Abstract
Large tracts of the human genome, known as gene deserts, are devoid of protein-coding genes. Dichotomy in their level of conservation with chicken separates these regions into two distinct categories, stable and variable. The separation is not caused by differences in rates of neutral evolution but instead appears to be related to different biological functions of stable and variable gene deserts in the human genome. Gene Ontology categories of the adjacent genes are strongly biased toward transcriptional regulation and development for the stable gene deserts, and toward distinctively different functions for the variable gene deserts. Stable gene deserts resist chromosomal rearrangements and appear to harbor multiple distant regulatory elements physically linked to their neighboring genes, with the linearity of conservation invariant throughout vertebrate evolution.
One of the major challenges of genomics is to understand how the genome is organized and, especially, which sequences and factors contribute to the complex and precise regulation of gene expression. These include cis-regulatory sequences controlling gene expression, insulators or boundary elements defining physical domains, and sequences that anchor genomic regions to specific nuclear locations (Dorsett 1999; Bell et al. 2001; Carter et al. 2002). The arrangement of these various regulatory elements (REs) has not been fully elucidated for any locus, and hence consistent patterns for multiple loci are not yet apparent, but these are the subjects of active current investigation.
One of the unexplained architectural asymmetries observed in the human genome sequence is the uneven distribution of genes (Lander et al. 2001; Venter et al. 2001). Specifically, it has been estimated that ∼25% of the human genome consists of gene deserts, defined as long regions containing no protein-coding sequences and without obvious biological functions (Venter et al. 2001). Some of these gene deserts have been shown to contain regulatory sequences that act at large distances to control the expression of neighboring genes (Nobrega et al. 2003; Kimura-Yoshida et al. 2004; Uchikawa et al. 2004). By contrast, other large gene-sparse regions are potentially nonessential to genome function, since they can be deleted without significant phenotypic effect (Russell et al. 1982; Rinchik et al. 1990; Nobrega 2004). It is possible that these differences reflect the existence of distinct categories of gene deserts, such that some deserts harbor sequence elements with critically important and conserved biological roles whereas others do not.
To investigate this possibility, we focused on sequence comparisons with the chicken genome, an organism strategically positioned between rodents and fish in the vertebrate evolutionary tree. By analyzing genomic structure, conservation patterns, and evolutionary relationships, we were able to classify gene deserts into two functionally different groups and to provide new insights regarding the functions of these intervals in the human genome.
Results
Identification of human gene deserts
The current human gene annotation (knownGenes mapped to the NCBI Build 34) (Karolchik et al. 2003) defines 18,134 distinct intergenic regions that cumulatively span 61.2% of the human genome (with subtelomeric and pericentromeric regions excluded from the analysis). The length of the intergenic intervals varies notably from a few base pairs to 5.1 Mb. The 3% longest intergenic intervals (545 genomic regions, with the shortest of them covering 640 kb) together span ∼25% of the sequenced human genome. This is consistent with previous estimates of gene desert coverage (Venter et al. 2001; Nobrega et al. 2003), and thus we have used this as the set of gene deserts in the current study. Remarkably, two small human chromosomes (HSA17 and HSA19) are distinct outliers, comprising almost entirely of genes surrounded by “regular” intergenic intervals (defined as 25%–75% of the intergenic intervals' length distribution curve and ranging from 6–72 kb in size). Each of these chromosomes contains only two gene deserts. In contrast, HSA4, HSA5, and HSA13 are heavily populated with gene deserts, corresponding up to 40% of the length of each chromosome (Fig. 1).
Figure 1.
Chromosome coverage by gene deserts (in blue) and regular intergenic regions (in red).
Gene deserts of these sizes are more frequent than might occur by chance if the placement of genes in the genome were random. A randomization study (see Methods) showed that, by chance alone, the probability of a gene desert reaching the observed maximal size of 5.1 Mb is below 10-4—the largest intergenic distance produced by randomizations was ∼2 Mb, a size exceeded by 76 of the observed gene deserts. The same study showed that, by chance alone, the probability of obtaining 545 deserts of size larger than 640 kb is, too, <10-4—the largest count of intergenic distances >640 kb produced by randomizations was only 75.
Compared with other genomic regions, gene deserts in general display a strikingly low G+C content, an elevated density of single nucleotide polymorphisms (SNPs), and a decrease in the fraction of conserved sequence between humans, chicken, and mouse (Table 1). The average repeat content of gene deserts is slightly higher than the genome average, but the fraction of DNA comprised of repetitive sequences ranges from 30% to 90%. This suggests that reduced levels of purifying selection pressure may be acting in gene deserts, furthering the hypothesis that these regions represent segments of relatively low biological activity, enriched in pseudogenes, repeats, and other nonfunctional sequences. Contrary to this hypothesis, however, it has been shown that some human gene deserts harbor distant gene REs that are deeply conserved in vertebrate species (Nobrega et al. 2003; Kimura-Yoshida et al. 2004).
Table 1.
Characteristic features of gene deserts, gene-rich regions, regular intergenic regions, and the average in the human genome, NCBI Build 34
| Region | Length (Mb) | G+C content | Chicken conservationa | Mouse conservationa | Repeat content | Density of SNPs |
|---|---|---|---|---|---|---|
| Gene deserts | 716 | 37.5% | 1.91% | 19.0% | 50.5% | 0.73/kb |
| Stable gene deserts | 207 | 38.3% | 4.28% | 25.6% | 46.9% | 0.69/kb |
| Variable gene deserts | 509 | 37.1% | 0.85% | 16.1% | 52.0% | 0.74/kb |
| Regular intergenic | 244 | 44.7% | 1.27% | 17.4% | 55.4% | 0.60/kb |
| Gene-rich | 285 | 47.4% | 4.35% | 28.0% | 48.9% | 0.57/kb |
| Average | 2842 | 40.9% | 2.98% | 22.4% | 48.5% | 0.66/kb |
Repeat content and SNP annotation were derived from the tabular genome annotation obtained from the UCSC Genome Browser utility.
