Abstract
We have carried out an initial analysis of the dynamics of the recent evolution of the splice-sites sequences on a large collection of human, rodent (mouse and rat), and chicken introns. Our results indicate that the sequences of splice sites are largely homogeneous within tetrapoda. We have also found that orthologous splice signals between human and rodents and within rodents are more conserved than unrelated splice sites, but the additional conservation can be explained mostly by background intron conservation. In contrast, additional conservation over background is detectable in orthologous mammalian and chicken splice sites. Our results also indicate that the U2 and U12 intron classes seem to have evolved independently since the split of mammals and birds; we have not been able to find a convincing case of interconversion between these two classes in our collections of orthologous introns. Similarly, we have not found a single case of switching between AT-AC and GT-AG subtypes within U12 introns, suggesting that this event has been a rare occurrence in recent evolutionary times. Switching between GT-AG and the noncanonical GC-AG U2 subtypes, on the contrary, does not appear to be unusual; in particular, T to C mutations appear to be relatively well tolerated in GT-AG introns with very strong donor sites.
Protein-coding genes are characteristically interrupted by introns in the genome of higher eukaryotic organisms. While intron function and origin has been debated at length (de Souza 2003; Fedorova and Fedorov 2003; Roy et al. 2003), recent comparative analyses show an abundance of conserved elements in intronic sequences (for instance, see Dermitzakis et al. 2002; Hare and Palumbi 2003). This strongly suggests that introns are rich in elements playing functional, probably regulatory, roles (Mattick 2001). Splicing of introns is found in all main branches of eukaryotes, that is, animals, plants, fungi, and protozoa, indicating an early origin of splicing within eukaryotes, or the existence, in the pre-eukaryotic world, of a precursor of splicing. Indeed, the two major molecular mechanisms by means of which splicing is produced, U2- and U12-dependent, seem to have evolved independently prior to the divergence of the animal and plant kingdoms (Burge et al. 1998; Zhu and Brendel 2003).
Within each of these two classes of splicing, sequence features involved in intron specification are essentially conserved across eukaryotes. In both classes, the sequence information needed to specify the 5′ and 3′ splice sites—hereafter also described as donor and acceptor sites respectively—is largely confined to their surrounding region (see Fig. 1). Conserved sequences in these regions interact with the splicing machinery to promote the assembly of the spliceosome and activate the biochemical pathway that leads to the production of the spliced mRNA (for review, see Burge et al. 1999). Despite the strong conservation, the sequence of splicing signals does not carry enough information to unequivocally specify introns in the large sequence of the pre-mRNA transcripts, occasionally hundreds of thousands of nucleotides long; and recent research suggests that signals other than those in the region of the splice sites play a role in the definition of the intron boundaries (for review, see Caceres and Kornblihtt 2002; Cartegni et al. 2002; Black 2003).
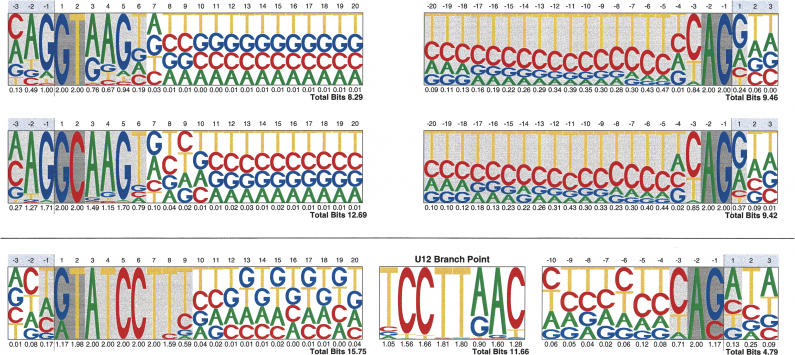
Figure 1.
Donor and acceptor sites' pictograms. Pictograms of the donor (left) and the acceptor (right) site sequences for the U2 (top) and U12 (bottom) splice sites. The sequence plots for GT-AG and GC-AG U2 introns are given separately. The conserved sequence of the U12 branch point is also shown. From human, mouse, rat, and chicken RefSeq genes, a total number of 337,336, 2506, and 935 splice-site sequences from CDS introns from Ensembl were included in GT-AG, GC-AG, and U12 splice site sets, respectively, to produce the corresponding pictograms.
Thus, in eukaryotic organisms, splicing introduces an additional level of decoding—prior to translation—on the sequence of the primary RNA transcript. There is a fundamental difference, however, between the genetic code—the mapping of nucleotide sequences (triplets) into 20 (or more) amino acids—and the splicing code—the mapping of nucleotide sequences into 3′ and 5′ intron boundaries. The genetic code is essentially deterministic; within a given species, a given triplet in the mRNA sequence results always in the same amino acid—the dual role in selenoproteins of the TGA triplet as stop and selenocysteine codon probably the most notable of all exceptions (for instance, see Kryukov et al. 2003). The splicing code, in contrast, is inherently stochastic; the probability of a splicing sequence in the primary transcript to participate in the definition of an intron boundary ranges from zero to one, and it is conditioned to very many different factors (which could be other sequences—maybe distant). The tissue-specific distribution of relative abundances of alternative splicing products (Xu et al. 2002; Yeo et al. 2004), for instance, reflects this nondeterministic nature of the splicing code.
