Abstract
A distinctive feature of the avian genome is the large heterogeneity in the size of chromosomes, which are usually classified into a small number of macrochromosomes and numerous microchromosomes. These chromosome classes show characteristic differences in a number of interrelated features that could potentially affect the rate of sequence evolution, such as GC content, gene density, and recombination rate. We studied the effects of these factors by analyzing patterns of nucleotide substitution in two sets of chicken-turkey sequence alignments. First, in a set of 67 orthologous introns, divergence was significantly higher in microchromosomes (chromosomes 11-38; 11.7% divergence) than in both macrochromosomes (chromosomes 1-5; 9.9% divergence; P = 0.016) and intermediate-sized chromosomes (chromosomes 6-10; 9.5% divergence; P = 0.026). At least part of this difference was due to the higher incidence of CpG sites on microchromosomes. Second, using 155 orthologous coding sequences we noted a similar pattern, in which synonymous substitution rates on microchromosomes (13.1%) were significantly higher than were rates on macrochromosomes (10.3%; P = 0.024). Broadly assuming neutrality of introns and synonymous sites, or constraints on such sequences do not differ between chromosomal classes, these observations imply that microchromosomal genes are exposed to more germ line mutations than those on other chromosomes. We also find that dN/dS ratios for genes located on microchromosomes (average, 0.094) are significantly lower than those of macrochromosomes (average, 0.185; P = 0.025), suggesting that the proteins of genes on microchromosomes are under greater evolutionary constraint.
Compared to mammals, avian genomes are small but contain a larger number of chromosomes. The chicken genome (Gallus gallus), which is characteristic of other bird genomes, contains 39 chromosome pairs (Burt 2002) but a total genome size of only 1.1 Gb (International Chicken Genome Sequencing Consortium [ICGSC] 2004). This compares with 20 chromosome pairs in a euchromatic genome of 2.5 Gb in mouse and 23 pairs in 2.9 Gb in humans (International Human Genome Sequencing Consortium [IHGSC] 2001; Mouse Genome Sequencing Consotium [MGSC] 2002). Avian chromosomes are also highly variable in size, leading to their classification into micro- and macrochromosomes. In this article, we follow the convention of the International Chicken Genome Sequencing Consortium (ICGSC 2004) by classifying chicken chromosomes into three classes: five macrochromosomes (GGA 1-5), measuring from ∼50 to 200 Mb in size, five intermediate chromosomes (GGA 6-10) ranging from 20 to 40 Mb, and 28 microchromosomes (GGA 11-38), on average ∼12 Mb long.
In addition to the differences in size, these three chromosome groups also exhibit marked differences in a number of other genomic features. Chicken microchromosomes are estimated to account for only 18% of the total female genome. Despite this they harbor ∼31% of all chicken genes (ICGSC 2004), giving them a gene dense structure with three to four times shorter intergenic sequences than on macrochromosomes. Furthermore, recombination rates are higher on microchromosomes (median rate, 6.4 cM/Mb) than on both intermediate (3.9 cM/Mb) and macrochromosomes (2.8 cM/Mb) (ICGSC 2004). It is also known that the GC content is higher on microchromosomes (Auer et al. 1987), as is the level of methylation (Grutzner et al. 2001), partly due to an increased number of CpG sites. Microchromosomes also are richer in CpG islands and replicate earlier than do macrochromosomes (McQueen et al. 1996, 1998; ICGSC 2004).
