Abstract
The identification of parasite genes controlling phenotypes such as drug resistance, virulence, immunogenicity, and transmission is vital to malaria research. Classical genetic methods have achieved these goals only rarely and with difficulty. We describe here a novel genetic method, Linkage Group Selection (LGS), which achieves rapid de novo location of genes encoding selectable phenotypes of malaria parasites. A phenotype-specific selection pressure is applied to the uncloned progeny of a genetic cross between two malaria parasites that differ in the relevant phenotype. Selected and unselected progeny are analyzed using genome-wide quantitative genetic markers. Markers of the “sensitive” parent, which are reduced after selection, are sequenced and located in genomic databases. They are expected to be closely linked to gene(s) determining the phenotype under selection. We have validated LGS with the rodent malaria parasite Plasmodium chabaudi chabaudi using a phenotype, pyrimethamine resistance, whose controlling gene, that encoding dihydrofolate reductase (dhfr), is known. We show that molecular markers closely linked to dhfr, and only those linked to this gene, were reduced or removed by pyrimethamine treatment in accordance with the expectations of LGS.
The identification of genes that control important parasite phenotypes such as drug resistance, growth rate, and strain-specific immunity, is of immense importance in the fight against malaria. Knowledge of the gene(s) controlling resistance to a specific antimalarial drug, for example, enables the monitoring of the spread of resistance, as well as potentially increasing the effectiveness of decisions concerning drug policy. Knowing which genes are involved in resistance to a drug can also help us to understand the molecular basis of drug resistance and aid in the design of new versions of drugs that are unaffected by the mutations causing parasite drug resistance (Yuvaniyama et al. 2003). Similarly, the identification of genes involved in the parasite's immunogenicity is crucial to the development of vaccines. Other parasite traits for which the genetic basis is unknown include virulence (i.e., what causes some parasites to be more harmful to a host than others) and transmissibility through particular vectors. Both would be better understood if the genes underlying these traits were known.
Existing methods in the genetic study of malaria parasites, such as linkage analysis, that attempt to locate genes controlling such traits are labor intensive and expensive. Without the generation of an extremely large number of recombinant clones (>1000), they have poor resolution, which makes the actual identification of the underlying genes extremely difficult unless strong candidates are already suspected. There is, in fact, an inverse relationship between the size of the locus within which possible target genes may be located and the number of recombinant clones that must be generated (Wellems et al. 1991; Su et al. 1997).
Linkage Group Selection (LGS), whose principles we validate here, was devised for application to malaria parasites in order to locate genes that control selectable phenotypes such as drug sensitivity, growth rate, and strain-specific immunity without the disadvantages of the huge inefficiency of classical linkage analysis. LGS (Fig. 1) uses a genetic cross between two unrelated parasites of the same species, one of which is sensitive and the other resistant to the relevant selection pressure (e.g., drug treatment). Following zygote formation, which in malaria parasites takes place in the mosquito, there is recombination between the parental genomes and the formation of haploid recombinant progeny. Each individual recombinant parasite will have inherited a random assortment of parental alleles. Half of the parasites will have inherited the “resistant” allele(s) of the gene(s) for the phenotype under investigation, and the remainder will have inherited the “sensitive” allele(s). This haploid recombinant parasite population is then exposed to the relevant selection pressure, as well as being passaged in the absence of the selection pressure. Alleles from both parents should be equally represented after selection, except at those loci linked to the gene(s) that determine(s) the parasites' response to this selection. At these loci alleles from the sensitive parent should have been reduced or eliminated in the “selected” population relative to the untreated population.
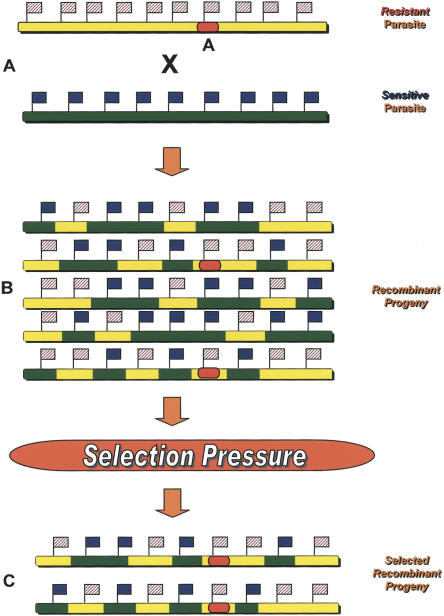
Figure 1.