Interspecies conservation describes the percentage of nonrepetitive sequence covered by the ECRs
SINE-type repetitive elements are depleted in gene deserts
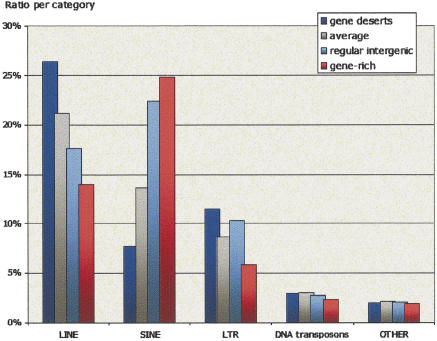
Although the relative density of repetitive elements in the gene deserts is comparable with the average distribution in the genome, the content of the various classes of repetitive elements is markedly different. The density of LINE elements is distinctly elevated and the density of SINE elements is decreased in gene deserts, when compared to averages for the human genome (Fig. 2). The opposite trend (relative LINE depletion accompanied by SINE enrichment) is observed for gene-rich (see Methods) and regular intergenic regions (Grover et al. 2003, 2004). These findings can be partially attributed to the decreased nonrepetitive G+C content in gene deserts that is known to be associated with LINE repetitive elements (Supplemental Fig. S1; Lander et al. 2001; Venter et al. 2001). However, the SINE content of the subset of regular intergenic regions having G+C ratio <40% is closer to the SINE content of gene-rich regions than that of gene deserts (Supplemental Table S1). This suggests that the G+C content is not the only factor for the observed imbalance in repeat families populating gene deserts. The accumulation of the observed imbalance in LINE versus SINE repetitive elements populating different genomic regions can be dated to the mammalian radiation. Ancient LINE-L2 repetitive elements contribute only minimally to the distribution of repeats (3.3% of the overall distribution in average), and their distribution does not show pronounced enrichment in any specific category of genomic interval. However, the LINE-L1 and SINE families of repetitive elements, which have expanded dramatically since the separation of rodent and primate lineages (Gibbs et al. 2004), are differentially distributed (Fig. 2).
Figure 2.
Ratio of different categories of repetitive elements populating different human genomic regions. Gene deserts are in blue, average counts for the human genome are in gray, regular intergenic regions are in light blue, and gene-rich regions are in red.
Dichotomy in evolutionary preservation of gene deserts
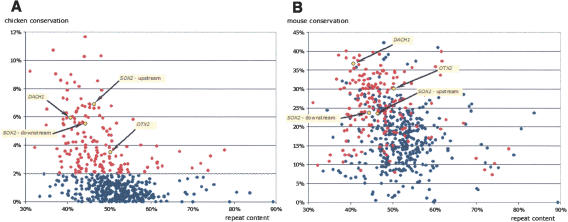
The average density of evolutionarily conserved regions (ECRs; for a definition, see Methods) detected in human/mouse (h/m) and human/chicken (h/c) alignments is similar in gene deserts and regular intergenic regions (with only a slight increase in density within gene deserts). However, there is a wide variation in ECR density among different gene deserts, which cannot be entirely attributed to the variation in repeat density (Fig. 3A). The distribution of h/c ECR content (hereafter referred to simply as conservation) in these regions ranges from 0%–12% and has an uneven shape, with many of the gene deserts having <2% of their sequence conserved (Fig. 3A). We used this arbitrary 2% h/c conservation cutoff to separate gene deserts into two categories, stable (172 regions; >2% conserved) and variable (373 regions; <2% conserved). This classification was initially used because empirically it highlights gene deserts that are well conserved throughout the time since the separation of mammalian and avian lineages; the usefulness of this estimated cutoff level was validated by later analyses (see below). Stable gene deserts have several critical properties indicating that they contain functional DNA elements. First, they include regions surrounding the DACH1, OTX2, and SOX2 genes, which have previously been shown to harbor long-distance transcriptional REs (Nobrega et al. 2003; Kimura-Yoshida et al. 2004; Uchikawa et al. 2004). Second, most stable gene deserts lie within a narrow window of repeat content, averaging 47.0% repetitive sequences. Interestingly enough, in contrast to the h/c comparisons, h/m conservation does not allow a clear differentiation between stable and variable gene deserts (Fig. 3B), suggesting a limitation of h/m comparisons in recapitulating the observed ancestral conservation pattern.
Figure 3.
Correlation of nonrepetitive conservation of the human gene deserts with chicken (A) and mouse (B) versus repeat content. Red color depicts the stable gene deserts that are >2% conserved with chicken throughout their length. Negative correlation of the nonrepetitive conservation level and repeat content is very weak in both chicken and mouse comparisons, with R2 reaching 0.06 in the case of mouse comparisons. Two stable gene deserts located upstream of the DACH1 and OTX2 gene and two other ones surrounding the SOX2 gene are in yellow.
To highlight the robustness of this partitioning of gene deserts, we also applied phylogenetic hidden Markov model (phastCons) annotation (Siepel and Haussler 2004a,b) to these regions (see Methods). Remarkably, stable gene deserts are effectively separated from the variable gene deserts in the phastCons conservation analysis (Supplemental Fig. S2); the criterion that at least 0.4% of the sequence is phastCons conserved is satisfied by 93% of stable gene deserts but only 10% of variable gene deserts.