Increasing amounts of evidence show that mutation rates vary over genomes on a variety of scales, including both within and between chromosomes (Wolfe et al. 1989; Matassi et al. 1999; Lercher et al. 2001; MGSC 2002; Smith et al. 2002; Hardison et al. 2003; Webster et al. 2004). Many genomic features that differ between micro- and macrochromosomes have been implicated in causing mutation rate variation (Caron et al. 2001; Eyre-Walker and Hurst 2001; IHGSC 2001; Lercher et al. 2002, 2003, 2004; MGSC 2002; Axelsson et al. 2004; Chuang and Li 2004; Webster et al. 2004). Studies in mammals report a positive correlation between GC content and both substitution rate (Eyre-Walker 1993; Bielawski et al. 2000; Williams and Hurst 2000; Smith et al. 2002) and levels of genetic variation (Sachidanandam et al. 2001). It is likely that the increased prevalence of hypermutable CpG dinucleotides in GC-rich sequences (Zhao and Boerwinkle 2002) is an important factor in increasing mutation rates in these regions. A further potential correlate of substitution rate is local recombination rate (Lercher and Hurst 2002; MGSC 2002; Hellmann et al. 2003a; Jensen-Seaman et al. 2004), which can be highly variable even at small scales on human chromosomes (McVean et al. 2004). This could be due to a direct causal effect, resulting from erroneous repair of double-strand breaks that initiate recombination (Strathern et al. 1995). However, local recombination rates also correlate with GC content (Kong et al. 2002), an observation argued to result from recombination driving the evolution of GC content (Meunier and Duret 2004). Recombination can affect levels of GC content through the process of biased gene conversion (BGC), which leads to the preferential fixation of AT→GC mutations (Eyre-Walker 1993; Galtier et al. 2001; Webster and Smith 2004). Depending on the evolutionary dynamics of GC content and the mutation pattern in a given genomic region, this process may also substantially alter substitution rates (Montoya-Burgos et al. 2003). Discerning which factors are directly responsible for causing mutations is therefore problematic due to complex interactions between different processes and genomic features.
With the recent advent of the chicken genome sequence, a variety of analyses of molecular evolution are possible by comparison with related avian species, such as turkey. The advantage of these comparisons over comparisons of the chicken sequence with mammalian genomes is first that more reliable estimates of substitution rates can be gained, as synonymous sites become saturated at large evolutionary distances. Second, there have been few chromosomal rearrangements between chicken and turkey, allowing a detailed analysis of the effect of karyotype on evolutionary rates (ICGSC 2004). To analyze variation in patterns of nucleotide substitution on macrochromosomes, intermediate chromosomes, and microchromosomes in birds, we generated two independent data sets. First, we generated ∼27.9 kb of orthologous noncoding sequence from 67 introns in chicken and turkey (Meleagris gallopavo) located across all chromosomal classes. Second, we constructed 155 chicken-turkey cDNA alignments (∼92.9 kb), also evenly spread across the genome, which we used to estimate both synonymous (dS) and nonsynonymous (dN) substitution rates.
Results
Full details of the genomic location, GC content, and levels of divergence in all alignments are available as Supplemental information. The averages of these statistics according to chromosomal class are presented in Tables 1 and 2 for intron and cDNA alignments, respectively. Note that genomic location was classified according to chicken chromosome. However, the chromosomal location in turkey is not expected to differ much from that in chicken considering the slow rate of chromosomal rearrangements in birds (Burt et al. 1999) and an estimated divergence time between the two species of 28 million years (Dimcheff et al. 2002). Only two large rearrangements are known and a cross-species FISH mapping study using >200 BAC clones did not reveal any additional interchromosomal rearrangements (ICGSC 2004).
Table 1.
Summary statistics for intron alignments
| Chromosome class | No. | Total bp aligned | Average divergence (95% CI) | GC content |
|---|---|---|---|---|
| Macro | 27 | 12138 | 0.099 (0.085–0.112) | 0.44 |
| Intermediate | 10 | 4840 | 0.095 (0.073–0.116) | 0.43 |
| Micro | 30 | 10907 | 0.117 (0.105–0.134) | 0.51 |
(95% CI) 95% confidence interval.
Table 2.
Summary statistics for cDNA alignments
| Chromosome class | No. | Total bp aligned | dN (95% CI) | dS (95% CI) | dN/dS (95% CI) | GC3 |
|---|---|---|---|---|---|---|
| Macro | 63 | 38958 | 0.016 (0.011–0.021) | 0.103 (0.090–0.125) | 0.185 (0.128–0.360) | 0.52 |
| Intermediate | 27 | 18573 | 0.011 (0.007–0.016) | 0.122 (0.097–0.158) | 0.105 (0.065–0.190) | 0.60 |
| Micro | 65 | 32847 | 0.012 (0.007–0.016) | 0.131 (0.111–0.156) | 0.094 (0.066–0.177) | 0.61 |
(95% CI) 95% confidence interval.
