Abstract
In the animal kingdom, genomic imprinting appears to be restricted to mammals. It remains an open question how structural features for imprinting evolved in mammalian genomes. The clustering of genes around imprinting control centers (ICs) is regarded as a hallmark for the coordinated imprinted regulation. Hence imprinted clusters might be structurally distinct between mammals and nonimprinted vertebrates. To address this question we compared the organization of the Beckwith Wiedemann syndrome (BWS) gene cluster in mammals, chicken, Fugu (pufferfish), and zebrafish. Our analysis shows that gene synteny is apparently well conserved between mammals and birds, and is detectable but less pronounced in fish. Hence, clustering apparently evolved during vertebrate radiation and involved two major duplication events that took place before the separation of the fish and mammalian lineages. A cross-species analysis of imprinting center regions showed that some structural features can already be recognized in nonimprinted amniotes in one of the imprinting centers (IC2). In contrast, the imprinting center IC1 is absent in chicken. This suggests a progressive and stepwise evolution of imprinting control elements. In line with that, imprinting centers in mammals apparently exhibit a high degree of structural and sequence variation despite conserved epigenetic marking.
Genomic imprinting describes mono-allelic gene expression in diploid organisms depending on the parental origin of the allele. Thus far, imprinting effects on gene expression have been observed mainly in mammalian species and in flowering plants (Grossniklaus et al. 2001; Reik and Walter 2001; http://cancer.otago.ac.nz/IGC/Web/home.html, http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html). In analyzed organisms only a small number of genes appears to be affected, whereas the majority is biallelically expressed. As imprinting effects are absent or marginal in other clades, it has been assumed that imprinting effects in mammals and plants may have evolved independently from each other. In the animal kingdom, genomic imprinting might be a mode of gene regulation specific for mammalian species and has been suggested to be associated with a specific linkage/clustering of these genes. Hence, the mammalian genome should either show a special arrangement of imprinted genes or carry special DNA elements (such as imprinting control elements, ICs) that are responsible for the regulation of imprinting and are presumably absent in other nonimprinted species. Comparing mammalian imprinted genes to their homologs in nonmammalian species might be helpful for the identification of such elements.
The Beckwith-Wiedemann syndrome (BWS) region currently represents the best investigated imprinting domain in the human and mouse genomes (Engemann et al. 2000; Ishihara et al. 2000; Onyango et al. 2000; Paulsen et al. 2000). In both species the region encompasses at least 10 imprinted genes. In human, the BWS region resides on chromosome 11p15.5 close to the telomere, whereas the orthologous region is at the very end of distal chromosome 7 in mouse. In human, the core region of the imprinting domain encompasses ∼800 kb including H19 at its telomeric end and PHLDA2 at the centromeric end (Onyango et al. 2000). In the mouse, size and gene organization are very similar; however, relative to the adjacent telomere the orientation of the domain is reversed in comparison to the human (Paulsen et al. 1998).
In the human and mouse BWS imprinting regions, two major elements for regulation of imprinted gene expression have been identified—the imprinting centers IC1 and IC2. IC1 is located upstream of H19 and has been shown to regulate reciprocal imprinting of the maternally expressed H19 and the paternally expressed Igf2, and Ins2 genes in mouse (Leighton et al. 1995; Forne et al. 1997; Olek and Walter 1997; Thorvaldsen et al. 1998; Khosla et al. 1999). IC2 is located in an intron of the maternally expressed Kcnq1 gene. Artificially introduced mutations in the mouse suggested that IC2 not only regulates imprinted gene expression of Kcnq1 but also affects in cis expression of neighboring genes such as Cdkn1c, Slc22a1l, Phlda2, Tssc4, and Ascl2 (Mitsuya et al. 1999; Smilinich et al. 1999; Horike et al. 2000; Cleary et al. 2001; John et al. 2001; Fitzpatrick et al. 2002). IC2 appears to be the promoter of the paternally expressed probably noncoding transcript Kcnq1ot1 (Lit1) that is oriented oppositely to Kcnq1 and overlaps with this gene. Similar to the Igf2r antisense transcript Air (Sleutels et al. 2002), the Kcnq1ot1 (Lit1) transcript may be involved in the regulation of imprinted gene expression.
In human and mouse, the BWS region includes a few genes that are biallelically expressed or exhibit incomplete or tissue-specific imprinting (Caspary et al. 1998; Lee et al. 1999; Enklaar et al. 2000; Horike et al. 2000; Paulsen et al. 2000; Prawitt et al. 2000). Among these are the human and mouse Trpm5, Tssc4, Cd81, and Phemx genes. In addition, the human ASCL2 and the murine Th genes appear to be biallelically expressed (Zhou et al. 1995; Miyamoto et al. 2002).
In our comparative analyses we included additional genes at the flanks of the BWS region between MUC2 and H19, and between PHLDA2 and MRGG. Among these were also the murine Tnfrsf22, Tnfrsf23, and Tnfrsf26 genes that do not possess human orthologs (Clark et al. 2002; Schneider et al. 2003).
We compared the gene organization within and around the BWS region to the homologous genes in chicken (Gallus gallus), pufferfish (Fugu rubripes), and zebrafish (Danio rerio). These nonmammalian species were chosen because they are the closest related nonmammalian species whose genomes are almost entirely sequenced (Aparicio et al. 2002; http://www.ensembl.org/Fugu_rubripes/, http://www.sanger.ac.uk/Projects/D_rerio/, http://www.ensembl.org/Gallus_gallus/). Whereas mammals and birds belong both to the amniotes, Fugu and zebrafish represent a most distantly related vertebrate clade. Although the genomic sequences of all three species are not completely assembled, the present status of sequence contigs allows the recognition of chromosomal linkage patterns and genomic arrangements (Smith et al. 2002).