Sequence conservation between species can result from purifying selection reflecting an active resistance to change or from a slower rate of neutral evolution in that region. Thus we investigated the estimated neutral substitution rates in gene deserts to ascertain whether they had a significantly slower neutral rate. The average substitutions per site in aligned ancestral repeats between human and mouse (tAR) has been used as a good estimate of the average substitutions per neutral site (Waterston et al. 2002; Hardison et al. 2003). We find that the value of tAR is higher in variable deserts (0.489 substitutions per site) than in stable deserts (0.476 substitutions per site). However, both of these are higher than the genome average of 0.462 substitutions per site. Thus gene deserts apparently accept neutral substitutions at a rate higher than the bulk of the genome. Likewise, the densities of SNPs and of interspersed repeats are elevated in gene deserts (Table 1), reflecting a robust rate of neutral change. Sites that have not changed are, therefore, potentially subject to purifying selection. The estimated neutral substitution rates for gene deserts are similar to those seen for regions of high noncoding conservation between chicken and human (International Chicken Genome Sequencing Consortium [ICGSC] 2004). They contrast markedly with the evolutionarily cold regions, which are characterized by low tAR values (Waterston et al. 2002; Hardison et al. 2003; Yang et al. 2004).
Inferences on the biological function of gene deserts
In order to address the biological function of gene deserts we investigated the Gene Ontology (GO) categories of genes that flank them. While the enrichment for genes in particular types was not very striking when all gene deserts were analyzed, some well-defined categories were highlighted once genes flanking only the stable gene deserts were considered. Specifically, we observed enrichment in genes coding for transcription factors, and genes involved in the regulation of transcription, DNA binding, regulation of metabolism, and development (Table 2). These observations are consistent with studies of Drosophila and nematode genes involved in development and those encoding transcription factors, which are also surrounded by much larger intergenic sequences than are housekeeping genes or genes involved in basic metabolic processes (Nelson et al. 2004). These observations raise the possibility that developmentally important genes, often endowed with complex expression patterns, are associated with regulatory units that during vertebrate evolution and the concomitant genome expansion have drifted apart from target genes to create some of the stable gene deserts now present in vertebrate genomes. A drastically different scenario emerged from similar analysis in variable gene deserts, pointing to genes with other functions. For example, genes involved in intercellular communication processes, receptor activity, neurophysiological processes, and organogenesis were found to be enriched in regions flanking variable gene deserts (Table 2).
Table 2.
Enrichment in Gene Ontology categories for stable and variable gene deserts
| Category | Enrichment | Classification |
|---|---|---|
| Stable gene deserts | ||
| Regulation of metabolism | 4.4 | biological process |
| Transcription factor activity | 4.2 | molecular function |
| Transcription coactivator activity | 4.0 | molecular function |
| Regulation of biosynthesis | 3.8 | biological process |
| Transcription regulator activity | 3.6 | molecular function |
| Transcription factor binding | 3.2 | molecular function |
| DNA binding | 2.8 | molecular function |
| Regulation of transcription | 2.8 | biological process |
| Transcription | 2.7 | biological process |
| Development | 2.0 | biological process |
| Variable gene deserts | ||
| Glutamate receptor activity | 7.8 | molecular function |
| Inotropic glutamate receptor activity | 7.7 | molecular function |
| Amine receptor activity | 6.2 | molecular function |
| Sulfotransferase activity | 4.2 | molecular function |
| Cell adhesion | 3.0 | biological process |
| Transmission of nerve impulse | 2.8 | biological process |
| Neuromuscular physiological process | 2.8 | biological process |
| Synaptic transmission | 2.7 | biological process |
| Calcium ion binding | 2.2 | molecular function |
| Organogenesis | 1.9 | biological process |
| Morphogenesis | 1.7 | biological process |
| Development | 1.7 | biological process |
| Cell communication | 1.6 | biological process |
The statistical significance of the reported numbers is supported by the P-values <10-5 as quantified in a comparison with the purely-by-chance expectations.
About 52% of all gene deserts are separated from another gene desert by at most 1 Mb and three genes. In particular, 33% (56 of 172) of stable gene deserts are paired in this manner with a stable partner, in what we call a conjoined stable gene desert. The genes interspersed between these conjoined stable gene deserts represent a unique class of loci that have evolved in a largely noncoding genomic environment from the times preceding the speciation event of mammals and birds. GO functional characterization of these genes indicates an enrichment in transcriptional gene regulatory functions and depletion in the response to stimulus category (P < 0.001). Other gene products function in skeletal development (BMP2), electron transport (COX7A3), muscle development (MEF2C), calcium ion binding (DGKB), apoptosis (FKSG2), and cell cycle (DBC1). Many of these genes are known or suspected to be involved in critical developmental steps or essential biochemical processes in vertebrates. The observed bias in genes in these interdesert regions indicates that noncoding elements regulating transcription of the transcription factors are kept under elevated levels of purifying selection throughout the evolution of vertebrates.
Robustness of the dichotomy of gene deserts
Comparative sequence analysis of human gene deserts with the homologous Fugu counterparts revealed that the density of human/Fugu (h/f) ECRs is 122-fold higher in stable gene deserts than in variable gene deserts. Moreover, the density of h/f ECRs in stable gene deserts was 3.5-fold higher then the average for the genome density of h/f ncECRs. Stable gene deserts harbor 98% of the h/f ECRs (760 out of 777) that are found in all gene deserts combined. This distinct partitioning in the evolutionary histories of the stable and variable gene deserts suggests that the arbitrary cutoff of 2% used to distinguish these regions does indeed identify two well-defined categories and suggests fundamentally different functions for stable and variable gene deserts.