Nucleotide substitution rates
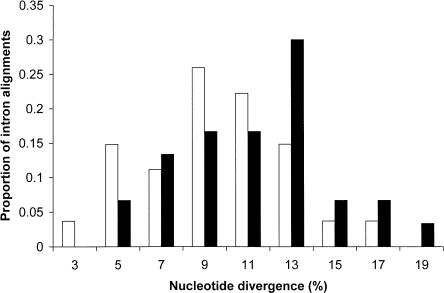
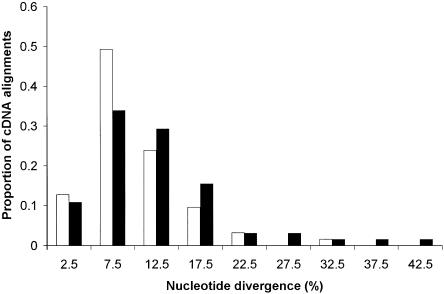
We found a clear difference in intronic divergence between the microchromosomes and the two other chromosome groups (Table 1; Fig. 1). When analyzing the divergences using the double bootstrapping method with 1000 replicates (see Methods), microchromosomes (11.7%) were found to have significantly higher divergence than did both macrochromosomes (9.9%; P = 0.016) and intermediate chromosomes (9.5%; P = 0.026). Macrochromosome and intermediate chromosome divergence values did not differ significantly. The same trend was observed at synonymous sites within cDNA alignments (Table 2; Fig. 2). Here, the difference between micro- (13.1%) and macrochromosomes (10.3%) was significant using the double bootstrapping procedure (P = 0.025), but divergence on intermediate chromosomes did not differ significantly with the other two chromosome classes.
Figure 1.
Histogram showing levels of chicken-turkey divergence in 27 macrochromosomal (white bars; chromosomes 1-5) compared with 30 microchromosomal introns (black bars; chromosomes 11-38). The X-axis shows the midpoint of each category, and the height of bars represents the relative proportion in each category.
Figure 2.
Histogram showing levels of chicken-turkey divergence at synonymous sites in 59 macrochromosomal (white bars) compared with 65 microchromosomal cDNA alignments (black bars). The X-axis shows the midpoint of each category, and the height of bars represents the relative proportion in each category.
Base composition and rate of sequence evolution
In line with observations from the complete chicken genome sequence (ICGSC 2004), the GC-content of intron alignments and GC3 in cDNA alignments on microchromosomes is higher than on macrochromosomes and intermediates (Tables 1, 2). Despite significant differences in intronic divergence between chromosomal classes, we find no clear correlation between intronic divergence and GC-content (Spearman's rank correlation, r = 0.13, P = 0.31). However, we observe a highly significant correlation between GC3 and synonymous substitution rates in genes (Spearman's r = 0.40; P < -10-6). When accounting for GC3, the significance of the difference in dS between micro- and macrochromosomes disappears (P = 0.33). There is a weak but nonsignificant negative correlation between intron length and divergence (Spearman's r = -0.24; P = 0.053).
Microchromosomes are known to have a higher incidence of hypermutable CpG dinucleotides. Considering the bias in the distribution of CpG sites and their fast mutation rate, we reanalyzed our intron data set, removing all mutations that had taken place in such sites. The mean divergence of macrochromosomes decreased to 8.6% and that of intermediates to 8.4%, while the divergence of microchromosomes reduced to 9.8%. There was no longer a significant difference in divergence between the chromosome classes (P = 0.117). In order to test that the lack of significance when excluding CpG mutations was not merely due to a reduction in the size of the data set, we reanalyzed the entire data set, randomly excluding the same number of substitutions as were identified as CpG substitutions. Divergence on microchromosomal introns was still significantly higher than on macrochromosomes (P = 0.029). It thus seems that mutations in CpG sites significantly contribute to the rate differences between small and large chromosomes in the chicken-turkey comparison.