A schematic representation of the genetic principles of Linkage Group Selection. (A) Two cloned haploid parasites that differ in their sensitivity to a particular selection pressure are genetically crossed. In this example, resistance to the selection pressure is conferred by a gene at locus A. The genome is represented by the colored bars, while the flags represent dense, genome-wide markers between the two strains. (B) The resulting recombinant progeny population will consist of thousands of parasites, each bearing a random assortment of parental alleles, and also a random assortment of parental markers. (C) This uncloned population is then subjected to the selection pressure under investigation, and the resulting selected population will consist solely of parasites that possess the resistant allele of the gene that controls the phenotype of interest, and therefore, any markers linked to this locus will be mainly from the resistant parent, their proportion increasing as the distance from the locus under selection decreases. We look for markers of the sensitive parent that are removed or reduced by the selection pressure; the greater the reduction in the intensity of a sensitive marker, the closer it is linked to the allele removed by the selection pressure.
By typing both the selected and the unselected recombinant populations with large numbers of quantitative, genome-wide markers, it is possible to identify those markers from the sensitive parent that are removed or reduced by the selection pressure. These markers are likely to be linked to the loci involved in controlling the response of the parasite to the selection pressure. The closer a marker is genetically linked to the target locus, the greater the reduction in its representation in the selected population. The markers can then be sequenced, and their positions located on the parasite genome in order to identify the genes to which they are linked.
LGS requires that the two parental clones are distinguished by a sufficient number of genetic markers to ensure that some will be linked to the genes of interest. For this purpose, we have used Amplified Fragment Length Polymorphism (AFLP), a PCR-based method for amplifying DNA fragments from genetically distinct cloned lines of parasites (see Methods for a description of the AFLP technique). We have previously shown that AFLP meets the requirements of LGS both as regards numbers of markers generated in different strains of Plasmodium chabaudi chabaudi (Grech et al. 2002) and their quantitation in a genetic mixture of the parasites (Martinelli et al. 2004).
Using AFLP for the generation of quantifiable genetic markers, we report here the successful validation of LGS in a study of pyrimethamine resistance in the rodent malaria parasite P. c. chabaudi.
Results
AJ and ASpyr1 are genetically distinct cloned lines of P. c. chabaudi originating from the same location in central Africa (Carter and Walliker 1975). ASpyr1 is a pyrimethamine-resistant line. In this parasite, pyrimethamine resistance is conferred by a single base mutation in codon 106 (S106N) of dhfr (the gene encoding dihydrofolate reductase) (Cheng and Saul 1994). The uncloned progeny of a genetic cross between these parasites were grown as blood infections in two groups of mice, either with or without pyrimethamine treatment. The surviving parasites from each treatment were harvested and DNA prepared and analyzed by AFLP, as previously described (Grech et al. 2002).
The Relative Intensity Indices (see Methods) of 206 AFLP markers unique to the sensitive (AJ) clone were compared between the pyrimethamine-selected and untreated cross progeny. Almost all AFLP markers showed similar band intensities in the material from the selected and unselected cross progeny. However, six markers from the AJ parent were significantly reduced in intensity. Indeed, one marker (AJAA05CA) was completely undetectable after drug selection.
It was possible to assign 120 of the AJ parental AFLP markers to a genetic linkage map of P. c. chabaudi generated from previous crosses between AS and AJ (see Methods for a description of how markers were assigned to this map). Figure 2 shows the relative intensities of these AJ markers in the drug-selected population (compared with the unselected population) arranged according to this genetic map. It was found that all of the AJ markers that were most reduced under pyrimethamine pressure lie on chromosome 7 of P. c. chabaudi. Moreover, when these markers are put into order according to the genetic map, they form a “selection valley” around the position of the marker (AJAA05CA), which had been eliminated by pyrimethamine treatment. The trough of this selection valley is closely linked to the position of pcdhfr on the genetic linkage map.