Although it is not possible to reliably determine whether a conserved element has regulatory activity using current computational techniques, Kolbe et al. (2004) recently developed an approach to quantify the regulatory potential of a genomic sequence. By using regulatory potential annotation of the human genome (see Methods), we compared the density of predicted REs in two categories of gene deserts. Remarkably, the RE density was found to be three times higher in stable gene deserts than in variable gene deserts. Also, a distinct separation of RE density within the two gene deserts' categories was observed (Supplemental Fig. S3). RE density was found to be <70 RE/1Mb for 84% of variable gene deserts and larger than this value for 82% of stable gene deserts.
By studying the average length of h/m and h/c ECRs, we found that ECRs in gene deserts are longer than those found in regular intergenic regions; the average h/m ECR length in gene deserts is 265 bp, whereas that in regular intergenic regions is 224 bp. Human and chicken alignments reveal even longer ECRs in gene desert regions, with an average h/c ECR of 282 bp, but shorter ECRs (222 bp) in the regular intergenic intervals. This difference is even more evident when stable gene deserts are considered; h/m ECRs average 288 bp in these regions, and h/c ECRs span 304 bp on average. In conjunction with our recent observation that a substantial fraction of known functional h/m noncoding ECRs (ncECRs) are >350 bp (Ovcharenko et al. 2004a,b), these data reiterate an enrichment of functional elements in the pool of ECRs that populate the stable gene deserts.
UTR conservation is amplified next to gene deserts
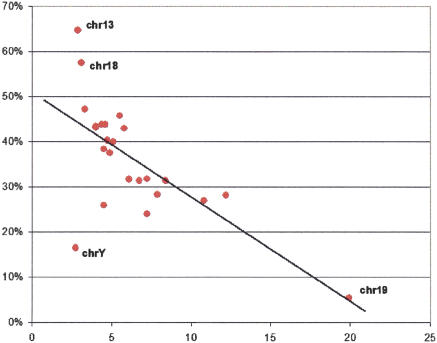
To study patterns of conservation in more detail, we classified ncECRs according to overlap with annotated knownGene 5′ and 3′-untranslated sequences, introns, and intergenic sequences (see Methods). We observed that the probability for a h/m ncECR (defined based on knownGene annotation) to also be conserved in chicken is significantly higher for UTRs than for all other noncoding elements, which is consistent with their higher average percent identity in h/m alignments (Waterston et al. 2002). While only 7.6% of h/m ncECRs were conserved in chicken, 24.7% of h/m ncECRs that overlap with either 5′ or 3′ UTRs were also conserved in chicken. We also analyzed the relationship between gene density and the probability of detecting h/c UTR-associated ECRs across different human chromosomes (Fig. 4). The analysis displayed a strong negative correlation between gene density and UTR conservation. For example, the most gene-rich human chromosome, HSA19, has the lowest percentage of genes with conserved UTRs, while in gene-poor HSA13 and HSA18, >55% of genes contain UTRs that are conserved between human and chicken. A detailed analysis of genes flanking gene deserts indicates that 50.1% of them contain UTRs conserved between human and chicken. Surprisingly, the human–chicken UTR conservation is very similar for both categories of gene deserts (variable gene deserts, 48.8%; stable gene deserts, 53.7%), while it is drastically decreased (to 13.4%) for genes located in gene-rich regions.
Figure 4.
Percentage of genes with UTRs conserved in chicken (vertical axis) versus the gene density (based on RefSeq annotation; in genes per 1 Mb of sequence as plotted at the horizontal axis). Red dots describe different human chromosomes.
This approximately fourfold difference between h/m ncECRs and h/m UTR-ECRs that are also conserved in chicken suggests that an increased selective pressure applies to UTRs and that functional elements lie within the conserved UTRs. For example, h/c conserved UTRs might preferably indicate genes with REs embedded in their untranslated regions, including potential enhancers or sequences involved in posttranscriptional regulatory mechanisms (Pesole et al. 2002). These data indicate that UTR sequences may play a more important role in the regulation of gene desert or regular intergenic loci expression than for genes residing in gene-rich domains.
Stable gene deserts are linked to neighboring genes
The availability of the near-complete sequence for the chicken genome allowed us to address an important question about the nature of gene deserts. Specifically, do gene deserts harbor functional elements directly associated with one or both flanking genes (such as transcriptional REs), or do they contain elements that function independently of neighboring genes (such as chromosome stability regions, matrix attachment regions, or noncoding RNAs)? If indeed gene deserts harbor distant regulatory sequences, this would strongly preclude the accumulation of synteny breakpoints within them, since rearrangement would be likely to separate a RE from the associated gene. To address the validity of this hypothesis, we analyzed the density of h/c and h/m syntenic breakpoints for different types of genomic intervals.
Remarkably, only two of the 172 identified human stable gene deserts are interrupted by a synteny breakpoint in h/c alignments (see Methods); four other deserts could not be reliably mapped to the chicken genome due to uncertainties associated with the current chicken sequence assembly. The remaining 166 human stable gene deserts appear to be conserved as single intact segments in chicken. The regions of contiguous ECR conservation spanned >80% of the length for 95% of these 166 stable gene deserts. Given the high frequency of syntenic rearrangements detected by human–chicken sequence alignments overall (ICGSC 2004), this finding suggests that stable gene deserts are functionally linked to at least one of the flanking genes. This observation is compatible with the hypothesis that gene deserts represent accumulations of critical gene REs that act at a distance. The elements' location, structural linearity, and integrity have been preserved throughout the evolution of vertebrates, highlighting a possibility that arrays of gene regulatory cis-elements are embedded throughout the length of stable gene deserts, resisting separation from each other and/or from the gene or genes that they regulate. This hypothesis has clearly been corroborated in certain gene deserts by recent studies of functional elements within those regions (Nobrega et al. 2003; Kimura-Yoshida et al. 2004; Uchikawa et al. 2004).
