI] [PubMed] [Google Scholar]
- Wilkinson, S.R., Taylor, M.C., Touitha, S., Mauricio, I.L., Meyer, D.J., and Kelly, J.M. 2002. TcGPXII, a glutathione-dependent Trypanosoma cruzi peroxidase with substrate specificity restricted to fatty acid and phospholipid hydroperoxides, is localised to the endoplasmic reticulum. Biochem. J. 364: 787-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., Murphy, T.D., Goldberg, M.L., and Karpen, G.H. 1998. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat. Genet. 18: 30-37. [DOI] [PubMed] [Google Scholar]
- World Health Organization. 2002. Control of Chagas disease. World Health Organization Technical Report Series 905, Geneva, Switzerland. [PubMed]
- Zilberman, D., Cao, X., and Jacobsen, S.E. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716-719. [DOI] [PubMed] [Google Scholar]