Results
Identification of homologous genes of the mammalian BWS region in chicken
For the investigation of the BWS gene region in chicken, we chose genes within the BWS region and also adjacent genes on human chromosome 11p15.5 and mouse distal chromosome 7. Imprinted genes were taken from the literature (Engemann et al. 2000; Onyango et al. 2000; Paulsen et al. 2000), and additional flanking genes were identified by transcript annotations of the chosen region using the Ensembl database (http://www.ensembl.org). In total, 28 (human) and 30 (mouse) genes were selected for comparative searches in the GenBank database (http://www.ncbi.nlm.nih.gov) or in the Ensembl database of assembled genomic shotgun sequences (http://www.ensembl.org). The selected segment started with MUC2 in the region flanking H19, and terminated at the opposite flank with MRGE where conservation of gene synteny in mammals ends. In human and mouse, the investigated genomic region is ∼2.1 Mb long. For identification of homologs in chicken, the peptide sequences encoded by the human genes and mouse genes were used for searches against the translated genomic chicken sequences. For almost all genes between Tnnt3 and Osbpl5, we found orthologs residing on two BAC contigs (contig 1: GenBank accession nos. BX640540, BX640401; contig 2: GenBank accession nos. BX649221, BX649222, BX640404, AP003796, AP003795, BX663531) (Fig. 1). In the Ensembl database of assembled shotgun sequences (http://www.ensembl.org), these contigs neighbor each other on chicken chromosome 5. In their neighborhood we localized orthologs for all protein-encoding genes between MUC2 and OSBPL5. We were not able to identify orthologs of MRGG and MRGE in the GenBank database or in the Ensembl database. Interestingly, the mouse Tnfrsf22, Tnfrsf23, and Tnfrsf26 genes do not possess orthologs in the human genome, but we found one ortholog in the chicken BWS region, which we named Tnfrsf22. Finally, we were not able to identify a potential homolog of the noncoding H19 gene in chicken by BLAST searches using the human and mouse H19 cDNA sequences as query sequences. In total, the region spanned by the orthologous chicken genes is ∼2 Mb. The size of the region and the order of genes appear to be almost identical to the mammalian BWS region (Fig. 1), including orthologs of 27 of 30 annotated mammalian genes of this region.
Figure 1.
Schematic map of orthologous genes in different species. Shown are maps of the human, chicken, zebrafish, and Fugu gene syntenies across the BWS region. The map is not to scale. Black bars indicate regions that are spanned by assembled genomic shotgun sequences and by BAC clones. For the remaining regions only shotgun assembled sequences were available. Interruptions of the horizontal lines indicate long distances between the genes. The chicken BWS region is present on two BAC contigs (contig 1: GenBank accession nos. BX640540, BX640401, contig 2: GenBank accession nos. BX649221, BX649222, BX640404, AP003796, AP003795, BX663531). The sequence contig in zebrafish is derived from five BAC sequences (GenBank accession nos. AL928843, AL929208, AL928880, BX001047, AL928628). The Fugu Igf2, Th, and Nap1l4 genes were also found in a cosmid sequence (GenBank accession no. AL021880).
Identification of homologous genes of the mammalian BWS region in zebrafish and Fugu
Similar to our strategy for chicken genes, we identified and mapped the BWS orthologs in zebrafish and Fugu sequences. Mapping was performed for the four best hits of each gene, thereby also identifying potential mapping positions of paralogs (see below).
Orthologous zebrafish genes were found for all BWS genes except for PHEMX, TSSC4, MRGG, and MRGE. Most BWS orthologs of KCNQ1, TRPM5, CDKN1C, and IGF2 map to five overlapping genomic zebrafish BAC sequences and an assembled whole-genome shotgun sequences contig (GenBank accession nos. AL928843, AL929208, AL928880, BX001047, AL928628) (Fig. 1, Supplemental Table 1). According to the current annotation, 11 genes are organized in five small linkage groups in maximal distance of 21 Mb to each other on zebrafish chromosome 7 (Fig. 1).
For Fugu, only assembled shotgun sequences (scaffolds) with no chromosomal assignment were available. Homologous genes in Fugu were identified by BLAST searches on all Fugu shotgun sequences of the Ensembl database. We identified homologs for all genes except for PHEMX, TSSC4, MRGG, and MRGE (Supplemental Table 2) and compiled their arrangement on genomic sequence scaffolds. Scaffold 9, comprising 615 Kb, contains orthologs of nine BWS cluster genes. Seven of them, the Fugu homologs of HCCA2, DUSP8, OSBPL5, PHLDA2, TH, IGF2, and MRPL23, match with their best similarity hit to sequence scaffold 9. IGF2, TH, and NAP1L4 were also found in a cosmid sequence (GenBank accession no. AL021880). However, genes of the central portion of the BWS cluster, such as CDKN1C, KCNQ1, TRPM5, CD81, and ASCL2 could not be assigned to this scaffold and are scattered on other sequence scaffolds. In summary, our analysis in zebrafish and Fugu suggests that the organization of the BWS cluster and flanking genes is partially recognizable in fish.