Dramatic differences in the density of synteny breakpoints were also observed between stable gene deserts, gene-rich regions, and average intergenic regions (Fig. 5). Interestingly, the density of synteny breakpoints was very high in gene-rich regions relative to the genome average for both h/m and h/c comparisons. One explanation may be that in sharp contrast to stable gene deserts, gene-rich regions have possibly evolved as hot spots of chromosomal rearrangements both before and after the primate–rodent radiation. However, these data also might suggest that the genes embedded within gene-rich segments are not as likely to be functionally linked to distant REs as are loci found in stable gene deserts.
Figure 5.
Density of synteny breakpoints per 1 Mb of sequence. Human–mouse comparisons are in orange; human–chicken in lilac.
Because variable gene deserts align poorly to the chicken genome, we could not reliably ascertain the frequency of syntenic h/c breakpoints in them. Interestingly, the frequency of h/m breakpoints is only slightly higher in variable deserts than in stable deserts (0.014 versus 0.01 per Mb); both are roughly 10-fold lower than the rates in gene-rich regions (0.16) and average in the genome (0.09).
Identification of gene deserts in the chicken and mouse genomes
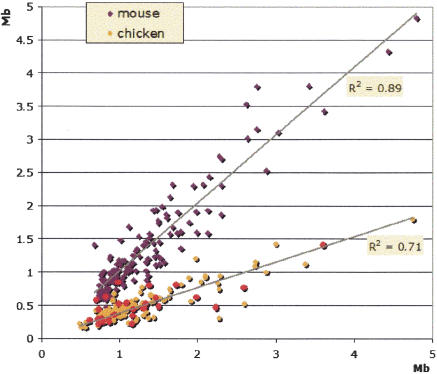
Restricting the preceding analysis of long-range synteny by searching for long linear chains of dense ECRs (see Methods) that span >80% of the length of the human stable gene deserts, we were able to carry out a direct and reliable mapping of orthologous regions in other species. Being based purely on nucleotide alignments, this method avoids uncertainties in defining gene deserts in chicken and mouse genomes that arise because gene annotation is not complete for these species. By requiring the original human and orthologous gene deserts to share boundary ECRs, we defined edge markers and consequently were able to reliably calculate the length for the corresponding gene deserts from different species. By using this approach, 149 human stable gene deserts were mapped to the mouse and chicken genomes as identified by a single contiguous sequence stretch in all three species per each such gene desert. By using this data set of h/m/c orthologous stable gene desert intervals, we compared their lengths in all three genomes (Fig. 6). No significant size differences were observed between individual human and mouse gene deserts beyond differences associated with minor mouse genome shrinkage. A strong correlation in lengths was also observed between human and chicken stable gene deserts (supported by R2 of 0.71). On average, chicken gene deserts were 0.39 times the size of their human counterparts, which is close to the average for these genomes; the chicken genome size is 0.37 of the human genome if the unplaced contigs are excluded from the consideration (ICGSC 2004). This suggests that events during mammalian evolution comparable to the inflation by repetitive elements had approximately the same rate in stable gene deserts and other genomic intervals.
Figure 6.
Length of orthologous stable gene desert counterparts in the chicken and mouse genomes compared with the human genome. Gene deserts from chicken microchromosomes are in red.
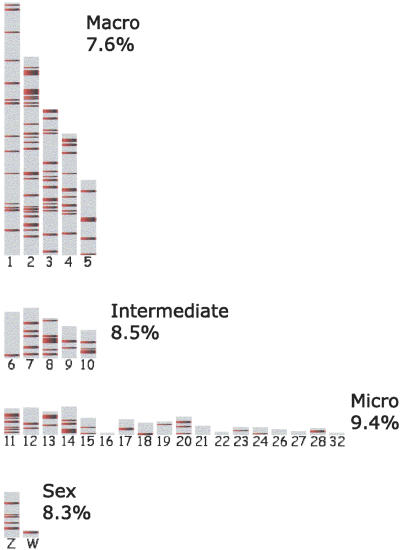
An interesting and unique feature of the chicken genome is an abundance of microchromosomes varying in size from 1.0 to 20.6 Mb. One might expect these to be depleted of long gene deserts, given the small size of the microchromosomes and also the possibility that microchromosomes may have evolved through multiple rearrangement events, while stable gene deserts tend to maintain their structural integrity and lack chromosomal breaks. In contrast to this expectation, we did not observe a decrease in the density or size of stable gene deserts on microchromosomes (Figs. 6, 7); rather the density of stable gene deserts was actually slightly higher in microchromosomes than in other chromosomal categories (Fig. 7). Thus, similarly to the pattern seen for human gene deserts (Fig. 1), the distribution of stable gene deserts in the chicken genome is largely independent of chromosome size. Also, the level of coverage of microchromosomes by stable gene deserts suggests that stable gene deserts do not have an obvious bias against appearance of synteny breaks in the surrounding regions, such as those that define the ends of these unusually small avian chromosomes.
Figure 7.
Distribution of stable gene deserts in the chicken genome (plotted as red lines). Chicken chromosomes are grouped into macro, intermediate, micro, and sex categories with the numerical characterization of average chromosome coverage by the stable gene deserts.