Recombination and rate of sequence evolution
In order to examine the potential effects of recombination on substitution rate, we utilized estimates of the average recombination rate for each chromosome by comparison of physical and genetic maps (ICGSC 2004). We find no significant correlation between average recombination rate and intronic divergence (Spearman's r = 0.20; P = 0.107) and no correlation between average recombination and dS from coding sequences (Spearman's r = 0.062, P = 0.45). However, as large variation in recombination rate has been observed on the fine scale (McVean et al. 2004) in humans, using rates of recombination averaged per chromosome is not optimal for this analysis.
To further understand the causes of variation in substitution rate, we compared frequencies of A↔T and G↔C substitutions on microchromosomes, intermediate chromosomes, and macrochromosomes. These substitutions are unlikely to be influenced by hypermutability at CpG sites or by BGC (Filatov 2004). However, if recombination is directly mutagenic, we expect all types of mutational change, including A↔T and G↔C to be elevated on microchromosomes. The level of divergence caused by A↔T substitutions on microchromosomes, intermediate chromosomes, and macrochromosomes are 1.57%, 1.55%, and 1.29%, respectively. These values for G↔C divergence are 1.85%, 1.29%, and 1.76%. None of these differences were significant using the double bootstrapping procedure. We therefore have no evidence to support a direct mutational effect of recombination on microchromosomes.
Purifying selection on microchromosomes
In order to investigate the relative effects of selection on all chromosome classes, we estimated and compared dN/dS ratios. Microchromosomes and intermediate chromosomes were both found to have reduced dN/dS ratios (0.094 and 0.105, respectively) compared with 0.185 on macrochromosomes. The difference between micro- and macrochromosomes was significant (P = 0.026). These figures are comparable to a previous estimate on avian proteins of dN/dS = 0.18 based on a comparison of 28 mainly macrochromosomal genes using a chicken-old world quail comparison (Keightley and Eyre-Walker 2000). We also find a weak but significant correlation between dN/dS and average chromosomal recombination rate (Spearman's r = -0.162, P = 0.048) but no significant correlation between dN/dS and GC3 (Spearman's r = -0.106, P = 0.19).
Discussion
In this study we analyzed substitution rates of avian chromosomes using chicken-turkey intron and coding sequence alignments. The results indicate an 18% higher average sequence divergence in introns on microchromosomes compared with macrochromosomes and a 26% higher average rate of synonymous substitutions in coding sequences on microchromosomes compared with macrochromosomes.
In several recent investigations, the neutrality of intron sequence evolution has been questioned (Hare and Palumbi 2003; Chamary and Hurst 2004; Halligan et al. 2004). The presence of extended splice-sites and potential regulatory elements means intron sequences could be subject to evolutionary constraint. However, selective constraint is unlikely to result in differences between intronic rates on micro- and macrochromosomes. In general, selective constraint is expected to homogenize differences caused by mutation rate variation. Furthermore, as introns on microchromosomes tend to be shorter (ICGSC 2004), a greater proportion of sequence is likely to be comprised of regulatory elements, which would result in a reduction in substitution rates on microchromosomes, which is the opposite of what we observe. The evidence for neutrality of silent sites in higher eukaryotes is not ambiguous either (Duret 2002; Hellmann et al. 2003b) but it seems unlikely that, for example, a codon usage bias would differ between chromosomal classes. Hence the observed differences in divergence on micro- and macrochromosomes are likely to result from variation in germ line mutation rates between chromosome classes.