Identification of paralogous genes in the human genome
Besides the BWS orthologous genes in Fugu and zebrafish we identified a number of paralogs in other chromosomal regions (Supplemental Tables 1, 2). In addition, paralogs have previously been described for a few imprinted BWS genes in human and mouse (Patton et al. 1998; Walter and Paulsen 2003). We next investigated whether paralogs of BWS genes were again linked in certain chromosomal regions indicating that clustering occurred before duplication.
In total we identified 21 human BWS gene paralogs by BLAST searches against the NCBI database of nonredundant protein sequences using the peptide sequences encoded by the human genes. Paralogs with scores lower than the best invertebrate hit were excluded. Most paralogs were located in small linkage groups on chromosomes 1, 11p15.1, 12, and 19 (Fig. 2, Supplemental Table 3). The most interesting concentration of paralogs was observed on chromosome 12. This chromosome harbors 12 paralogs. The gene order along the chromosome IGF1 - PAH - ASCL1 and PHDLA1 - NAP1L1 - OSBPL8 resembles the condensed organization of the BWS cluster on human chromosome 11 (IGF2 - TH - ASCL2 and PHLDA2 - NAP1L4 - OSBPL5). An additional paralog cluster on human chromosome 19 consists of three closely linked genes, TNNT1, TNNI3 and KIAA1811 which are paralogs of the BWS flanking genes TNNT3, TNNI2, STK29. Surprisingly this paralog cluster resides in only ∼1.7 Mb distance to the imprinted PEG3 gene. A group of paralogs (MRGX1–4, LOC340990) is located on human chromosome 11p15.1. This includes TPH, which was identified as a paralog of TH by Patton et al. (1998).
Figure 2.
Organization of BWS genes and their paralogs in human and Fugu. Gene organization is shown as schematic maps of the human chromosomes and Fugu scaffolds (not to scale; for precise positions on the human chromosomes see Supplemental Table 3). Interruptions of the vertical gray lines representing human chromosomes indicate distances longer than 4 Mb between genes. Human chromosomes (Hs) are labeled by their numbers, as are Fugu sequence scaffolds (Fr). Initially selected genes on human chromosome 11p15.5 (see Fig. 1) are boxed, and their paralogs are shown in black. Additional genes are labeled in gray.
A similar clustering of paralogs was observed in Fugu. We identified four sequence scaffolds whose paralogous genes showed homologous arrangement to the human chromosomes 12, 19, and 1, respectively (Fig. 2). Fugu scaffold 253 contained orthologs of genes on human chromosomes 12 and 19, indicating that these might have been linked in early vertebrates. In conclusion, the conserved linkage of some paralogs in fish and human suggests that the duplications of genes within the BWS imprinting cluster predates the radiation of fish and other vertebrates.
Sequence conservation around the BWS IC2 in vertebrates
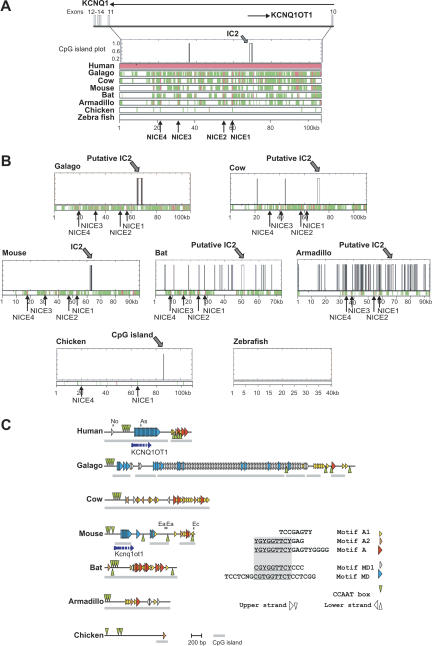
The similar gene organization of the BWS region in mammals and chicken suggests that the mammalian gene synteny was already fixed before radiation of the mammalian and avian lineages, that is, before imprinting was established. In mammals, a key element to control imprinted expression in the cluster is the imprinting center IC2 within the Kcnq1 gene which largely overlaps with a CpG island (Smilinich et al. 1999; Engemann et al. 2000). IC2 is located in intron 10 of the KCNQ1/Kcnq1 gene in human and mouse. We therefore examined how well sequences or structural features within or around the IC2 are conserved in imprinted mammalian species and nonimprinted organisms such as chicken. We compared the corresponding genomic sequences of human, galago, cow, mouse, bat, armadillo, chicken, and zebrafish (http://www.nisc.nih.gov/projects/zooseq/comp_seq_org.cgi, http://www.ensembl.org/Gallus_gallus/, http://www.sanger.ac.uk/Projects/D_rerio). Fugu could not be analyzed because of incomplete sequences. In all analyzed mammalian species and chicken, the Kcnq1 gene structure is nicely conserved. The same holds for zebrafish, with the exception of exons 1b, 1c, 2a, and 9. The size of the region between exon 10 and 11 is highly similar among all of the species, ranging from 70 kb in bat to 110 kb in chicken and cow. In zebrafish intron 10 is considerably shorter, encompassing 40 kb.