Figure 2.
The comparative intensities of AJ AFLP markers in the pyrimethamine-selected progeny, compared with unselected progeny, of a cross between P. c. chabaudi clones ASpyr1 (pyrimethamine resistant) and AJ (pyrimethamine sensitive). The AFLP markers are arranged in their genetic order on individual chromosomes of P. c. chabaudi. The chromosomes themselves are numbered at the top of the graph. The reductions in the intensities of markers in chromosome 7 are inversely related to their approximate (genetic or physical) distance from the locus for pcdhfr, the expected target of selection under pyrimethamine.
In order to confirm that pcdhfr was within this selection valley, we were able to measure the proportions of pcdhfr parental alleles in the treated and untreated progeny directly using the method of proportional sequencing (see Methods). The proportion of the AJ allele of pcdhfr in the untreated population was 78%, while in the pyrimethamine-selected population, it was reduced to undetectable levels (<2%). This shows that pyrimethamine treatment had removed virtually all parasites carrying the sensitive (AJ) allele of pcdhfr from the cross progeny.
In order to confirm their physical linkage with dhfr, the six AFLP markers that showed intensity reductions in the selected population, and which formed the selection valley around pcdhfr, were sequenced and their homologs located in the Plasmodium falciparum genome (http://www.sanger.ac.uk/Projects/P_falciparum/) wherever possible. This was necessary because the P. c. chabaudi genome database is incomplete. Therefore, in order to physically map these markers, we exploited the gene synteny between P. falciparum (physical map) and P. c. chabaudi (genetic map) (Hunt et al. 2004). We were able to identify P. falciparum homologs for three of these markers, all of which lay within 120 kb of the P. falciparum dhfr locus (pfdhfr) (Table 1). The homolog of the AJ AFLP marker (AJAA05CA), which was undetectable following pyrimethamine selection, lay only 10 kb from the location of pfdhfr, the predicted target locus of pyrimethamine pressure.
Table 1.
The relative intensities of markers under pyrimethamine selection, their positions on the P. c. chabaudi linkage map, and the positions of their homologs on P. falciparum chromosome 4, the location of pfdhfr
| Marker namea | Comparative marker intensityb | Genetic distance (cM) from dhfr (P. c. chabaudi)c | Physical distance (kb) from dhfr (P. falciparum)d |
|---|---|---|---|
| AJAA05CA | 0.00 | 5.3 | 10 |
| AJAG02CA | 0.13 | 9.5 | n/a |
| AJTG01AG | 0.06 | 13.4 | 35 |
| AJTG01GA | 0.08 | 13.4 | n/a |
| AJAC02TT | 0.42 | 33.0 | 119 |
| AJAA03AT | 0.60 | 44.5 | n/a |
| AJAA01GAe | 1.02 | 53.5 | n/a |
The comparative intensities of the markers are seen to decrease with increasing proximity to the gene under selection.
n/a = not available.
AFLP markers were named as follows: the first two letters identify the clone to which a marker is specific (AJ), the next two letters indicate the EcoRI primer selective bases (e.g., AA), the numbers identify the marker for that clone and primer combination in order of its molecular size, and the last two letters identify the Msel selective bases
Comparative Marker Intensity is a measure of the intensity of a marker in the selected population compared to its intensity in the unselected population
Markers were assigned to a linkage map between AS and AJ as described in Methods
P. c. chabaudi marker sequences were blasted against the P. falciparum genome to determine the positions of their homologs
This marker is not under pyrimethamine selection, and is included as an example of a flanking marker to give an indication of the size of the selection valley
Discussion
Linkage Group Selection (LGS) is the application of a specific selection pressure (such as drug treatment) to the entire recombinant progeny of a genetic cross between two members of the same species that differ phenotypically in their response to the selection pressure, and evaluation of the effects of the selection upon the progeny at loci across the entire genome using quantitative genetic markers. The object and the endpoint of this exercise is the identification of specific genes that control the phenotype under investigation. The LGS approach is ideally suited to organisms such as the malaria parasites whose genetics are conventionally Mendelian, but which are in a haploid state for most of their life cycle when phenotype-specific selection pressures can be applied. These circumstances apply to all of the protozoan parasites of the phylum Apicomplexa that include not only the malaria parasites, but also their relatives Theileria, Babesia, Toxoplasma, and Coccidia, all of which are important pathogens of humans and/or animals.