Mutations in CpG dinucleotides are an important factor for explaining the high divergence of microchromosomal intron sequences. Methylated CpG sites easily deaminate, resulting in C→T transitions possibly 10 times more frequent than other mutations (Robertson and Wolffe 2000). Removal of mutations that had taken place in these dinucleotides clearly reduces the divergence difference between the chromosomal classes. A number of further effects could result in differences in substitution rates between avian autosomes, including recombination being mutagenic. Base composition may directly alter regional substitution rates if global rates of AT→GC and GC→AT mutations differ, although the effect is largely dependent on the equilibrium GC content a sequence is evolving toward (Piganeau et al. 2002). In addition, BGC may influence substitution rates by altering the pattern of nucleotide fixation, although the effect of this process is also dependent on equilibrium GC content, and little is known about the evolutionary dynamics of base composition in the avian genome. Unfortunately, reliable estimates of fine scale variation in recombination rate across the chicken genome are not yet available, so the relationship between recombination and substitution rate is unclear.
By analysis of dN/dS ratios, we demonstrated that the proteins of genes located on microchromosomes are more evolutionarily conserved. This echoes findings from a mouse-rat comparison (Williams and Hurst 2000) in which the dN/dS ratio was found to be negatively correlated with GC content (and hence recombination rate). Two potential hypotheses have been invoked to explain this observation. Firstly, it could represent local variation in the efficacy of natural selection, which is known to covary with recombination rate (Hill and Robertson 1966; Nordborg et al. 1996). Second, it could indicate that microchromosomes are enriched for slowly-evolving genes that fulfill conserved functions, such as housekeeping genes. This has been suggested to be the case in GC-rich regions of the human genome (Lercher et al. 2003), although no evidence for a clustering of genes with similar expression patterns according to chromosomal class has been found in the chicken genome (ICGSC, in prep.). Simulations indicate that the efficacy of selection is unlikely to vary much due to recombination in species with high average levels of recombination, such as birds (Nordborg et al. 1996). It is therefore unclear whether variation in patterns of evolutionary conservation reflects a fundamental feature of genome architecture or whether it is a consequence of variation in recombination rate.
The observation of a high rate of sequence evolution on microchromosomes in chicken and turkey represents evidence for a difference in mutation rate between two distinct autosomal classes in a eukaryotic genome. This should, for example, influence the use of molecular clocks in birds as it means there will be lineage specific effects due to differences in karyotype. It is also interesting to note that the small genome size of birds, contributed to by the small size of microchromosomes, if selectively advantageous as proposed by some (Burt 2002), comes with a cost of a high mutational load.
Methods
Collection of intron sequence data
Chicken and turkey intron sequences used in this study were taken from four sources: the chicken genome sequence (www.ensembl.org/Gallus_gallus), GenBank (www.embl.org), a previous study (Axelsson et al. 2004), or sequencing (for details, see Supplemental information). To identify intronic sequence, we first BLASTed chicken cDNA sequences against the draft human genome at National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov). Large gaps in the chicken sequence produced in such BLAST alignments were judged to be the positions of putative introns. For all genes analyzed in this study, this approach revealed putative chicken introns at precisely the same positions as in orthologous human genes. Using this information, we designed exonic PCR primers for amplification and sequencing of both chicken and turkey introns (for details, see Supplemental information). Sequences <100 bp were excluded from the study to reduce the potential effects of conserved regulatory elements. Sequence data generated for this article have been deposited in GenBank under accession numbers AY685058-62, AY685066-94, and AY685097-111.
Sequence analysis of intron data
We used the program Sputnik (www.espressosoftware.com) to remove microsatellites from the sequences and RepeatMasker (www.repeatmasker.org) to mask interspersed repeats. Orthologous chicken and turkey introns were then aligned with ClustalW using default settings (Thompson et al. 1994) and adjusted by eye. Pairwise distances were estimated using the baseml program in PAML 3.11 (Yang 1997), with the Tamura-Nei (Tamura and Nei 1993) model of sequence evolution. Distances were estimated assuming that all sites evolve at the same rate (i.e., no among site rate variation). CpG mutations were defined as being CG in one species and either TG or CA in the other (same applied to coding sequences). The genomic location of all introns was determined by BLAST searches against the chicken genome; this confirmed results from searches against the human genome. The program seqtool (Mikael Brandström) was used to graphically represent BLAST hits and identify regions of ambiguous orthology, such as could be present in recently expanded gene families. No such instances were observed.