The number of CpG islands within intron 10 varies significantly. However, in armadillo, cow, galago, and bat we could identify CpG islands at positions homologous to the experimentally identified IC2 CpG islands in human and mouse. Surprisingly, chicken also contains a single short CpG island at an IC2 equivalent position (Fig. 3), whereas CpG islands are entirely absent in zebrafish. The size of the IC2-like CpG islands ranges from 202 and 2360 bp, and their sequence similarity is not very pronounced (Fig. 3, see also Engemann et al. 2000). However, sequences upstream and downstream of IC2 are highly conserved in pairwise alignments between mammals, whereas the overall similarity to chicken and zebrafish is rather low or absent. In a multiple alignment of all mammalian species, four highly conserved elements, NICE1–4 (= neighboring IC elements) ranging from 141 to 500 bp were detected with a sequence identity of >70% (Fig. 3, Supplemental Table 4). BLAST searches against the genomic sequences of mouse and human revealed that the NICE sequences are unique in both genomes (data not shown). NICE1 was found to be contained in two bovine EST sequences (GenBank accession nos. AV592964, CN440620) in which it is spliced to exon 11 of Kcnq1. This suggests that NICE1 may be a part of an alternative transcript of the bovine Kcnq1 gene. However, thus far no other matching ESTs from any other amniote can be found in EST databases. Two of the NICE elements (NICE1 and NICE4) are even well conserved in chicken, suggesting that they may represent ancient regulatory elements or rudiments of an ancient transcript in this region.
Figure 3.
Sequence conservation in Kcnq1 intron 10 in vertebrates. (A) Multiple alignments of Kcnq1 intron 10: the genomic human sequence was taken as reference sequence and compared to the genomic galago, cow, mouse, bat, armadillo, chicken, and zebrafish sequences. Before alignment, repetitive elements were masked using RepeatMasker software. Aligned regions are shown in green, highly conserved elements in red (>70% identity, >100 bp length). NICE1–NICE4 are highly conserved in all analyzed mammals. NICE1 and NICE4 are conserved in chicken (>60% identity, >100 bp length). The position of the IC2 CpG island in the human sequence is indicated by the CpG island plot above the multiple alignment. The CpG island plot shows CpG islands that fulfil the definition of a CpG island (length >200 bp, G+C content >50%, CpGobserved/CpGexpected >0.6, http://www.ebi.ac.uk/emboss/cpgplot/). The given scale bar is related to the human sequence. (B) The distributions of CpG islands in Kcnq1 in different vertebrate species. In pairwise alignments, the vertebrate sequences were used as reference sequences and the human sequence as second sequence. Scale bars are related to the reference sequence in each alignment. (C) Arrangements of repeated conserved sequence motifs in the putative IC2 in mammals. The consensus sequences of conserved motifs are listed. Segments that are conserved in overlapping motifs are underlined. The arrangements of these motifs in the different species are shown by different triangles. Motif MD was identified by Mancini-DiNardo et al. (2003). For the identified motifs the following numbers of mismatches to the consensus sequence were allowed: motif A, three mismatches; motifs MD1 and A2, two mismatches; motif A1, one mismatch; motif MD, six mismatches; CCAAT boxes, no mismatches. In some species the analyses were extended to regions flanking the CpG islands that are highlighted by gray bars. For the mouse and human sequences, the transcriptional start sites of Kcnq1ot1 (Du et al. 2004) are depicted by broken arrows indicating that the 3′extension of the transcript is not known. In mouse and human, location of restriction sites (No, NotI; As, AscI; Ea, EagI; Ec, EcoRI) that have been used for characterization of the IC2 in other studies (Du et al. 2003, 2004; Mancini-diNardo et al. 2003) are indicated.
In contrast to the similarities of the IC2 region between mammals and chicken, we could not detect the H19 gene in chicken or fish or any significant homologies to the IC1 5′ of the H19 gene, whereas the Igf2 gene and the Mrpl23 gene flanking the H19-IC1 region can easily be identified. The region between Igf2 and Mrpl23 in chicken does not contain any CpG-rich region (Supplemental Fig. 1). In contrast, we found several CpG islands associated with the chicken Igf2 gene: two small CpG islands are located in the last intron and last exon of the gene. These positions correspond to the differentially methylated region (DMR2) in the human and mouse Igf2 genes.
In summary, our analysis shows that some structural features of the imprinting control center 2 (IC2) such as the presence of a CpG island and conservation of flanking sequences (NICE elements) can already be recognized in chicken. In contrast, the second imprinting center (IC1) is apparently absent in chicken.
Identification of conserved repeated motifs in the mammalian IC2 CpG islands
Despite the striking conservation in sequences flanking the IC2, the IC2 CpG islands of mouse and human do not exhibit pronounced sequence conservation (Mancini-DiNardo et al. 2003). This is surprising, because deletion experiments in both organisms suggest a functional equivalence of the dissimilar CpG islands (Horike et al. 2000; Fitzpatrick et al. 2002). The only conserved feature described for both CpG islands concerns the appearance of repeated motifs (Mancini-DiNardo et al. 2003). In a comparison of all available mammalian IC2 sequences, we carefully searched for the conservation of such repeated structures. Using a combination of different software tools (see Methods) we identified a number of conserved repeated sequence motifs within the IC2 of mammals (Fig. 3C). Our analysis shows that the originally described repeated sequence motif (Mancini-DiNardo et al. 2003), which we called motif MD, can be detected in human, galago, and mouse, but not in cow, bat, or armadillo. Nevertheless, we identified three repeated motifs which were similar to MD. All of these motifs share a central core motif, 5′YGYGGTTCY3′, but differ in the sequences 5′ or 3′ of it (Fig. 3C). However, the number, sequence variation, and structural arrangement of all of the motifs vary significantly among the mammalian species. The most impressive arrangement is a 98-times repetition of motif MD1 in galago. The only conserved structural feature of all putative IC2s appears to be the concentration of A1, A2, and A motifs at its 3′ end. To test the IC2-specific enrichment of such motifs, we examined the occurrence of motifs A, A1, and A2 in randomly selected control CpG islands. We found all motifs to be significantly more frequent in the putative IC2 CpG islands than in randomly selected control CpG islands (P < 0.05, t-test). Hence the presence of these specific repeats appears to be a hallmark of the IC2 region. Among the conserved motifs, we also found one copy of motif A2 in the chicken CpG island.