Using the rodent malaria parasite P. c. chabaudi, and applying the LGS method, we have shown here that a group of linked AFLP markers were reduced in intensity when the uncloned progeny of a genetic cross between a pyrimethamine-resistant and a pyrimethamine-sensitive clone were subjected to pyrimethamine treatment. These markers were, moreover, linked on a P. c. chabaudi genetic linkage map to pcdhfr, the gene already known to determine prymethamine resistance in the parental line used in the cross. The homologs of these markers were also found to be physically linked to pfdhfr, the corresponding gene in P. falciparum. The data demonstrate the existence, under pyrimethamine treatment, of a selection valley with the dhfr locus at its apex, and have, therefore, validated LGS as a method for locating a gene controlling a selectable phenotype such as drug resistance.
The extent to which we can expect to be able to map an unknown gene by LGS depends (1) upon the density of markers across the genome, (2) our ability to quantitate the proportions of parasites carrying specific markers, (3) the strength of the specific phenotypic selection pressure itself, and (4) the number of independent recombinants in the cross progeny. For each of these, the requirements of LGS have been met in the present experiments. We have previously shown that an extremely high density of genetic markers is potentially available in P. c. chabaudi. Our recent estimates show polymorphisms between any two cloned isolates of P. c. chabaudi at an average frequency of about 1 per 100 bp (Grech et al. 2002). We have also been able to measure with sufficient accuracy, the proportions of parasites carrying parental alleles in a genetically complex population using the AFLP method. The procedure used for selection at the target locus, namely pyrimethamine treatment, was highly effective for the purposes of LGS. Finally, our data, by defining a clear selection valley around the locus under selection, indicate that we achieved sufficiently large numbers of independent recombinant progeny in our cross.
The results of the present work have demonstrated that it is easily possible by LGS to detect a single controlling gene for a given selectable phenotype. However, the method is also expected to be effective where several loci are involved. By calculation from known genetic recombination rates in malaria parasites (Walker-Jonah et al. 1992; Su et al. 1999), and from our own experience to date, individual LGS selection valleys have widths of around half to 1 Mb, so that the Plasmodium genome can accommodate at least 20 nonoverlapping selection valleys. This means that if individual loci involved in controlling a phenotype do not interact with each other, then at least 20 selection valleys, and the equivalent number of controlling loci, could readily be detected by LGS. If, however, as may often be the case, the controlling loci do interact (i.e., they depend upon each other for the strengths of their effects), then the selection valleys associated with each locus would be shallower, depending upon the number of interacting loci and the strength of their mutual dependency. Even if loci were totally interdependent for their effect, at least two to three such loci should be readily detected by LGS with current methods of allele quantification. Lower levels of interdependence and more sensitive quantification methods would increase the number of potentially detectible interacting loci.
While LGS is ideally suited to organisms that are haploid at the point at which recombinant progeny are subjected to a selection pressure, it could also be used to identify the genetic loci controlling selectable phenotypes in diploid organisms. In this case, however, it would be necessary to produce an F2 generation from a cross in order to generate progeny carrying reassorted alleles at each polymorphic locus, and upon which to apply the selection pressure. In the diploid state, genes can be recessive, dominant, or codominant, and depending upon which of these situations applies to the genes under selection, the degree of reduction in the proportion of a sensitive allele after selection will vary. Except where a resistant allele is recessive, there will be incomplete elimination of sensitive allele(s) in a selected population. There will, however, always be some reduction in the proportion of sensitive alleles compared with neutral alleles after selection, and given appropriately sensitive techniques for measuring differences in marker proportion between selected and unselected populations, LGS will be able to resolve loci containing the genes that control the phenotype.