Discussion
Gene deserts are large intergenic regions that collectively cover 25% of the human genome. We show that they have distinct evolutionary histories and sequence signatures that set them apart from the rest of the genome. In particular, different types of repetitive elements are not uniformly represented; human gene deserts are enriched in LINE elements, while regular intergenic regions have preferably accumulated SINE elements. These data are compatible with previous studies that have shown differences in repeat content in gene-rich and gene-poor domains (Medstrand et al. 2002; Grover et al. 2003, 2004). The large differences in categories of repetitive sequences in various genomic fractions suggest a purifying selection against the accumulation of SINE elements in gene deserts and LINE elements in regular intergenic intervals and gene-rich regions. A possible explanation for this selective pressure preventing SINE accumulation in gene deserts could be attributed to the unusually CpG-rich nature of SINE elements, which makes them potential targets for genomic methylation (Yoder et al. 1997). These regions could act as methylation nucleation centers and extend this effect out onto the neighboring nontransposable regions (Hasse and Schulz 1994; Rubin et al. 1994). Alu-originated methylation, which is associated with suppression of gene transcription in imprinted regions (Greally 2002), might also function to block distant gene regulation by disrupting REs scattered throughout the gene deserts. If this is the case, evolutionary forces could work against overpopulating gene deserts that contain distant REs with SINE repetitive elements. Another possible explanation to the observed SINE depletion in gene deserts relates to the Alu-associated recombination effects capable of removing or repositioning gene regulatory domains (Medstrand et al. 2002).
Comparative sequence analysis of the human gene deserts and orthologous chicken regions effectively separates gene deserts into two categories—stable and variable. Stable gene deserts display high levels of sequence similarity in human and chicken, while the variable deserts appear to be specific to the mammalian lineage. Stable gene deserts display lower repeat density and an amount of h/m sequence conservation comparable to that of the gene-rich regions of the human genome, suggesting that considerable degrees of purifying pressure are acting over these stable gene deserts. A third of the stable gene deserts are conjoined; i.e., they cluster in pairs surrounding a small number of genes. These conjoined deserts create long loci in the genome with minimum gene density, which are much more effectively preserved throughout the evolution of vertebrates than the rest of the genome. Perhaps not surprisingly, the majority of genes that are either flanked by stable gene deserts or are neighboring these highly conserved intervals are functionally related to core biochemical processes such as regulation of transcription, skeletal and muscle development, DNA binding, and regulation of metabolism.
The density of h/f ECRs is negligibly small across variable gene deserts and is simultaneously strongly elevated in stable gene deserts, suggesting a separation in the biological function and evolutionary importance for these two categories of gene deserts. Stable gene deserts are thus prime candidates for regions with key distant gene REs in the human genome. The function of variable gene deserts is more ambiguous. They possibly represent recently evolved regions that have not yet been fixed; alternatively they may lack important function and represent genomic “junkyards.” This dichotomy potentially reconciles the apparent disparity in studies showing that while certain human gene deserts are rich in gene REs (Nobrega et al. 2003; Kimura-Yoshida et al. 2004; Uchikawa et al. 2004) some of these regions have no phenotypic impact when deleted from the mouse genome (Nobrega et al. 2004).
In support of the idea that stable gene deserts are enriched in long-range regulators, we detected a threefold higher density of computationally predicted REs in stable gene deserts than in the variable gene desert regions. The syntenic stability of stable gene deserts also suggests that distinct types of evolutionary events have shaped gene deserts and gene-rich regions. While gene-rich regions accumulate synteny breakpoints twice as fast as the average intergenic regions, stable gene deserts are depleted of synteny breakpoints. Ninety-six percent of stable gene deserts are represented as a single syntenic block in the genomes of humans, mice, and chicken despite their large size. The almost absolute preservation of chromosomal integrity of stable deserts suggests that the regulation of genes flanking them differs from that in gene-rich regions. We hypothesize that genes flanking stable gene deserts are most likely to be associated with distant gene REs that cannot be separated from coding sequences by recombination events, while the regulation of the genes within gene-rich genomic regions typically takes place through promoters and/or intronic elements. Strong enrichment of the h/c UTR conservation of genes flanking gene deserts suggests that these genes might require evolutionary preservation of both transcriptional and posttranscriptional control.
By using contiguous synteny relationships for the human genome with the genomes of mice and chicken, we were able to identify stable gene deserts in chicken and mice without requiring a reliable gene annotation for these two species. Human and mouse stable gene deserts are very similar in length, and the difference in length between specific human and chicken gene deserts agrees with the human genome expansion coefficient. The uniform expansion of individual stable gene deserts over the course of mammalian evolution implies that the function of distant REs is largely independent of the absolute distance between neighboring REs, or between the REs and the corresponding genes. However, vertebrate evolution has kept these components in a fixed relative order and at considerable distances from one another, suggesting that distant spacing of elements and their relative orders within the deserts and flanking genes is also important to function. Finally, the distribution of stable gene deserts in the chicken genome is not diminished in microchromosomes, suggesting that desert-associated chromosomal stability may disappear not far beyond the boundaries of the gene deserts and their adjacent genes. Although much remains to be explained about the function of gene deserts in general, these findings provide some potential new insights to distant regulatory activity. Our evolutionary analysis emphasizes the importance of stable gene deserts and suggests that they are likely to play a critical biological role in vertebrates.