The stochasticity of the splicing code offers opportunities for evolution that are absent in the highly deterministic genetic code. The availability of an increasing number of eukaryotic genomes makes it possible to investigate such an evolutionary process. Here, we report on findings obtained by comparing a large collection of orthologous introns (introns occurring at equivalent locations in orthologous genes) and their defining splice sites in human, mouse, rat, and chicken. Our results provide insights into the dynamics of the evolution of splice-site sequences during the most recent period of the history of life on earth.
Results
In this section, we first report results concerning interconversion between the two major classes of introns, U2 and U12, and subtype switching within each class. Then, we report on the comparison of splice-site sequences in human, rodents, and chicken. We have compared the overall sequence patterns of splice sites and investigated the level of sequence conservation between orthologous splice sites.
The analyses described here are very sensitive to the identification of true orthologous introns, as well as to the prediction of correct splice boundaries, particularly in the case of the noncanonical U12 introns. Because U12 introns constitute only a tiny fraction of all eukaryotic introns, computational gene prediction methods ignore them. Therefore, in absence of good cDNA coverage, computational gene catalogs are likely to heavily misrepresent them. Such is the case in the chicken genome. In an effort to conciliate the amount of data with reliability, we have resorted to different data sets to perform different types of analyses. Gene predictions from the RefSeq collection (Pruitt et al. 2003)—a collection of genes with good cDNA support—have been used for interspecific analysis of splice-site sequence patterns and for the identification and analysis of mammalian U12 introns. However, there are very few chicken genes in RefSeq. The larger—but strongly biased toward GT-AG canonical U2 introns—Ensembl collection (Birney et al. 2004; http://www.ensembl.org) has been used for interspecific comparison of splice-site patterns. A set of mammalian-avian curated orthologous introns—referred to as the HMRG set in this work (see Methods section)—has been used for the comparison of orthologous splice-site sequences. Table 1 describes the sizes of the data sets used in this study.
Table 1.
Summary of initial data and filtered orthologs sets.
| (A) Initial data sets
| ||||||
|---|---|---|---|---|---|---|
| Ensembla
|
UCSC genome browserb RefSeqc
|
|||||
| Species | Version | Genes | Introns | Version | Genes | Introns |
| humand | v19.34a | 33,633 | 284,125 | HGv16/NCBI34 | 21,744 | 206,814 |
| mousee | v19.30 | 30,665 | 218,163 | MGSCv4/NCBI32 | 17,988 | 139,258 |
| ratf | v19.3a | 28,545 | 192,459 | RGSCv3.1 | 4877 | 43,393 |
| chickeng | v22.1.1 | 28,491 | 252,226 | CGSCv2 | 1496 | 12,632 |
| (B) Filtered orthologs
| ||||
|---|---|---|---|---|
| Sets | Genes | Introns | ||
| Total | human | 6043 | 48,939 | (out of 51,876) |
| mouse | 5680 | 45,543 | (out of 47,193) | |
| rat | 1847 | 13,929 | (out of 14,245) | |
| Orthologs | human/mouse | 5550 | 44,119 | |
| human/rat | 1737 | 13,259 | ||
| mouse/rat | 1416 | 9655 | ||
| Triads | human/mouse/rat | 1283 | 8895 | |
(A) Initial data sets: the initial pool of genes/introns from which we filtered all the data sets for this work (aBirney et al. 2004; bKarolchik et al. 2003; cPruitt et al. 2003; dLander et al. 2001; eWaterston et al. 2002; fRat Genome Sequencing Project Consortium 2004; gInternational Chicken Genome Sequencing Consortium 2004).
(B) Filtered orthologs: the number of RefSeq orthologous genes and introns derived from these data sets.
Intron classes
Two distinct types of pre-mRNA introns are found in most higher eukaryotic organisms (Sharp and Burge 1997). They differ in the spliceosome complex that excise them during RNA processing. More than 99% of eukaryotic introns are spliced by the U2 spliceosome, while a minor class are spliced by the U12 spliceosome. U2 and U12 introns differ in the conserved sequences flanking their splice sites (see Fig. 1). Vertebrate U2 introns are characterized by the highly variable consensus [CA]AG/GT[AG]AGT at the donor (5′) site, (where [CA] means C or A, and / denotes the exon-intron boundary) and by a polypyrimidinerich stretch between the acceptor site and a poorly conserved branch point. The branch point and the acceptor site are usually separated by 11-40 nucleotides, although cases are known where they can be over 100 nucleotides apart (Helfman and Ricci 1989; Smith and Nadal-Ginard 1989). U2 introns almost always exhibit the conserved GT and AG dinucleotides at the 5′ and 3′ intron boundaries, respectively. The only remarkable exception is the existence of U2 GC-AG introns, which appears with a frequency <1% (Burset et al. 2001).