LGS has much greater efficiency than the classical linkage-analysis methods previously used for gene identification in malaria and other Apicomplexan parasites. Classical linkage analysis requires the expensive and time-consuming processes of cloning independent recombinant parasites and the subsequent characterization of the genotype and phenotype of each clone. Without very great effort, the resolution of the location of a gene of interest is poor by classical genetic methods. For example, the target locus for chloroquine resistance in P. c. chabaudi was defined within a region of 250 kb after the characterization of 28 independent recombinant clones of this parasite (Hunt et al. 2004). Similarly, the identification of a locus underlying chloroquine resistance in P. falciparum was first defined to within 400 kb using 16 clones and 85 RFLP markers (Wellems et al. 1991). Only after 6 yr of further work, was the relevant locus defined to within 36 kb using 1120 recombinant progeny (Su et al. 1997).
LGS achieves an equivalent degree of genetic resolution much more rapidly and at much lower cost. As is the case in a classical genetic analysis, the resolution of an LGS analysis depends upon the number of independent recombinants arising from a genetic cross. However, instead of analyzing each recombinant genotype separately, LGS treats the entire cross progeny in a single analysis. Its genetic resolution depends upon the sensitivity of the quantitative assays used to measure the representation of each marker. A method that could measure the intensity of any marker in a selected cross progeny to an accuracy of ±1% relative to its intensity in the unselected progeny, would resolve its genetic distance from the target locus of the applied selection pressure to an accuracy of ±1 cM. In the case of malaria parasites, 1 cM represents a physical distance along a chromosome of ∼20 kb (as determined from the studies of Walker-Jonah et al. 1992; Carlton 1995; Su et al. 1999). Assuming a uniform recombination rate across an entire Plasmodium genome (unlikely to be strictly true, but a reasonable first approximation), such a method should locate any marker to an accuracy of ±20 kb from a target locus; a method with an accuracy of ±3% would locate a marker with an accuracy of ±60 kb, and so on. AFLP itself appears to have a resolution of 5%–10% (Martinelli et al. 2004). However, once the region within which a target gene lies has been located to within a few 100 kb, more accurate methods, such as proportional sequencing (P. Hunt, R. Fawcett, R. Carter, and D. Walliker, in prep.) and RTQ–PCR (Cheesman et al. 2003) can be applied to rapidly narrow the region within which the target of selection is to be found.
While the classical genetic approach can, as we have discussed, be very powerful, it is also extremely laborious, time-consuming, and therefore, costly. LGS achieves an equivalent resolution vastly more rapidly and at correspondingly lower cost because it is applied directly to the uncloned progeny of a genetic cross. In the case of malaria parasites, such crosses readily generate many thousands of independent recombinant parasite lines. In a single experimental operation, the application of the relevant selection pressure to these uncloned progeny provides the material for a single genome-wide screening with the available genetic markers. In contrast, to achieve the same degree of genetic resolution, the classical approach involves the screening of the equivalent number (thousands) of individual cloned progeny in as many separate screening operations.
Rodent malaria parasites are ideally suited to LGS analysis because of the ease with which they allow the full Plasmodium life cycle to be achieved. However, any host/parasite system that allows the completion of the full life-cycle of an Apicomplexan parasite is potentially open to investigation by LGS. Because of its efficiency, LGS can also be practically applied to systems that have been previously impossible or extremely expensive to investigate by genetic means. Crosses between strains of P. falciparum have been obtained under laboratory conditions using chimpanzees (Wellems et al. 1991), and crosses of Plasmodium knowlesi and Plasmodium cynomolgi (closely related to the human malaria parasite, Plasmodium vivax) can be generated in Rhesus monkeys. These now can be readily and profitably investigated using LGS analysis.
There are many potential applications of LGS in the field of malaria parasite genetics. It is, for example, largely unknown which genes control important parasite traits such as differences in growth rates between strains, or encode antigens responsible for strain-specific immunity in malaria. LGS can address these questions by applying the relevant selection pressure to the trait under investigation. In the case of growth rate, for example, uncloned recombinant progeny of a cross between a faster-growing and a slower-growing strain of malaria parasite need only to be grown in the vertebrate host to select for those parasites that possess the allele(s) that underlies the growth rate of the faster-growing parental strain. To investigate strain-specific immunity, individual hosts immunized with either of two antigenically distinct parasite strains can be challenged with the uncloned recombinant progeny of a cross between them. In this way, those recombinants that possess alleles that code for a target antigen specific to the immunizing parental strain will be selected against, and the surviving population will thus be monoallelic at loci that encode strain-specific antigens.