In addition, we searched for consensus sequences of CTCF and YY1 binding sites (Bell and Felsenfeld 2000; Du et al. 2003; Kim et al. 2003) and other specific motifs of unknown function which were discussed as potential signatures of imprinting centers in the literature (Wang et al. 2004). Based on consensus sequences, none of these motifs was found to be conserved in the putative mammalian IC2 CpG islands.
In addition to highly repeated motifs, some repeated CCAAT boxes have been described for human and mouse. They are located 5′ of the repeated motifs MD, and appear to initiate the transcription of Kcnq1ot1 (Lit1) (Du et al. 2004). These CCAAT boxes are apparently well conserved in all mammalian species at similar positions either within the CpG island or close to it (Fig. 3C). Interestingly, the chicken CpG island is also flanked by a pair of CCAAT boxes.
Discussion
Clustering and duplication
In this paper we show that the chromosomal arrangement of genes in the mammalian BWS region is well conserved in chicken and can even be partially recognized in fish. The arrangement of genes in the BWS region was apparently fixed before divergence of birds and mammals, hence, predating the fixation of imprinting mechanisms.
In addition to the phylogenetic conservation of the BWS gene clusters in vertebrates, clustering of their nearest paralogs is also apparently conserved. This indicates that at least two major duplication events happened in the course of the evolution of the BWS region before divergence of the mammalian and fish lineages.
Evolution of imprinting centers and DMRs
It remains unclear whether important imprinting elements such as the CpG island containing imprinting centers were already present in such ancestral vertebrate clusters. We have not yet detected CpG islands at the corresponding positions in fishes, but we identified a CpG island at a position corresponding to the mammalian IC2 CpG islands in chicken. This chicken CpG island contains one copy of motif A2 and a pair of CCAAT boxes, both features of the mammalian IC2 CpG islands. It remains to be determined whether this CpG island has promoter activity and is linked to a Kcnq1ot1 (Lit1)- like transcript in chicken. It will be also of interest to test whether the IC2-like CpG island in chicken confers allele-specific expression of the neighboring Cdkn1c or Kcnq1 genes. As in mammals, both genes possess pronounced CpG islands at their 5′ end (data not shown).
In addition, we found another CpG island in chicken at a position corresponding to the differentially methylated region 2 (DMR2) of the mammalian Igf2 gene. In mammals, the DMR2 CpG island overlaps with the last two exons of the gene and has been shown to be involved in expression control by mediating interactions with the IC1 imprinting center (Lopes et al. 2003). As a corresponding IC1 is missing in chicken, the importance of this DMR2-like region for expression control in chicken is questionable. In addition, the apparent absence of IC1 in chicken is consistent with the absence of parentally imprinted expression of the Igf2 gene in this organism (Koski et al. 2000; O'Neill et al. 2000; Nolan et al. 2001; Yokomine et al. 2001).
The mammalian IC2
Pronounced CpG islands that are likely to represent the imprinting center IC2 are present in all of the mammalian species we analyzed, ranging from armadillo to human. However, the DNA sequences of these CpG islands are only weakly conserved, suggesting that functional conservation does not depend on strong sequence conservation. The only common feature of all IC2 CpG islands is the presence of several distinct short conserved motifs with an overlapping consensus sequence. Repeated motifs (named MD here) within IC2 were described by Mancini-DiNardo et al. (2003) and were shown to be part of a silencer element in the murine and human IC2 (Du et al. 2003; Mancini-DiNardo et al. 2003; Thakur et al. 2003). Based on these findings one might assume that their structure is conserved in mammals. We did not find evidence for the presence of the complete motif MD in all of the mammalian species studied. However, the MD motif contains a core sequence, 5′YGYGGTTCY3′, which we found repeated in other motifs (A, A1, and A2) that are present in all of the analyzed mammalian species, indicating that this segment might be relevant for the assumed silencer function.
In summary, our data support a progressive evolution of the BWS region, beginning with the fixation of gene order, subsequent formation of the IC2 CpG island, variation and amplification of repeat motifs, and late, mammalian-specific appearance of H19 and the neighboring IC1.
Methods
cDNA and peptide sequences
Genes were selected from the genomic DNA segment between MUC2 and MRGE irrespective of whether they were imprinted or not. The GenBank accession nos. of selected cDNA sequences are given in Supplemental Table 1. Peptide sequences were taken as annotated in the GenBank data files.