U12 introns are characterized by a strong consensus/ [AG]TATCCTT at the donor site, and TCCTT[AG]AC at the branch point. They also lack the polypyrimidine tract upstream of the acceptor site, characteristic of U2 introns. Also, in contrast to U2 introns, the distance between this acceptor site and the branch point is consistently short, between 10 and 20 nucleotides (Dietrich et al. 2001). Although initially discovered because of the unusual AT and AC dinucleotides at the 3′ and 5′ splice sites (Jackson 1991; Hall and Padgett 1994), it was later shown that U12 introns can exhibit a variety of terminal dinucleotides, the vast majority, however, are GT-AG or AT-AC (Dietrich et al. 1997; Sharp and Burge 1997; Levine and Durbin 2001; Zhu and Brendel 2003). Subtype switching within U12 introns, as well as conversion from U12 to U2 introns, has been documented (Burge and Karlin 1998), although amazing stability has been reported for U12 introns over very large evolutionary times (Zhu and Brendel 2003).
We have used the U12 donor site and branch point patterns above to identify U12 introns in the human and rodent RefSeq collections (see Methods). Table 2 lists the resulting frequencies of the different splice classes, and subtypes within each class. Numbers are consistent with those previously published (Burset et al. 2001; Levine and Durbin 2001). Identification of U12 introns was not attempted in chicken because of the small size of the RefSeq database for this organism. Figure 1 uses sequence pictograms to display the consensus for GT-AG U2 splice signals in mammals and chicken. It also displays the mammalian consensus for GC-AG U2 and U12 splice sites. In sequence pictograms (Schneider and Stephens 1990; Burge et al. 1999) the frequencies of the four nucleotides at each position along the signal are represented by the heights of their corresponding letters. The information content (intuitively, the deviation from random composition) is computed at each position, and summed up along the signal. The larger the information content, the more conserved the signal.
Table 2.
Intron class and subclass frequencies in mammals
| Human | Mouse | Rat | ||
|---|---|---|---|---|
| U2 | GT-AG | 48,212 (98.9%) | 44,817 (98.8%) | 13,707 (98.7%) |
| GC-AG | 355 (0.7%) | 330 (0.7%) | 96 (0.7%) | |
| Other | 184 (0.4%) | 218 (0.5%) | 80 (0.6%) | |
| Total | 48,751 | 45,365 | 13,883 | |
| U12 | GT-AG | 131 (69.7%) | 128 (71.9%) | 36 (78.3%) |
| AT-AC | 51 (27.1%) | 47 (26.4%) | 9 (19.6%) | |
| Other | 6 (3.2%) | 3 (1.7%) | 1 (2.2%) | |
| Total | 188 | 178 | 46 |
Intron class conversion
Orthologous mapping revealed that in all cases, orthologous mouse-rat and human-rodent introns—from the RefSeq data set—were either both U12 or both U2. A few cases were initially classified as instances of intron conversion. After close inspection, however, we realized that all of these cases could be explained either by misprediction of the intron boundaries or by splice sequence patterns slightly off consensus. (See Supplemental materials for the cross-species alignments at the intron boundaries of all predicted U12 introns). Remarkably, therefore, not one single convincing case of U12 to U2 conversion or vice-versa has occurred since the divergence of the human and rodent lineages. To investigate whether conservation of intron class extends beyond the mammalian lineage, we have mapped the 412 human, mouse, and rat U12 introns from Table 2, which correspond to 202 unique orthologs, into the chicken genome. The mapping was obtained by comparing, using exonerate (G. Slater, unpubl.), the two exons harboring the intron against the chicken genome sequence (see Methods). A total of 38 mammalian U12 introns were unequivocally mapped into the chicken genome. (See Supplemental material for cross-species alignments at the intron boundaries of the mammalian U12 introns mapped into the chicken genome). The 38 chicken introns had the typical donor-site sequence of U12 introns, and 36 had the typical U12 branch point. In the other two cases, sequences reminiscent of the U12 branch point could still be found, although departing clearly from the consensus. Since these two cases are both of the GT-AG U12 subtype, it is tempting to speculate that they may correspond to intermediates in the interconversion pathway between U12 and U2 introns. Against this hypothesis, however, is the fact that no strong polypyrimidine tract, suggestive of U2 function, can be found upstream of the acceptor site. With the exception of these two cases, the branchpoint sequence was extremely conserved between mammals and chicken, showing no more than two mismatches, but often being identical. The position of the branch point has also been conserved; with only one exception, the larger displacement observed was of 4 nucleotides. These results strongly argue that U2 and U12 introns have evolved independently, at least since the split of mammals and birds.
Subtype switching
Although subtype switching between GT-AG and AT-AC U12 introns has been documented (Burge et al. 1998), we have not found any such case within rodents, between human and rodents, or between mammals and chicken in our set of U12 orthologous introns. It appears that this phenomenon occurs at a very slow rate over evolutionary time (see cross-species alignments of orthologous U12 introns in the Supplemental material).
Within U2 introns, on the contrary, switching between GC-AG and GT-AG subclasses, and vice-versa, is not unusual. Table 3A lists the pairwise frequency of subtype switching within U2 introns, and subtype distribution within orthologous mammalian triads. Because of the limited number of cases available in the RefSeq collection, we have ignored chicken genes in this analysis. A total of 190 of the 290 human (66%) and 289 mouse (66%) GC-AG introns are conserved in both species. Similar proportions are observed between human and rat. Within rodents, 60 of the 68 mouse (88%) and 67 rat (90%) GC-AG introns are conserved in both species. The availability of orthologous introns from three organisms allows the investigation of the dynamics of subtype switching within U2 introns (see Table 3B). We have divided GC-AG introns' orthologous triads into (1) “ancient”; the intron is GC-AG subtype in the three species, and thus it is likely to predate the split of human and rodents; (2) “modern”; the intron is GC-AG subtype in either human or rodents. Because of the lack of a reference out-group, however, we cannot distinguish here those ancient GC-AG introns that have reverted to GT-AG in one of the two lineages from those modern GC-AG introns that have arisen in one of the lineages; and (3) “recent”; the intron is of GC-AG subtype only in one of the rodent species. The most parsimonious hypothesis is that the switch to GC-AG has occurred after the split of mice and rats.