Before the development of LGS, its underlying concepts had been used at a population level to track genes of malaria parasites that encode the targets of a specific selection pressure. Thus, specific haplotypes have been identified surrounding a target locus following population-level selective sweeps under drug selection in malaria parasites (Wootton et al. 2002; Nair et al. 2003). It has also been pointed out that alleles of polymorphic genes encoding antigens under a strain-specific immune selection should be present at lower frequency in individual infections in endemic populations under natural malaria transmission than are alleles of neutral polymorphic genes (Hastings 1996). Both concepts are implicit in LGS. However, in contrast to LGS, these population-based investigations have depended upon the collection and individual analysis of large numbers of natural isolates of the parasites. The LGS approach described here applies these principles in controlled genetic crosses to the rapid and efficient location of the gene(s) that determine a specific phenotype.
Methods
Parasites, mice, and mosquitoes
The two cloned strains of P. c. chabaudi used in this investigation were ASpyr1 (pyrimethamine resistant) and AJ (pyrimethamine sensitive). These clones were derived from two genetically distinct lines of P. c. chabaudi isolated from thicket rats, Thamnomys rutilans, in the Central African Republic (Carter and Walliker 1975). They were cloned by limiting dilution of blood forms and ASpyr1 was derived from AS (pyrimethamine sensitive) after selection for pyrimethamine resistance (Walliker et al. 1975). The mice used were inbred CBA females, aged 6–8 wk at the time of infection. Mosquitoes were Anopheles stephensi from a laboratory colony.
Generation of recombinant progeny from genetic crosses between P. c. chabaudi clones ASpyr1 and AJ
Single-clone infections of ASpyr1 and AJ were grown individually in donor mice. Blood from these mice was then extracted and mixed together to produce an inoculum containing equal proportions of the two clones. This inoculum was administered intraperitoneally to three mice, so that each mouse received a total of 2 × 106 parasites (1 × 106 of each clone). Six days post-inoculation, the presence of gametocytes was confirmed in the mice by thin blood-smear microscopy. The mice were then anaesthetized, and each was placed on top of a mosquito cage containing ∼200 female mosquitoes 5–7 d post-emergence from pupae. The mosquitoes were allowed to feed on the mice for 30 min. Ten mosquitoes from each cage were dissected 7 d post-feed, and the midguts were checked for the presence of oocysts. Sporozoites were collected from each cage of mosquitoes at day 14 post-infection by dissection of the salivary glands. These were pooled and injected intraperitoneally into five mice, and the resulting infections followed by thin blood-smear microscopy.
Selection of the ASpyr1 × AJ P. c. chabaudi cross progeny under pyrimethamine pressure
When the sporozoite-induced infections reached peak parasitaemia (10%–15%), the parasites were harvested, pooled, and inoculated into a group of five mice. Each mouse in the group received 1 × 106 parasites. This initial experimental group was left untreated, and the parasites were harvested at peak parasitaemia for AFLP analysis to provide a reference point for markers analyzed in the subsequent treatment groups. Parasites were pooled and subinoculated from this group into two further groups of five mice, one of which was treated with pyrimethamine, and the other left untreated. These two groups provided the material for the comparison of markers between drug-treated and untreated parasite populations.
Pyrimethamine was administered orally at a dose of 10 mg/kg of mouse body weight daily at 24-h intervals for 4 d, starting 3 h after parasite challenge.
Both the pyrimethamine-treated and the untreated blood-stage cross progeny were allowed to grow to peak parasitaemia (30%–40% for untreated, and 15%–20% for treated), at which point the blood was harvested. Two samples of parasite DNA were prepared for AFLP and other molecular analyses by pooling separately the blood from the treated and untreated mice.
Generation and analysis of AFLP markers
DNA extracted from the pyrimethamine-selected, and the non-selected cross-progeny was analyzed by AFLP as previously described (Grech et al. 2002). Briefly, DNA was digested with two enzymes, EcoRI and MseI, and the resulting randomly sized fragments were ligated to adaptors. Primers were specifically designed to bind to these adaptors and used to amplify the DNA fragments. Two additional nucleotides extending into the actual genomic sequence were added at the 3′-end of primers to reduce the number of bands to be analyzed. AFLP bands were visualized on an acrylamide gel by radiolabeling of one of the primers. Polymorphic bands could then be visualized on the gel as being present in one strain and absent in the other.