Identification of homologous genes in different species
Peptide sequences of selected genes were taken for BLASTP searches on annotated peptide sequences or translated genomic sequences in GenBank (http://www.ncbi.nlm.nih.gov) and Ensembl databases (http://www.ensembl.org; Fugu database: Release March 3, 2003, version 18.2.1; zebrafish database: release 3, November 27, 2003; chicken database: version 22.1.1, release May 26, 2004). Similar BLASTP searches were performed on translated annotated genes or genomic sequences in GenBank (sections: nonredundant sequences, and high through-put genomic sequences). Only peptide sequences longer than 100 amino acids were taken as query sequence. Matching sequences with probability values lower than e-7 were selected if the sequence alignment encompassed at least one-third of the query sequence (McLysaght et al. 2002). Matching sequences that showed higher probability values than the best match to nonvertebrate sequences were excluded from further analysis. In order to simplify further analysis, the number of selected homologs per gene was limited to the four sequences with highest similarities to the query sequences. The genomic positions of identified homologous genes were estimated by BLAST searches using the cDNA sequence as query sequence against the genomic DNA sequences in the Ensembl database.
Identification of Kcnq1 exons and CpG islands
For the human genomic Kcnq1 DNA sequence, exon positions were taken from the annotation of a genomic sequence (GenBank accession no. AJ006345, Neyroud et al. 1999). Kcnq1 exons in other species were identified by BLAST comparison (http://www.ncbi.nlm.nih.gov/BLAST/) of the human KCNQ1 protein sequence to the translated genomic DNA sequence of the species of interest. Genomic sequences with the following GenBank accession nos. were used: AJ271885 (mouse), AC147396 (cow), AC146964 (bat), AC147392.2 (galago), AC148124.2 (galago), AC147402 (armadillo), AL928843 (zebrafish). The sequence of intron 10 in chicken was downloaded from the Ensembl database. CpG islands were identified using the CpG plot software provided by the European Bioinformatics Institute (http://www.ebi.ac.uk/emboss/cpgplot/) using default parameters. Pairwise alignment to the human master sequence the positions of interspersed repeats were estimated using RepeatMasker (A.F.A. Smit and P. Green, unpub., http://repeatmasker.org). Pairwise alignments of genomic DNA sequences were generated by the PipMaker software (Schwartz et al. 2000, http://bio.cse.psu.edu/pipmaker).
Identification of conserved sequence motifs
Analyzed genomic sequences that contain IC2-like CpG islands in different species are listed in Supplemental Table 5. Because the galago sequence contains a large array of tandem repeats that might falsify motif searches, this sequence was excluded from the primary analyses using the freely accessible MEME software (Bailey and Elkan 1994, http://bioweb.pasteur.fr/seqanal/motif/meme/). The analyses were variegated allowing different motif lengths (15, 20, 30, and 50 bp). Motifs that appeared in more than three of the six species were chosen for further analyses. The frequencies of these motifs in the selected CpG islands including the galago and chicken CpG islands were determined using fuzznuc software (http://bioweb.pasteur.fr/seqanal/interfaces/fuzznuc.html). Searches were performed on the upper and lower DNA strands, allowing variegating numbers of mismatches. The frequencies of the motifs were compared to their frequencies in randomly selected CpG islands in mouse and human. These control groups consisted of 10 CpG islands of either human or murine origin (Supplemental Table 6). The significance of different motif densities in the putative IC2 CpG islands and in control groups was tested with t-tests.
Acknowledgments
This study was supported by Deutsche Forschungsgemeinschaft grant #WA1029/3-1.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2689805. Article published online before print in December 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A. et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28-36. [PubMed] [Google Scholar]
- Bell, A.C. and Felsenfeld, G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482-485. [DOI] [PubMed] [Google Scholar]
- Caspary, T., Cleary, M.A., Baker, C.C., Guan, X.J., and Tilghman, S.M. 1998. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol. Cell. Biol. 18: 3466-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, L., Wei, M., Cattoretti, G., Mendelsohn, C., and Tycko, B. 2002. The Tnfrh1 (Tnfrsf23) gene is weakly imprinted in several organs and expressed at the trophoblast-decidua interface. BMC Genet. 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, M.A., van Raamsdonk, C.D., Levorse, J., Zheng, B., Bradley, A., and Tilghman, S.M. 2001. Disruption of an imprinted gene cluster by a targeted chromosomal translocation in mice. Nat. Genet. 29: 78-82. [DOI] [PubMed] [Google Scholar]
- Du, M., Beatty, L.G., Zhou, W., Lew, J., Schoenherr, C., Weksberg, R., and Sadowski, P.D. 2003. Insulator and silencer sequences in the imprinted region of human chromosome 11p15.5. Hum. Mol. Genet. 12: 1927-1939. [DOI] [PubMed] [Google Scholar]