Table 3.
Observed cases of U2 subtype switching within mammals
| (A) Orthologous pairs
| ||||
|---|---|---|---|---|
| GT, GT | GC, GC | GC, GT | GT, GC | |
| human/mouse | 38,922 | 190 | 100 | 99 |
| human/rat | 11,693 | 61 | 33 | 23 |
| mouse/rat | 8441 | 60 | 8 | 7 |
| (B) Orthologous triads
| ||||||
|---|---|---|---|---|---|---|
| Human | Mouse | Rat | Occurrences | |||
| “ancient” GT-AG | ||||||
| GT | GT | GT | 7784 | |||
| “ancient” GC-AG | ||||||
| GC | GC | GC | 45 | |||
| “moderate” GC-AG | ||||||
| GC | GT | GT | 23 | |||
| GT | GC | GC | 11 | |||
| “recent” GC-AG | ||||||
| GT | GT | GC | 8 | |||
| GT | GC | GT | 5 | |||
| “ancient” GC-AG, “recent” GC → GT | ||||||
| GC | GC | GT | 2 | |||
| GC | GT | GC | 1 | |||
| Total | 95 | |||||
(A) Orthologous pairs: occurrence of donor site dinucleotide pairs at intron boundaries of orthologous intron pairs. For instance, we have found 65 instances in which the orthologous donor site is GC in human and GT in mouse.
(B) Orthologous triads: occurrence of donor site dinucleotides at intron boundaries in orthologous intron triads. For instance, we have found 23 cases in which the donor site is GC in human, but GT in both mouse and rat.
According to this classification, 47% (45) of the GC-AG introns are ancient, 36% (34) are modern, and 14% (13) are recent. Because human introns act as a reference out-group, we can establish (under the most parsimonious hypothesis) the direction of the GT/GC switch between mouse and rat orthologous introns. Although the numbers are too small to draw definitive conclusions, we observe more GT to GC than GC to GT substitutions (13 vs. 3). This is obviously mostly due to the overwhelmingly larger number of GT-AG than GC-AG introns, but indicates that switching from GT to GC in the donor site of U2 introns is not completely unfavorable. In this regard, it is interesting to note that GC-AG introns' exhibit a stronger and less variable donor-site sequence than GT-AG introns (Fig. 1). Indeed, the information content of GC-AG donor sites is 12.4, while that of GT-AG donor sites is only 8.2. Probably, the substitution GT→GC, less favorable energetically, needs to be compensated by stronger complementarity in the rest of the site. Indeed, while GC-AG introns make up only 0.7% of all U2 introns (see Table 2), when considering only those U2 introns whose donor-site sequence is the perfect complement to the U1 snRNA 5′ end sequence ([AGC]AG/G[CT]AAGT), then, the percentage of GC-AG introns rises to 11.35% (317 of 2792).
Comparison of splice site sequence patterns
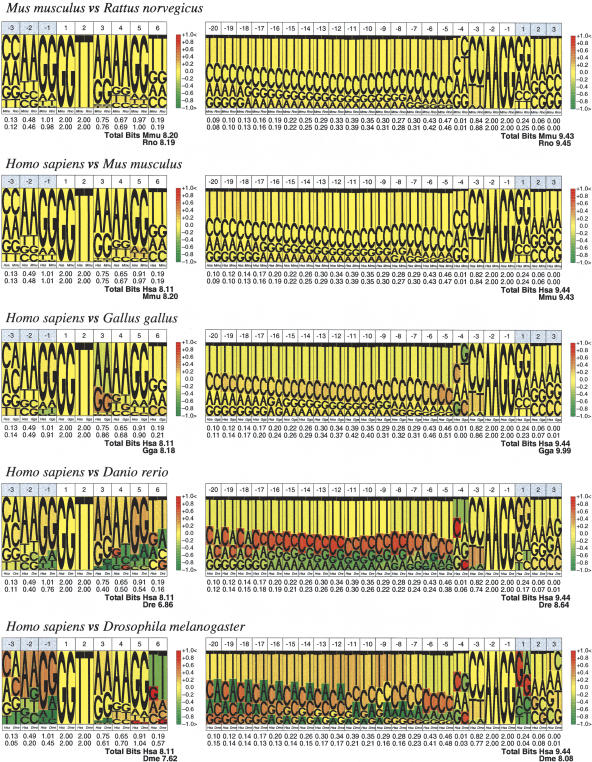
We have investigated here whether the splice-site sequence patterns have changed appreciably since the mammalian and avian split. One way to investigate the variation is to visually compare pictograms or logos (Fig. 1) obtained from collections of sites from different species, derived from the Ensembl database. To facilitate this task, we have extended sequence pictograms into comparative pictograms. In these, the nucleotide distributions of the two species at each position are represented side by side, and the ratio of the nucleotide proportions indexes a range of colors from green to red, indicating nucleotide overrepresentation in one of the two species (see Methods and Supplemental material). Figure 2 shows the comparative pictograms for mouse and rat, human and mouse, and human and chicken. For reference, we have also computed them for human and zebrafish and human and fly. As it is possible to see, comparative pictograms suggest that splice sequence patterns are largely homogeneous within tetrapoda (the pictograms are mostly yellowish), but noticeably distinct from those of other vertebrate and invertebrate taxa. Statistical analysis in which we have explicitly computed the distances between splice-site sequence patterns, using a variety of methods, supports this interpretation (see Supplemental material).