Collection of coding sequence data
We downloaded all available expressed turkey cDNA sequences from EMBL (www.embl.org) and BLASTed them against all predicted genes in chicken. We used an expect value of <e-100 as cut off value when extracting reliable hits from the BLAST run. For each turkey entry, if more than one chicken sequence scored better than the threshold value, only the best hit was selected for further analysis. In order to remove regions where orthology was ambiguous, we then BLASTed all matching chicken cDNAs against the chicken genomic sequence and visually inspected all hits using seqtool (see above). Gene prediction data from the chicken genome project were used to extract the coding sequence from most of the selected chicken genes. In cases in which the positions of start and stop codons were not annotated, we searched for possible open reading frames within the sequence. In all such cases only one possible open reading frame covering the whole sequence was found. Thirty-three of the 253 turkey sequences with chicken homologs were annotated as complete coding sequences. For these sequences, we relied on the accompanying turkey annotation for identifying the correct reading frame.
Sequence analysis of coding data
The 253 cDNA alignments were created using default settings in ClustalW (Thompson et al. 1994). Each alignment was checked by eye, and a small number of unreliable segments were discarded by manual editing. A number of the turkey ESTs proved to contain only 5′ or 3′ UTR sequence and hence no longer matched the trimmed chicken coding sequence. These sequences were removed from the analysis. In other alignments, stop codons were detected in the turkey sequence, possibly due to artificial frame shifts introduced during the alignment process, errors introduced during sequencing, or the turkey sequence being collected from a pseudogene. These alignments were also removed from the analysis. A total of 155 high-quality autosomal alignments were kept for the subsequent divergence analysis. The codeml program of the PAML package (Yang 1997) was used to estimate dN, dS, and dN/dS ratios, implementing the Goldman and Yang model of sequence evolution (Goldman and Yang 1994).
Hypothesis testing
For both of the data sets, we used a nonparametric bootstrapping method termed double bootstrapping (Axelsson et al. 2004) to estimate confidence intervals and perform hypothesis testing for noncoding and synonymous divergence. Starting with the intron data set, for each of the three chromosomal classes (microchromosomes, intermediate chromosomes, and macrochromosomes) we first bootstrapped by introns, randomly sampling introns with replacement to give the same total number of introns as in the original data set, and then for each of the intron alignments we bootstrapped by sites, randomly sampling sites with replacement to generate alignments of the same length as the originals. The coding data set was treated similarly with one exception: Instead of bootstrapping by sites in the second step of the procedure, we now bootstrapped by codons to randomly generate alignments of the same length as the originals. The first stage of the bootstrapping procedure accounts for rate variation between different sequences, as may be due to regional variation in mutation (Ellegren et al. 2003). The second stage of the bootstrapping accounts for random variation in divergence at individual sites. Pairwise distances were calculated for each of the alignments after the double bootstrapping, and the unweighted mean of these distances was calculated.
The bootstrapping process was repeated 1000 times, thereby giving 1000 sets of distance estimates for the three chromosome classes from which to estimate rate statistics. The standard deviation of the bootstrap values gives an estimate of the standard error of the bootstrapped statistic (Sokal and Rohlf 1995). Hypothesis testing requiring the comparison of rate statistics was performed by direct comparison of randomized bootstrap values.
Acknowledgments
We would like to thank Johanna Sandling for valuable help in generating intron sequence data for this study. We acknowledge support from the Swedish Research Council. H.E. is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from the Knut and Alice Wallenberg Foundation.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3021305. Article published online before print in December 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AY685058-AY685062, AY685066-AY685094, and AY685097-AY685111.]