The intensities of AFLP markers can be directly measured using imaging software. The Relative Intensity Indices of AFLP markers from the drug-treated and untreated groups were calculated as previously described (Martinelli et al. 2004). Relative Intensity Index is defined as the intensity of a polymorphic band divided by the intensity of a specific nonpolymorphic band (Intensity Index), divided by the equivalent index of the same bands measured from the pure parental strain. We have previously shown that the Relative Intensity Index of an individual AFLP marker is quantitatively related to the proportion of parasites carrying that marker in a population (Martinelli et al. 2004).
Assignment of AFLP markers to locations in Plasmodium genomes
AFLP markers were ordered on a genetic linkage map of P. c. chabaudi (Martinelli 2003) obtained from previously generated crosses between AS-derived clones and AJ strains of P. c. chabaudi (Walliker et al. 1975; Rosario 1976; Carlton et al. 1998). A total of 674 AFLP markers were typed for each of 28 cross-progeny clones, and were subsequently assigned to linkage groups using the Map Manager QTX software (Manly et al. 2001). A total of 44 RFLP markers characterized in a previous study (Carlton et al. 1998) were used as genetic anchors to allow the assignment of the various linkage groups to chromosomes. In total, 11 chromosomes could be identified, while 12 linkage groups of as yet unknown assignment remain, which include the three remaining chromosomes (i.e., chromosomes 2, 4, and 14).
Some markers were also mapped, wherever necessary and possible, to locations in the P. falciparum genome. This was done by sequencing the markers of interest and identifying their homologs in the P. falciparum database.
Proportional sequencing
To directly quantify the proportions of the sensitive and resistant alleles of pcdhfr present in the selected and unselected cross progeny, DNA was amplified by nested PCR at the pcdhfr locus using primers common to nonpolymorphic sequences from both parental alleles. The resulting PCR products were purified and sequenced. Sequencing results were analyzed using the SeqED v 1.0.3 software (Applied Biosystems, Inc., 1992), which allows the visualization of chromatograms of sequenced DNA and the quantitation of individual fluorescent peaks. The relative heights of peaks at the polymorphic sites in pcdhfr can be used as an index of the relative proportions of the dhfr alleles from AJ and ASpyr1 in each sample. With five replicate samples, and with reference to a calibrated series of mixtures between ASpyr1 and AJ (Cheesman et al. 2003), it is possible to estimate the percentage of parasites carrying the ASpyr1 or AJ alleles of the pcdhfr gene with a standard error of <3% (P. Hunt, R. Fawcett, R. Carter, and D. Walliker, in prep.).
Acknowledgments
We thank the Wellcome Trust and the Medical Research Council of the U.K. for financial support for this work, Les Steven for technical assistance, and Deborah Charlesworth, Sandra Cheesman, Margaret Mackinnon, and Sittiporn Pattaradilokrat for discussion.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2866205.
References
- Carlton, J. 1995. “The genetics of chloroquine resistance in the rodent malaria parasite Plasmodium chabaudi.” Ph.D. thesis, University of Edinburgh, UK.