Figure 2.
Comparative pictograms for donor and acceptor splice sites. Comparative pictograms of donor and acceptor sites for pairwise comparisons between species at different phylogenetic distances. At each position, the nucleotide distribution of the two species is displayed, the height of the letters corresponding to their relative frequency at the position. The color in the background of the letters indicates the underrepresentation (green) or overrepresentation (red) of a given nucleotide in the second species (right) with respect to the first (left).
Sequence conservation of orthologous U2 splice sites
In this section, we investigate sequence conservation at orthologous splice sites. Here, we have used the HMRG set of curated mammalian-avian orthologous introns (Methods). In two ways, Figure 3 displays comparisons of orthologous splice sites, the percentage of sequence identity at each nucleotide position in the splice sites and at an intronic region 10 nucleotides long adjacent to the sites. Identity has been computed after aligning the orthologous splice-site sequences at the intron boundaries. Because these alignments are ungapped, the characteristic geometric decay of conservation within the intron observed for mouse-rat and for human-rodent comparisons is suggestive of significant sequence conservation between orthologous introns at this phylogenetic distance. In contrast, for mammalian and chicken comparisons, the ungapped alignment shows an almost abrupt decay right after the splice site—very similar to that observed when comparing unrelated sites.
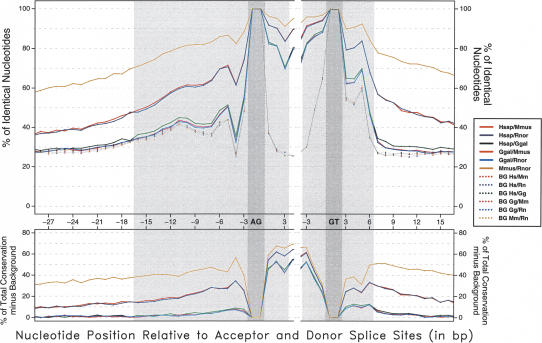
Figure 3.
Sequence conservation level of orthologous GT-AG splice sites. Shaded gray areas correspond to the typical sequence span of splice-site signals. The average identity between the orthologous sequences is plotted across the splice signals (see Discussion). Background identity has been estimated from pairs of nonorthologous sites. (Bottom) The result of subtracting background conservation from total conservation.
To investigate what fraction of sequence conservation in splice sites is due to splicing function, we computed background sequence conservation between pairs of (randomly chosen) nonorthologous sites. As expected, background identity is ∼25% outside of the splice signals. Within the splice signals, background conservation at each position roughly correlates with the information content at that position. Interestingly, at the acceptor site, it exhibits a bimodal shape—consistent with the polypyrimidine tract appearing at two different preferential locations. There is also a slow decay of background conservation upstream of the acceptor site—suggesting that the boundaries of this site are not precisely defined.
As shown in Figure 3, orthologous splice-site sequences are more conserved than expected solely from their role in splicing. Interestingly, this additional conservation is larger than that obtained at adjacent intronic sites for mammalian-chicken comparisons, but not for human-rodent and mouse-rat comparisons (Fig. 3, bottom). The abrupt decay of background conservation right after the donor site allows us to quantify this observation at these sites. This is less obvious in acceptor sites, because their boundaries are not as sharply defined. Indeed, we have computed the average sequence identity in the four rightmost intronic positions of the donor site (positions +3 to +6 in Fig. 1), and at four adjacent positions outside of the site (+7 to +10). The values of background conservation in these two regions are ∼50% and 26%-27%, respectively, for all pairs of species. For mouse-rat orthologous comparisons, the values are 89% and 76%, respectively, for human-mouse, 78% and 53%, respectively, and for human-chicken, 62% and 31%, respectively. That is, conservation due to nonsaturation is smaller at the donor site than at adjacent positions (89 - 50 = 39% vs. 74 - 26 = 48%) for comparisons within rodents, similar for human-rodent comparisons (27% vs. 26%) and larger for human-chicken comparisons (12% vs. 4%). While it cannot be ruled out that this additional conservation reflects the existence of a small class of donor sites conserved beyond the generic consensus, a simpler explanation is that the reaching of saturation (understood here as the level of conservation at which orthologous sites are as conserved as unrelated sites, 27% identity at intronic sites, 50% at donor sites) is slower at sites under functional constraints. In the case of splicing, nucleotide substitutions at the splice sites may impair splice function. Thus, while the substitution process since the divergence of the mammalian and avian lineages has lead to almost complete saturation in proximal intronic sites (31% identity), donor sites (62% identity) are still far from saturation.