References
- Auer, H., Mayr, B., Lambrou, M., and Schleger, W. 1987. An extended chicken karyotype, including the NOR chromosome. Cytogenet. Cell Genet. 45: 218-221. [DOI] [PubMed] [Google Scholar]
- Axelsson, E., Smith, N.G.C., Sundstrom, H., Berlin, S., and Ellegren, H. 2004. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol. Biol. Evol. 18: 1538-1547. [DOI] [PubMed] [Google Scholar]
- Bielawski, J.P., Dunn, K.A., and Yang, Z. 2000. Rates of nucleotide substitution and mammalian nuclear gene evolution: Approximate and maximum-likelihood methods lead to different conclusions. Genetics 156: 1299-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, D.W. 2002. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 96: 97-112. [DOI] [PubMed] [Google Scholar]
- Burt, D.W., Bruley, C., Dunn, I.C., Jones, C.T., Ramage, A., Law, A.S., Morrice, D.R., Paton, I.R., Smith, J., Windsor, D., et al. 1999. The dynamics of chromosome evolution in birds and mammals. Nature 402: 411-413. [DOI] [PubMed] [Google Scholar]
- Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M.C., van Asperen, R., Boon, K., Voute, P.A., et al. 2001. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science 291: 1289-1292. [DOI] [PubMed] [Google Scholar]
- Chamary, J.V. and Hurst, L.D. 2004. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: Evidence for selectively driven codon usage. Mol. Biol. Evol. 21: 1014-1023. [DOI] [PubMed] [Google Scholar]
- Chuang, J.H. and Li, H. 2004. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2: E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimcheff, D.E., Drovetski, S.V., and Mindell, D.P. 2002. Phylogeny of Tetraoninae and other galliform birds using mitochondrial 12S and ND2 genes. Mol. Phylogenet. Evol. 24: 203-215. [DOI] [PubMed] [Google Scholar]
- Duret, L. 2002. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 12: 640-649. [DOI] [PubMed] [Google Scholar]
- Ellegren, H., Smith, N.G.C., and Webster, M.T. 2003. Mutation rate variation in the mammalian genome. Curr. Opin. Genet. Dev. 13: 562-568. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A. 1993. Recombination and mammalian genome evolution. Proc. R. Soc. Lond. B 252: 237-243. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A. and Hurst, L.D. 2001. The evolution of isochores. Nat. Rev. Genet. 2: 549-555. [DOI] [PubMed] [Google Scholar]
- Filatov, D.A. 2004. A gradient of silent substitution rate in the human pseudoautosomal region. Mol. Biol. Evol. 21: 410-417. [DOI] [PubMed] [Google Scholar]
- Galtier, N., Piganeau, G., Mouchiroud, D., and Duret, L. 2001. GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics 159: 907-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, N. and Yang, Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725-736. [DOI] [PubMed] [Google Scholar]
- Grutzner, F., Zend-Ajusch, E., Stout, K., Munsche, S., Niveleau, A., Nanda, I., Schmid, M., and Haaf, T. 2001. Chicken microchromosomes are hypermethylated and can be identified by specific painting probes. Cytogenet. Cell Genet. 93: 265-269. [DOI] [PubMed] [Google Scholar]
- Halligan, D.L., Eyre-Walker, A., Andolfatto, P., and Keightley, P.D. 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14: 273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Connor, M., Kolbe, D., et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13: 13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, M.P. and Palumbi, S.R. 2003. High intron sequence conservation across three mammalian orders suggests functional constraints. Mol. Biol. Evol. 20: 969-978. [DOI] [PubMed] [Google Scholar]
- Hellmann, I., Ebersberger, I., Ptak, S.E., Paabo, S., and Przeworski, M. 2003a. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, I., Zollner, S., Enard, W., Ebersberger, I., Nickel, B., and Paabo, S. 2003b. Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 13: 831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W.G. and Robertson, A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269-294. [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (ICGSC). 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature (in press). [DOI] [PubMed]