- Carlton, J., Mackinnon, M.J., and Walliker, D. 1998. A chloroquine resistance locus in the rodent malaria parasite, Plasmodium chabaudi. Mol. Biochem. Parasitol. 93: 57-72. [DOI] [PubMed] [Google Scholar]
- Carter, R. and Walliker, D. 1975. New observations on the malaria parasites of rodents of the Central African Republic—Plasmodium vinckei petteri subsp. nov. and Plasmodium chabaudi Landau, 1965. Ann. Trop. Med. Parasitol. 69: 187-196. [DOI] [PubMed] [Google Scholar]
- Cheesman, S.J., de Roode, J.C., Read, A.F., and Carter, R. 2003. Real-time quantitative PCR for analysis of genetically mixed infections of malaria parasites: Technique validation and applications Mol. Biochem. Parasitol. 131: 83-91. [DOI] [PubMed] [Google Scholar]
- Cheng, Q. and Saul, A. 1994. The dihydrofolate reductase domain of rodent malarias: Point mutations and pyrimethamine resistance. Mol. Biochem. Parasitol. 65: 361-363. [DOI] [PubMed] [Google Scholar]
- Grech, K., Martinelli, A., Pathirana, S., Walliker, D., Hunt, P., and Carter, R. 2002. Numerous, robust genetic markers for Plasmodium chabaudi by the method of amplified fragment length polymorphism. Mol. Biochem. Parasitol. 123: 95-104. [DOI] [PubMed] [Google Scholar]
- Hastings, I.M. 1996. Population genetics and the detection of immunogenic and drug-resistant loci in Plasmodium. Parasitology 112: Part 2, 155-164. [DOI] [PubMed] [Google Scholar]
- Hunt, P., Martinelli, A., Fawcett, R., Carlton, J., Carter, R., and Walliker, D. 2004. Gene synteny and chloroquine resistance in Plasmodium chabaudi. Mol. Biochem. Parasitology 136: 157-164. [DOI] [PubMed] [Google Scholar]
- Manly, K.F., Cudmore Jr., R.H., and Meer, J.M. 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930-932. [DOI] [PubMed] [Google Scholar]
- Martinelli, A. 2003. “Strain-specific immunity in malaria: A molecular and genetic approach.” Ph.D. thesis, University of Edinburgh, UK.
- Martinelli, A., Hunt, P., Cheesman, S.J., and Carter, R. 2004. The application of Amplified Fragment Length Polymorphism to the genetic analysis of selectable phenotypes in malaria parasites. Mol. Biochem. Parasitol. 136: 117-122. [DOI] [PubMed] [Google Scholar]
- Nair, S., Williams, J.T., Brockman, A., Paiphun, L., Mayxay, M., Newton, P.N., Guthmann, J.P., Smithuis, F.M., Hien, T.T., White, N.J., et al. 2003. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol. Biol. Evol. 20: 1526-1536. [DOI] [PubMed] [Google Scholar]
- Rosario, V.E. 1976. Genetics of chloroquine resistance in malaria parasites. Nature 261: 585-586. [DOI] [PubMed] [Google Scholar]
- Su, X-Z., Kirkman, L.A., Fujioka, H., and Wellems, T.E. 1997. Complex polymorphisms in an ∼330 kDa protein are linked to chloroquine-resistant P. falciparum in south-east Asia and Africa. Cell 91: 593-603. [DOI] [PubMed] [Google Scholar]
- Su, X., Ferdig, M.T., Huang, Y.M., Huynh, C.Q., Liu, A., You, J.T., Wootton, J.C., and Wellems, T.E. 1999. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science 286: 1351-1353. [DOI] [PubMed] [Google Scholar]
- Walker-Jonah, A., Dolan, S.A., Gwadz, R.W., Panton, L.J., and Wellems, T.E. 1992. An RFLP map of the Plasmodium falciparum genome, recombination rates and favored linkage groups in a genetic cross Mol. Biochem. Parasitol. 51: 313-320. [DOI] [PubMed] [Google Scholar]
- Walliker, D., Carter, R., and Sanderson, A. 1975. Genetic studies on Plasmodium chabaudi: Recombination between enzyme markers. Parasitology 70: 19-24. [DOI] [PubMed] [Google Scholar]
- Wellems, T.E., Walker-Jonah, A., and Panton, L.J. 1991. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc. Natl. Acad. Sci. 88: 3382-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton, J.C., Feng, X.R., Ferdig, M.T., Cooper, R.A., Mu, J.B., Baruch, D.I., Magill, A.J., and Su, X-Z. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418: 320-323. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama, J., Chitnumsub, P., Kamchonwongpaisan, S., Vanichtanankul, J., Sirawaraporn, W., Taylor, P., Walkinshaw, M.D., and Yuthavong, Y. 2003. Insights into antifolate resistance from malarial DHFR-TS structures Nat. Struct. Biol. 10: 357-365. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.sanger.ac.uk/Projects/P_falciparum/; Sequence data for P. falciparum chromosome 4.