Discussion
Thanks to the availability of genome sequences for a number of mammalian and one avian species, we have been able to investigate the dynamics of the evolution of splice-site sequences in recent evolutionary times. Our results confirm that the splicing code is under evolution, albeit very slow. Indeed, while differences between overall splice-site sequence patterns correlate well with phylogenetic distance, they have remained largely homogeneous within tetrapoda, showing noticeable differences only at larger phylogenetic distances—such as those separating tetrapoda from fish.
Even though the splicing code appears to have remained quite constant within tetrapoda, our results also indicate that specific splice-site sequences may suffer significant changes during evolution and remain functional. Figure 3 displays the percentage of sequence identity at each nucleotide position across orthologous splice sites within rodents, between human and rodents, and within mammals and chicken. At all distances, orthologous splice-site sequences are more conserved than unrelated splice sites, but they have significantly diverged, showing an intermediate level of conservation between that of exon and intron sequences. The existence of additional sequences enhancing or repressing the recognition of the splice sites (for instance, see Caceres and Kornblihtt 2002; Cartegni et al. 2002; Black 2003) may partially explain the robustness of the exonic structure in front of changes in the splice-site sequences.
The greater conservation observed in mammalian chicken orthologous splice sites than in unrelated sites indicates that nucleotide substitution since the mammalian avian split has not yet reached saturation at these sites (estimated at ∼50% identity at donor sites). At this phylogenetic distance, however, saturation has been reached at intronic sites, showing a level of conservation similar to that of unrelated sequences. This is the most likely explanation for the excess conservation over background observed in splice sites for comparisons between mammals and chicken, but absent in comparisons within mammals—where saturation has not been reached either at intronic sites.
In any case, the characteristic conservation of orthologous splice sites suggests that comparative prediction of splicing— through the modeling of the conservation in orthologous sites— could improve over methods based on the analysis of a single genome. Comparative prediction of splice sites could be particularly relevant to the prediction of alternative splicing—a problem still poorly solved—since it appears that a large fraction of alternative splicing events are conserved between related species, such as human and mouse (Thanaraj et al. 2003).
The availability of a large collection of orthologous intron sequences has also allowed us to investigate the evolutionary relationship between the minor U12 splicing class, and the major U2 class. Our results seem to indicate that U12 and U2 introns have evolved independently after the split of mammals and birds, since we have not been able to document a single convincing case of conversion between these two types of introns in our data sets. Certainly, because we have used a rather stringent criteria of U12 membership, it cannot be completely ruled out that such cases exist—maybe associated with dramatic changes in exonic structure, which our analysis cannot detect. On the other hand, although subtype switching between GT-AG and AT-AC U12 introns has been documented (Burge et al. 1998), we have not found any such case in our sets of U12 orthologous introns. In contrast, switching between the minor GC-AG and the major GT-AG subtypes within U2 introns is not unusual, and appears to be relatively well tolerated in introns with very strong donor sites. Comparison of orthologous introns has also allowed us to refine the sequences involved in the specification of the U12 introns (see Methods and Fig. 1). These sequences, while more conserved than signals involved in U2 intron specification, are more degenerate that previously thought.
Splicing remains an intriguing phenomenon. The results presented here, however, indicate that the increasing availability of sequences from genomes at different evolutionary distances will greatly contribute to the understanding of splicing, in particular, to understanding its history and its fundamental coding characteristics.
Methods
All of the statistical analyses were performed with the R package (Ihaka and Gentleman 1996; http://www.r-project.org/) using ad hoc scripts for the preparation of exploratory data analysis plots.
RefSeq genes and introns
Assembled chromosomal sequences and their associated annotations were downloaded from the UCSC Genome Browser (Kent et al. 2002; Karolchik et al. 2003; http://genome.cse.ucsc.edu/). The results described in this work were obtained on the assemblies listed in Table 1.
RefSeq genes interrupted with stop codons, or for which the amino acid sequence derived from the genomic coordinates had a difference of more than three amino acids in length or more than five gaps in the alignment when compared with the original amino acid sequence, were discarded. After this filtering step, 16,803 genes from the 21,744 annotated genes of the human HGv16 data set, 9734 genes from the 17,988 of the mouse MGSCv4, and 2783 genes from the 4877 of the rat RGSCv3.1 were retained.
Orthologous mammalian RefSeq introns
Gene sets
The set of homologous gene pairs was downloaded from the NCBI's HomoloGene database (Zhang et al. 2000; http://www.ncbi.nlm.nih.gov/HomoloGene/). From 369,338 homolog pairs, there were 46,522 pairs corresponding to human-mouse, human-rat, or mouse-rat orthologous genes. Redundancy was removed in order to keep only unique putative ortholog pairs. Only those gene pairs in which the two members were in the final gene set resulting after the filtering process above were taken into account. Ternaries of human, mouse, and rat genes were built when possible. Otherwise, the gene pairs were considered.
This process yielded 1283 human-mouse-rat triads. In addition, 4267 human-mouse ortholog pairs, 454 human-rat pairs, and 133 mouse-rat pairs were obtained. These numbers correspond to 6043, 5680, and 1847 unique RefSeq genes for human, mouse, and rat, respectively. When performing pairwise comparisons, the corresponding genes in the triads were included in the set of pairs. Thus, the resulting extended pair-wise sets contained 5550 human-mouse, 1737 human-rat, and 1416 mouserat pairs. All data sets, as well as graphical displays of sequence comparisons of the orthologous sequences are available from http://genome.imim.es/datasets/hmrg2004/.