Methods
Randomization study of the gene deserts' distribution in the human genome
If positions within known genes (exons or introns) are not counted, the human genome assembly from July 2003 consists of 51 segments that are bounded by a telomere, a centromere, or an assembly gap (unassembled region) of size >250 kb, totaling 1.75 Gb. Within those segments there are 18,134 intergenic regions— these contain a total of 286 gaps, each of under 250 kb. While some of the intergenic regions have size 0, many have considerable length; in particular, the largest of these regions measures 5.1 Mb, and 545 of the regions exceed 640 kb. In order to evaluate the likelihood that such wealth of gene deserts could occur by chance, we computed empirical P-values as follows. We derived a “null” set of intergenic distances, by randomly selecting positions (duplicates possible) from a set of 51 intervals having the sizes of the above-mentioned 51 genomic segments, avoiding positions corresponding to the 286 short gaps. Sufficiently many positions were selected as to create 18,134 interposition distances, and we then determined (1) the maximum interposition distance and (2) the number of interposition distances >640 kb. This process was repeated 1000 times, generating 1000 maximum distances and 1000 counts of distances >640 kb. The largest interposition distance in all trials was 2,033,165 (so none of the 1000 maxima exceeded 2.1 Mb), and in none of the trials were there >75 interposition distances in excess of 640Kb. Thus, empirical P-values for both observed maximum and count are <10-4.
Identification of gene-rich regions
In order to define gene-rich regions, we first identified all the gene clusters in the human genome separated by intergenic regions >100 kb. Out of the 3581 clusters fitting these criteria, 144 clusters contained ≥20 genes. The three most gene-rich regions were located on HSA19, HSA17, and HSA16, each spanning >4 Mb of sequence and comprising >140 genes. Some segments of the most gene-rich regions correspond to the expansion of zinc-finger transcription factors, Kallikreins, Keratins, and other tandemly duplicated gene families (Dehal et al. 2001; Shannon et al. 2003). However, other gene-rich intervals are densely packed with unique genes of many different functional classes. In total, these gene-rich regions covered 285 Mb of the human genome, with 15 clusters originating from the most gene-rich human chromosome HSA19.
Identification of ECRs
The analysis of syntenic relationships and conservation profiles was done through the annotation of ECRs in the alignments of genomes. We employed the BLASTZ-based genome alignments generated by the ECR Browser (http://ecrbrowser.dcode.org) (Ovcharenko et al. 2004a). A genomic interval was annotated as an ECR if it was >100 bp and >70% identity as defined by the number of nucleotide matches in a sliding window; 184k ECRs were identified in h/c alignments and 1268k ECRs in h/m alignments. Sixteen percent of h/m and 59% of h/c ECRs overlapped exons of known genes, creating a noticeable imbalance of coding over noncoding components of h/c nucleotide conservation.
Sixty-six thousand h/f ncECRs were identified as described (Ovcharenko et al. 2004a,b). A deeper filtering of known and putative transcripts, pseudogenes, mRNAs, as well as proximal promoter sequences, resulted in 2968 h/f ncECRs that lack protein-coding activity and are distantly positioned from the transcriptional start site of adjacent genes.
PhastCons conservation
We utilized the phylogenetic hidden Markov model (phastCons) conservation profile (Siepel and Haussler 2004a,b) of the human genome calculated from the human/chimp/mouse/rat/chicken multiz alignments as available from the UCSC Genome Browser database (Karolchik et al. 2003). PhastCons conservation assigns a conservation score to every base pair in the alignment that “loosely reflects % identity ratio” (description of the annotation database at http://genome.ucsc.edu). We scanned through the phastCons conservation profile of the human genome and identified all the genomic segments that consist of bases with at least 0.7 identity score that are >100 bp; there were 111,950 such regions. We mapped 15,402 of them to the human gene deserts and calculated the percentage of gene deserts' sequence enclosed in one of these regions.
Predicting REs
Three-way regulatory potential annotation computed from human–mouse–rat alignments (Blanchette et al. 2004; Kolbe et al. 2004), as available from the UCSC Genome Browser (Karolchik et al. 2003), was utilized to predict REs in the human genome. We utilized the 0.0002 threshold from a calibration study investigating sensitivity and specificity of three-way RP scores on the hemoglobin β gene cluster (http://hgdownload.cse.ucsc.edu/goldenPath/hg16/regPotential/) as a minimal value for each base pair in a cluster. Also, a cluster was required to be at least 100 bp long. Using this approach, 326,830 REs were predicted in the human genome.
Large blocks of conserved synteny
In order to create a map of genome synteny based on nucleotide-type alignments, we scanned the data set of all triplets of ECRs consecutively present in both species (two neighboring ECRs were selected as consecutively located only if they were separated by <100kb in both genomes). These ECR triplets defined anchors of genome similarity and were used to construct long syntenic blocks by clustering ECR triplets together using the 100-kb threshold again. A filtering out of regions that cover <50 kb in one of the species created a data set of long regions of conserved synteny with an exclusion of minor breakpoints that can be associated with evolutionary micro-rearrangements, such as retrotransposition or sequence reshuffling guided by transposable elements. Subsequent joining of these long regions of synteny created longer regions of synteny if the separation of a pair of syntenic segments was <1 Mb in both genomes.
Using this approach, large-scale similarity of the human and mouse genomes was modeled with ∼300 and 500 synteny breakpoints between human and mouse and between human and chicken, respectively. (Chicken chromosomes Un, random, and several others representing unassembled chicken sequence were excluded from consideration.) Due to the longer evolutionary separation of birds from humans than of rodents from humans, we observed different levels of genome coverage by syntenic blocks in human–mouse and human–chicken comparisons. H/m large syntenic blocks covered ∼96% of mammalian genomes. H/c large syntenic blocks covered 90% of the chicken genome and 78% of the human genome.