- Du, M., Zhou, W., Beatty, L.G., Weksberg, R., and Sadowski, P.D. 2004. The KCNQ1OT1 promoter, a key regulator of genomic imprinting in human chromosome 11p15.5. Genomics 84: 288-300. [DOI] [PubMed] [Google Scholar]
- Engemann, S., Strodicke, M., Paulsen, M., Franck, O., Reinhardt, R., Lane, N., Reik, W., and Walter, J. 2000. Sequence and functional comparison in the Beckwith-Wiedemann region: Implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet. 9: 2691-2706. [DOI] [PubMed] [Google Scholar]
- Enklaar, T., Esswein, M., Oswald, M., Hilbert, K., Winterpacht, A., Higgins, M., Zabel, B., and Prawitt, D. 2000. Mtr1, a novel biallelically expressed gene in the center of the mouse distal chromosome 7 imprinting cluster, is a member of the Trp gene family. Genomics 67: 179-187. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, G.V., Soloway, P.D., and Higgins, M.J. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32: 426-431. [DOI] [PubMed] [Google Scholar]
- Forne, T., Oswald, J., Dean, W., Saam, J.R., Bailleul, B., Dandolo, L., Tilghman, S.M., Walter, J., and Reik, W. 1997. Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc. Natl. Acad. Sci. 94: 10243-10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Spillane, C., Page, D.R., and Kohler, C. 2001. Genomic imprinting and seed development: Endosperm formation with and without sex. Curr. Opin. Plant Biol. 4: 21-27. [DOI] [PubMed] [Google Scholar]
- Horike, S., Mitsuya, K., Meguro, M., Kotobuki, N., Kashiwagi, A., Notsu, T., Schulz, T.C., Shirayoshi, Y., and Oshimura, M. 2000. Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum. Mol. Genet. 9: 2075-2083. [DOI] [PubMed] [Google Scholar]
- Ishihara, K., Hatano, N., Furuumi, H., Kato, R., Iwaki, T., Miura, K., Jinno, Y., and Sasaki, H. 2000. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in Igf2/H19 imprinting. Genome Res. 10: 664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, R.M., Ainscough, J.F., Barton, S.C., and Surani, M.A. 2001. Distant cis-elements regulate imprinted expression of the mouse p57(Kip2) (Cdkn1c) gene: Implications for the human disorder, Beckwith-Wiedemann syndrome. Hum. Mol. Genet. 10: 1601-1609. [DOI] [PubMed] [Google Scholar]
- Khosla, S., Aitchison, A., Gregory, R., Allen, N.D., and Feil, R. 1999. Parental allele-specific chromatin configuration in a boundary-imprinting-control element upstream of the mouse H19 gene. Mol. Cell. Biol. 19: 2556-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kollhoff, A., Bergmann, A., and Stubbs, L. 2003. Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum. Mol. Genet. 12: 233-245. [DOI] [PubMed] [Google Scholar]
- Koski, L.B., Sasaki, E., Roberts, R.D., Gibson, J., and Etches, R.J. 2000. Monoalleleic transcription of the insulin-like growth factor-II gene (Igf2) in chick embryos. Mol. Reprod. Dev. 56: 345-352. [DOI] [PubMed] [Google Scholar]
- Lee, M.P., Brandenburg, S., Landes, G.M., Adams, M., Miller, G., and Feinberg, A.P. 1999. Two novel genes in the center of the 11p15 imprinted domain escape genomic imprinting. Hum. Mol. Genet. 8: 683-690. [DOI] [PubMed] [Google Scholar]
- Leighton, P.A., Ingram, R.S., Eggenschwiler, J., Efstratiadis, A., and Tilghman, S.M. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375: 34-39. [DOI] [PubMed] [Google Scholar]
- Lopes, S., Lewis, A., Hajkova, P., Dean, W., Oswald, J., Forne, T., Murrell, A., Constancia, M., Bartolomei, M., Walter, J., et al. 2003. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 12: 295-305. [DOI] [PubMed] [Google Scholar]
- Mancini-DiNardo, D., Steele, S.J., Ingram, R.S., and Tilghman, S.M. 2003. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum. Mol. Genet. 12: 283-294. [DOI] [PubMed] [Google Scholar]
- McLysaght, A., Hokamp, K., and Wolfe, K.H. 2002. Extensive genomic duplication during early chordate evolution. Nat. Genet. 31: 200-204. [DOI] [PubMed] [Google Scholar]
- Mitsuya, K., Meguro, M., Lee, M.P., Katoh, M., Schulz, T.C., Kugoh, H., Yoshida, M.A., Niikawa, N., Feinberg, A.P., and Oshimura, M. 1999. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum. Mol. Genet. 8: 1209-1217. [DOI] [PubMed] [Google Scholar]
- Miyamoto, T., Hasuike, S., Jinno, Y., Soejima, H., Yun, K., Miura, K., Ishikawa, M., and Niikawa, N. 2002. The human ASCL2 gene escaping genomic imprinting and its expression pattern. J. Assist. Reprod. Genet. 19: 240-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyroud, N., Richard, P., Vignier, N., Donger, C., Denjoy, I., Demay, L., Shkolnikova, M., Pesce, R., Chevalier, P., Hainque, B., et al. 1999. Genomic organization of the KCNQ1 K+ channel gene and identification of C-terminal mutations in the long-QT syndrome. Circ. Res. 84: 290-297. [DOI] [PubMed] [Google Scholar]
- Nolan, C.M., Killian, J.K., Petitte, J.N., and Jirtle, R.L. 2001. Imprint status of M6P/IGF2R and IGF2 in chickens. Dev. Genes Evol. 211: 179-183. [DOI] [PubMed] [Google Scholar]
- O'Neill, M.J., Ingram, R.S., Vrana, P.B., and Tilghman, S.M. 2000. Allelic expression of IGF2 in marsupials and birds. Dev. Genes Evol. 210: 18-20. [DOI] [PubMed] [Google Scholar]