Introns sets
We devised a protocol to extract orthologous intron pairs and triads from the above set of orthologous genes. First, all of the pairs of consecutive exons for each gene were aligned with t_coffee (Notredame et al. 2000; http://igs-server.cnrs-mrs.fr/cnotred/Projectshomepage/tcoffeehomepage.html) using default parameters against all of the exonic pairs from the corresponding orthologous genes. This step ensured that we were working with the most accurate set of orthologous introns, despite changes in the exonic structure of orthologous genes (such as missing exons due to misannotations or gaps in the assemblies). Second, the exonic structure of the gene was projected onto the alignments. Third, from orthologous gene pairs or ternaries, only those exon pairs in which all intron positions occurred at conserved positions in the alignment and the intron phases were conserved and retained. Plots on which the exonic structures have been projected onto the alignments can be accessed at http://genome.imim.es/datasets/hmrg2004/.
Orthologous HMRG introns
A set of human, mouse, rat, and chicken 1:1:1:1 confident orthologous introns was taken from International Chicken Genome Sequence Consortium (2004) (P. Bork and I. Letunic, pers. comm.). The set consisted of 1041 orthologous genes, totalizing 9110 orthologous introns. After mapping those genes into the annotations for the newer assemblies used in this analysis, 863 genes and 6524 introns remained in the four species orthologous set. The sequences 75 bp upstream and downstream of the signal core nucleotides (GT and AG for instance) were used in the orthologous splice-sites' sequence conservation analysis.
Intron class
U12 introns were searched, relying on the conserved donor-site sequence and the acceptor-site branch point. Mammalian introns were initially considered to be U12 if (1) they matched the motif [AG]TATCCTT (where [AG] means A or G) from position +1 at the donor splice site; and (2) they matched the motif TCCT T[AG]A[CT] at the region from -5 to -20 upstream the acceptor splice site. When looking for the U12 branch point, up to two mismatches were allowed, and the hit was accepted if at least one adenine was found in position 6 or 7 of the motif—to avoid branch point hits without biological sense. Visual inspection of introns orthologous to U12 introns, but which initially failed to meet this criteria, suggested that this initial definition is too stringent. Therefore, we searched only for the presence of a strong branch point signal at the appropriate location in orthologous introns. After inspection of all of those cases in which the two orthologous introns contain such a signal, we found a few additional cases in which the donor-site sequences strongly resemble the characteristic U12 donor site sequence, but failed to match the consensus above. Indeed, we have found that only the nucleotides at positions +2 (T), +3 (A), +4 (T), and +5 (C) within the intron are absolutely conserved in U12 donor-site sequences (TATC). Position +6, thought to be an invariable C (Burge et al. 1999), may also be a T, and positions +7 and +8 can actually be occupied by any nucleotide. This more degenerate pattern was the one used to identify chicken U12 introns, where, at most, a gap (in addition to one mismatch) was also allowed to match the branch-point consensus. These results, which help to characterize the sequences that define U12 introns, illustrate the power of comparative genomics to refine our knowledge of the functional sequences encoded in eukaryotic genomes.
Mapping of mammalian U12 introns into the chicken genome
DNA sequences of the exon-pairs delimiting each U12 intron were mapped into chicken genomic sequences using exonerate (http://www.ebi.ac.uk/guy/exonerate/). Only those alignments that preserved the mammalian splice site were taken into account. Introns obtained in that way were classified into U2/U12 classes following the same criteria as in the above section.
Comparison of splice site sequence patterns
We have quantified the different use of nucleotides in splice sites by different species and represent it by comparative pictograms. A comparative pictogram is a graphical representation of the nucleotide proportions observed in two different sets of aligned sequences. In this article, these sets are splice sites of different species and the proportions are calculated for every position along the splice site. As in sequence pictograms, the sizes of nucleotides scale with their observed proportions, but here the nucleotides of the two sets are put side by side to ease their comparison. Moreover, the background occupied by each nucleotide is colored with the ratio of the proportions (the relative risk). Further details are given in the Supplemental material.
We have further analyzed the different nucleotide usage in splice sites of different species by two kinds of comparisons as follows: (1) by building confidence intervals for the relative risks and counting how many of them include a ratio value of 1 (i.e., no difference of nucleotide usage), and (2) by assessing the site species dependence, that is, the extent to what the occurrences of the observed splice sites depend, statistically speaking, on the species to which they belong to. Further details are given in the Supplemental material also.
Acknowledgments
We thank the International Chicken Genome Sequencing Consortium for providing the genomic sequences, as well as the Rat and Mouse Consortia from past collaborations. We are particularly grateful to Ivica Letunic and Peer Bork for providing the set of HMRG orthologous introns with which some of the analyses were performed. Juan Valcárcel, Genís Parra, Eduardo Eyras, Webb Miller, David Haussler, Robert Baertsch, Chris Ponting, Alberto Roverato, Kim Worley, and two anonymous referees are gratefully acknowledged for advice and helpful comments. We also thank òscar González for keeping the database mirrors up to date. Special thanks to Jan-Jaap Wesselink and Charles Chapple for their suggestions when proofreading this document. J.F.A. is supported by a predoctoral fellowship from the “Fundació IMIM” (Spain). This research is supported by grant BIO2000-1358-C02-02 from “Plan Nacional de I+D” (Spain), and grant ASD from the European Commission.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3108805. Article published online before print in December 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: P. Bork and I. Letunic.]