- International Human Genome Sequencing Consortium (IHGSC). 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman, M.I., Furey, T.S., Payseur, B.A., Lu, Y., Roskin, K.M., Chen, C.F., Thomas, M.A., Haussler, D., and Jacob, H.J. 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14: 528-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley, P.D. and Eyre-Walker, A. 2000. Deleterious mutations and the evolution of sex. Science 290: 331-333. [DOI] [PubMed] [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241-247. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J. and Hurst, L.D. 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337-340. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Williams, E.J., and Hurst, L.D. 2001. Local similarity in evolutionary rates extends over whole chromosomes in human-rodent and mouse-rat comparisons: Implications for understanding the mechanistic basis of the male mutation bias. Mol. Biol. Evol. 18: 2032-2039. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., and Hurst, L.D. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31: 180-183. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., Pavlicek, A., and Hurst, L.D. 2003. A unification of mosaic structures in the human genome. Hum. Mol. Genet. 12: 2411-2415. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Chamary, J., and Hurst, L.D. 2004. Gene expression and genomic regionality in rates of evolution. Genome Res. 14: 1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi, G., Sharp, P.M., and Gautier, C. 1999. Chromosomal location effects on gene sequence evolution in mammals. Curr. Biol. 9: 786-791. [DOI] [PubMed] [Google Scholar]
- McQueen, H.A., Fantes, J., Cross, S.H., Clark, V.H., Archibald, A.L., and Bird, A.P. 1996. CpG islands of chicken are concentrated on microchromosomes. Nat. Genet. 12: 321-324. [DOI] [PubMed] [Google Scholar]
- McQueen, H.A., Siriaco, G., and Bird, A.P. 1998. Chicken microchromosomes are hyperacetylated, early replicating, and gene rich. Genome Res. 8: 621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean, G.A., Myers, S.R., Hunt, S., Deloukas, P., Bentley, D.R., and Donnelly, P. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581-584. [DOI] [PubMed] [Google Scholar]
- Meunier, J. and Duret, L. 2004. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 21: 984-990. [DOI] [PubMed] [Google Scholar]
- Montoya-Burgos, J.I., Boursot, P., and Galtier, N. 2003. Recombination explains isochores in mammalian genomes. Trends Genet. 19: 128-130. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium (MGSC). 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., Charlesworth, B., and Charlesworth, D. 1996. The effect of recombination on background selection. Genet. Res. 67: 159-174. [DOI] [PubMed] [Google Scholar]
- Piganeau, G., Mouchiroud, D., Duret, L., and Gautier, C. 2002. Expected relationship between the silent substitution rate and the GC content: Implications for the evolution of isochores. J. Mol. Evol. 54: 129-133. [DOI] [PubMed] [Google Scholar]
- Robertson, K.D. and Wolffe, A.P. 2000. DNA methylation in health and disease. Nat. Rev. Genet. 1: 11-19. [DOI] [PubMed] [Google Scholar]
- Sachidanandam, R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933. [DOI] [PubMed] [Google Scholar]
- Smith, N.G.C., Webster, M.T., and Ellegren, H. 2002. Deterministic mutation rate variation in the human genome. Genome Res. 12: 1350-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R.R. and Rohlf, F.J. 1995. Biometry. Freeman and Company, New York.
- Strathern, J.N., Shafer, B.K., and McGill, C.B. 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140: 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. and Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512-526. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. Clustal-W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, M.T. and Smith, N.G.C. 2004. Fixation biases affecting human SNPs. Trends Genet. 20: 122-126. [DOI] [PubMed] [Google Scholar]
- Webster, M.T., Smith, N.G.C., Lercher, M.J., and Ellegren, H. 2004. Gene expression, synteny, and local similarity in human noncoding mutation rates. Mol. Biol. Evol. 21: 1820-1830. [DOI] [PubMed] [Google Scholar]
- Williams, E.J. and Hurst, L.D. 2000. The proteins of linked genes evolve at similar rates. Nature 407: 900-903. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Sharp, P.M., and Li, W.H. 1989. Mutation rates differ among regions of the mammalian genome. Nature 337: 283-285. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Zhao, Z. and Boerwinkle, E. 2002. Neighboring-nucleotide effects on single nucleotide polymorphisms: A study of 2.6 million polymorphisms across the human genome. Genome Res. 12: 1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]