- Olek, A. and Walter, J. 1997. The pre-implantation ontogeny of the H19 methylation imprint. Nat. Genet. 17: 275-276. [DOI] [PubMed] [Google Scholar]
- Onyango, P., Miller, W., Lehoczky, J., Leung, C.T., Birren, B., Wheelan, S., Dewar, K., and Feinberg, A.P. 2000. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 10: 1697-1710. [DOI] [PubMed] [Google Scholar]
- Patton, S.J., Luke, G.N., and Holland, P.W. 1998. Complex history of a chromosomal paralogy region: Insights from amphioxus aromatic amino acid hydroxylase genes and insulin-related genes. Mol. Biol. Evol. 15: 1373-1380. [DOI] [PubMed] [Google Scholar]
- Paulsen, M., Davies, K.R., Bowden, L.M., Villar, A.J., Franck, O., Fuermann, M., Dean, W.L., Moore, T.F., Rodrigues, N., Davies, K.E., et al. 1998. Syntenic organization of the mouse distal chromosome 7 imprinting cluster and the Beckwith-Wiedemann syndrome region in chromosome 11p15.5. Hum. Mol. Genet. 7: 1149-1159. [DOI] [PubMed] [Google Scholar]
- Paulsen, M., El-Maarri, O., Engemann, S., Strodicke, M., Franck, O., Davies, K., Reinhardt, R., Reik, W., and Walter, J. 2000. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum. Mol. Genet. 9: 1829-1841. [DOI] [PubMed] [Google Scholar]
- Prawitt, D., Enklaar, T., Klemm, G., Gartner, B., Spangenberg, C., Winterpacht, A., Higgins, M., Pelletier, J., and Zabel, B. 2000. Identification and characterization of MTR1, a novel gene with homology to melastatin (MLSN1) and the trp gene family located in the BWS-WT2 critical region on chromosome 11p15.5 and showing allele-specific expression. Hum. Mol. Genet. 9: 203-216. [DOI] [PubMed] [Google Scholar]
- Reik, W. and Walter, J. 2001. Genomic imprinting: Parental influence on the genome. Nat. Rev Genet. 2: 21-32. [DOI] [PubMed] [Google Scholar]
- Schneider, P., Olson, D., Tardivel, A., Browning, B., Lugovskoy, A., Gong, D., Dobles, M., Hertig, S., Hofmann, K., Van Vlijmen, H., et al. 2003. Identification of a new murine tumor necrosis factor receptor locus that contains two novel murine receptors for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J. Biol. Chem. 278: 5444-5454. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels, F., Zwart, R., and Barlow, D.P. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810-813. [DOI] [PubMed] [Google Scholar]
- Smilinich, N.J., Day, C.D., Fitzpatrick, G.V., Caldwell, G.M., Lossie, A.C., Cooper, P.R., Smallwood, A.C., Joyce, J.A., Schofield, P.N., Reik, W., et al. 1999. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc. Natl. Acad. Sci. 96: 8064-8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.F., Snell, P., Gruetzner, F., Bench, A.J., Haaf, T., Metcalfe, J.A., Green, A.R., and Elgar, G. 2002. Analyses of the extent of shared synteny and conserved gene orders between the genome of Fugu rubripes and human 20q. Genome Res. 12: 776-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, N., Kanduri, M., Holmgren, C., Mukhopadhyay, R., and Kanduri, C. 2003. Bidirectional silencing and DNA methylation-sensitive methylation-spreading properties of the Kcnq1 imprinting control region map to the same regions. J. Biol. Chem. 278: 9514-9519. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen, J.L., Duran, K.L., and Bartolomei, M.S. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes & Dev. 12: 3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, J. and Paulsen, M. 2003. The potential role of gene duplications in the evolution of imprinting mechanisms. Hum. Mol. Genet. 12 Spec. No. 2: R215-R220. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Fan, H., Yang, H.H., Hu, Y., Buetow, K.H., and Lee, M.P. 2004. Comparative sequence analysis of imprinted genes between human and mouse to reveal imprinting signatures. Genomics 83: 395-401. [DOI] [PubMed] [Google Scholar]
- Yokomine, T., Kuroiwa, A., Tanaka, K., Tsudzuki, M., Matsuda, Y., and Sasaki, H. 2001. Sequence polymorphisms, allelic expression status and chromosome locations of the chicken IGF2 and MPR1 genes. Cytogenet. Cell. Genet. 93: 109-113. [DOI] [PubMed] [Google Scholar]
- Zhou, Q.Y., Quaife, C.J., and Palmiter, R.D. 1995. Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature 374: 640-643. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information.
- http://www.ensembl.org/Fugu_rubripes/; Sanger Institute Fugu Genome Browser.
- http://www.ensembl.org; Sanger Institute Ensembl Genome Browser.
- http://cancer.otago.ac.nz/IGC/Web/home.html; Imprinted Gene Catalogue, University of Otago.
- http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html; Mammalian Genetics Unit, Harwell UK.
- http://www.sanger.ac.uk/Projects/D_rerio; Sanger Institute The Danio rerio Sequencing Project.
- http://www.ensembl.org/Gallus_gallus/; Sanger Institute Chicken Genome Browser.
- http://www.ebi.ac.uk/emboss/cpgplot; European Bioinformatics Institute.
- http://bio.cse.psu.edu/pipmaker; PipMaker.
- http://www.nisc.nih.gov/projects/zooseq/comp_seq_org.cgi; NIH Intramural Sequencing Center.
- http://bioweb.pasteur.fr/seqanal/interfaces/fuzznuc.html; Institute Pasteur.
- http://bioweb.pasteur.fr/seqanal/motif/meme; Institute Pasteur.
- http://repeatmasker.org; RepeatMasker.