References
- Birney, E., Andrews, T.D., Bevan, P., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cuff, J., Curwen, V., Cutts, T., et al. 2004. An overview of Ensembl. Genome Res. 14: 925-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291-336. [DOI] [PubMed] [Google Scholar]
- Burge, C.B. and Karlin, S. 1998. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 8: 346-354. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Padgett, R.A., and Sharp, P.A. 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2: 773-785. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.S. 1999. Splicing precursors to mRNAs by the spliceosomes. In The RNA world (eds. R.F. Gesteland et al.), pp. 525-560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Burset, M., Seledtsov, I., and Solovyev, V. 2001. SpliceDB: Database of canonical and noncanonical mammalian splice sites. Nucleic Acids Res. 29: 255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186-193. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285-298. [DOI] [PubMed] [Google Scholar]
- de Souza, S.J. 2003. The emergence of a synthetic theory of intron evolution. Genetica 118: 117-121. [PubMed] [Google Scholar]
- Dermitzakis, E.T., Reymond, A., Lyle, R., Scamuffa, N., Ucla, C., Deutsch, S., Stevenson, B.J., Flegel, V., Bucher, P., Jongeneel, C.V., et al. 2002. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420: 578-582. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.C., Incorvaia, R., and Padgett, R.A. 1997. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell 1: 151-160. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.C., Peris, M.J., Seyboldt, A.S., and Padgett, R.A. 2001. Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol. 21: 1942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova, L. and Fedorov, A. 2003. Introns in gene evolution. Genetica 118: 123-131. [PubMed] [Google Scholar]
- Hall, S.L. and Padgett, R.A. 1994. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol. 239: 357-365. [DOI] [PubMed] [Google Scholar]
- Hare, M.P. and Palumbi, S.R. 2003. High intron sequence conservation across three mammalian orders suggests functional constraints. Mol. Biol. Evol. 20: 969-978. [DOI] [PubMed] [Google Scholar]
- Helfman, D.M. and Ricci, W.M. 1989. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 17: 5633-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka, R. and Gentleman, R. 1996. R: A language for data analysis and graphics. J. Computat. Graph. Stat. 5: 299-314. [Google Scholar]
- International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature (in press). [DOI] [PubMed]
- Jackson, I.J. 1991. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 19: 3795-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., Baertsch, R., Diekhans, M., Furey, T.S., Hinrichs, A., Lu, Y.T., Roskin, K.M., Schwartz, M., Sugnet, C.W., Thomas, D.J., et al. 2003. The UCSC genome browser database. Nucleic Acids Res. 31: 51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov, G., Castellano, S., Novoselov, S., Lobanov, A., Zehtab, O., Guigó;, R., and Gladyshev, V. 2003. Characterization of mammalian selenoproteomes. Science 300: 1439-1443. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Levine, A. and Durbin, R. 2001. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 29: 4006-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J.S. 2001. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2: 986-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., and Heringa, J. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205-217. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D., Tatusova, T., and Maglott, D.R. 2003. NCBI reference sequence project: Update and current status. Nucleic Acids Res. 31: 34-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Roy, S.W., Fedorov, A., and Gilbert, W. 2003. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc. Natl. Acad. Sci. 100: 7158-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, T. and Stephens, R. 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18: 6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P. and Burge, C. 1997. Classification of introns: U2-Type and U12-Type. Cell 91: 875-879. [DOI] [PubMed] [Google Scholar]
- Smith, C.W. and Nadal-Ginard, B. 1989. Mutually exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location: Implications for constitutive splicing. Cell 56: 749-758. [DOI] [PubMed] [Google Scholar]
- Thanaraj, T., Clark, F., and Muilu, J. 2003. Conservation of human alternative splice events in mouse. Nucleic Acids Res. 31: 2544-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Modrek, B., and Lee, C. 2002. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 30: 3754-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G., Holste, D., Kreiman, G., and Burge, C. 2004. Variation in alternative splicing across human tissues. Genome Biol. 5: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203-214. [DOI] [PubMed] [Google Scholar]
- Zhu, W. and Brendel, V. 2003. Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res. 31: 4561-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://genome.imim.es/datasets/hmrg2004/; further supplemental materials for this study.
- http://genome.cse.ucsc.edu/; UCSC Genome Browser, from which the human, mouse, rat and chicken feature annotations and genome assemblies used in this study were downloaded.
- http://www.ensembl.org/; Ensembl Genome Browser, from which a larger set of human, mouse, rat and chicken gene annotation sets were retrieved.
- http://www.ncbi.nlm.nih.gov/HomoloGene/; NCBI's HomoloGene database, from where initial RefSeq orthologous pairs were obtained.
- http://igs-server.cnrs-mrs.fr/cnotred/Projectshomepage/tcoffeehomepage.html; a multiple sequence alignment package.
- http://www.ebi.ac.uk/guy/exonerate/; a generic tool for sequence comparison.
- http://www.r-project.org/; the R project for statistical computing.