Acknowledgments
We would like to thank the anonymous reviewers for their valuable comments, Francesca Chiaromonte for suggestions about the randomization study, and John Karro and Shan Yang for determining rates of neutral evolution. W.M. and R.H. were supported by NHGRI grant HG02238 and NIDDK grant DK065806; G.G.L. was supported by LLNL LDRD-04-ERD-052 grant; and I.O. was in part supported by DOE SCW0345 grant. The work was performed under the auspices of the United States Department of Energy by the University of California, Lawrence Livermore National Laboratory Contract No. W-7405-Eng-48.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3015505. Article published online before print in December 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Bell, A.C., West, A.G., and Felsenfeld, G. 2001. Insulators and boundaries: Versatile regulatory elements in the eukaryotic. Science 291: 447-450. [DOI] [PubMed] [Google Scholar]
- Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F., Roskin, K.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14: 708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, D., Chakalova, L., Osborne, C.S., Dai, Y.F., and Fraser, P. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32: 623-626. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. [DOI] [PubMed] [Google Scholar]
- Dorsett, D. 1999. Distant liaisons: Long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9: 505-514. [DOI] [PubMed] [Google Scholar]
- Gibbs, R.A. Weinstock, G.M., Metzker, M.L., Muzny, D.M., Sodergren, E.J., Scherer, S., Scott, G., Steffen, D., Worley, K.C., Burch, P.E., et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Greally, J.M. 2002. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. 99: 327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover, D., Majumder, P.P., Rao, C.B., Brahmachari, S.K., and Mukerji, M. 2003. Nonrandom distribution of Alu elements in genes of various functional categories: Insight from analysis of human chromosomes 21 and 22. Mol. Biol. Evol. 20: 1420-1424. [DOI] [PubMed] [Google Scholar]
- Grover, D., Mukerji, M., Bhatnagar, P., Kannan, K., and Brahmachari, S.K. 2004. Alu repeat analysis in the complete human genome: Trends and variations with respect to genomic composition. Bioinformatics 20: 813-817. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Connor, M., Kolbe, D., et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13: 13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasse, A. and Schulz, W.A. 1994. Enhancement of reporter gene de novo methylation by DNA fragments from the α-fetoprotein control region. J. Biol. Chem. 269: 1821-1826. [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (ICGSC). 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature (in press). [DOI] [PubMed]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Yoshida, C., Kitajima, K., Oda-Ishii, I., Tian, E., Suzuki, M., Yamamoto, M., Suzuki, T., Kobayashi, M., Aizawa, S., and Matsuo, I. 2004. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development 131: 57-71. [DOI] [PubMed] [Google Scholar]
- Kolbe, D., Taylor, J., Elnitski, L., Eswara, P., Li, J., Miller, W., Hardison, R., and Chiaromonte, F. 2004. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 14: 700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Medstrand, P., van de Lagemaat, L.N., and Mager, D.L. 2002. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 12: 1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, C.E., Hersh, B.M., and Carroll, S.B. 2004. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol. 5: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega, M.A., Zhu, Y., Plajzer-Frick, I., Afzal, V., and Rubin, E.M. 2004. Megabase deletions of gene deserts result in viable mice. Nature 431: 988-993. [DOI] [PubMed] [Google Scholar]
- Nobrega, M.A., Ovcharenko, I., Afzal, V., and Rubin, E.M. 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Ovcharenko, I., Nobrega, M.A., Loots, G.C., and Stubbs, L. 2004a. ECR Browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32: W280-W286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko, I., Stubbs, L., and Loots, G.G. 2004b. Interpreting mammalian evolution using Fugu genome comparisons. Genomics 84: 890-895. [DOI] [PubMed] [Google Scholar]
- Pesole, G., Liuni, S., Grillo, G., Licciulli, F., Mignone, F., Gissi, C., and Saccone, C. 2002. UTRdb and UTRsite: Specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs: Update 2002. Nucleic Acids Res. 30: 335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchik, E.M., Carpenter, D.A., and Selby, P.B. 1990. A strategy for fine-structure functional analysis of a 6- to 11-centimorgan region of mouse chromosome 7 by high-efficiency mutagenesis. Proc. Natl. Acad. Sci. 87: 896-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, C.M., VandeVoort, C.A., Teplitz, R.L., and Schmid, C.W. 1994. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. 22: 5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, L.B., Montgomery, C.S., and Raymer, G.D. 1982. Analysis of the albino-locus region of the mouse, IV: Characterization of 34 deficiencies. Genetics 100: 427-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, M., Hamilton, A.T., Gordon, L., Branscomb, E., and Stubbs, L. 2003. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 13: 1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A. and Haussler, D. 2004a. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 11: 413-428. [DOI] [PubMed] [Google Scholar]
- ———. 2004b. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol. Biol. Evol. 21: 468-488. [DOI] [PubMed] [Google Scholar]
- Uchikawa, M., Takemoto, T., Kamachi, Y., and Kondoh, H. 2004. Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech. Dev. 121: 1145-1158. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Yang, S., Smit, A.F., Schwartz, S., Chiaromonte, F., Roskin, K.M., Haussler, D., Miller, W., and Hardison, R.C. 2004. Patterns of insertions and their covariation with substitutions in the rat, mouse, and human genomes. Genome Res. 14: 517-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, J.A., Walsh, C.P., and Bestor, T.H. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13: 335-340. [DOI] [PubMed] [Google Scholar]
Web site references
- http://ecrbrowser.dcode.org; ECR Browser.
- http://genome.ucsc.edu; UCSC Genome database.
- http://hgdownload.cse.ucsc.edu/goldenPath/hg16/regPotential/; data used to generate the regulatory potential tracks.