Abstract
Misfolding and aggregation of amyloidogenic polypeptides lie at the root of many neurodegenerative diseases. Whilst protein aggregation can be readily studied in vitro by established biophysical techniques, direct observation of the nature and kinetics of aggregation processes taking place in vivo is much more challenging. We describe here, however, a Förster resonance energy transfer sensor that permits the aggregation kinetics of amyloidogenic proteins to be quantified in living systems by exploiting our observation that amyloid assemblies can act as energy acceptors for variants of fluorescent proteins. The observed lifetime reduction can be attributed to fluorescence energy transfer to intrinsic energy states associated with the growing amyloid species. Indeed, for α-synuclein, a protein whose aggregation is linked to Parkinson’s disease, we have used this sensor to follow the kinetics of the self-association reactions taking place in vitro and in vivo and to reveal the nature of the ensuing aggregated species. Experiments were conducted in vitro, in cells in culture and in living Caenorhabditis elegans. For the latter the readout correlates directly with the appearance of a toxic phenotype. The ability to measure the appearance and development of pathogenic amyloid species in a living animal and the ability to relate such data to similar processes observed in vitro provides a powerful new tool in the study of the pathology of the family of misfolding disorders. Our study confirms the importance of the molecular environment in which aggregation reactions take place, highlighting similarities as well as differences between the processes occurring in vitro and in vivo, and their significance for defining the molecular physiology of the diseases with which they are associated.
Keywords: amyloid beta-peptides, biosensors, FLIM, protein folding, synuclein
1. Introduction
The phenomenon of protein misfolding and aggregation is one of the most important topics in current biomedical research.[1] The failure of proteins to fold correctly, or to remain correctly folded, lies at the centre of a growing number of increasingly prevalent pathological conditions, referred to collectively as misfolding disorders, that range from Parkinson’s and Alzheimer’s diseases to type II diabetes, and cystic fibrosis.[1] One of the most common pathogenic consequences of protein misfolding is the formation of aggregates that results in the deposition of the protein concerned as amyloid fibrils and plaques.[2] These species are associated with the most common forms of neurodegenerative disorders, particularly those associated with ageing, and therefore an understanding of their formation and behaviour in vivo is of paramount importance in the modern world.[1, 3] A key requirement in developing such an understanding in the context of disease is the ability to observe directly the nature and kinetics of the aggregation process both in vitro and in vivo. This objective has, however, proved to be extremely challenging to achieve and indeed the physiological roles of the variety of misfolded species formed during the process of aggregation in situ are not yet understood in any detail.[4] Traditional imaging techniques, such as transmission electron microscopy (TEM) and atomic force microscopy (AFM), are powerful methods for studying protein aggregation in excised tissue or in vitro,[1] but because of their invasive nature they are not readily applicable to living organisms. Optical techniques do not suffer from such limitations and have made it possible to visualise, non-invasively and in vivo, events taking place on a molecular scale.[5]
Herein we report on a novel FRET (Förster resonance energy transfer) sensor for studying the oligomerisation of proteins and peptides in vitro and in vivo, and apply it to α-synuclein (AS), a small protein localised in the presynaptic terminal of neurons, and whose aggregation is linked to the pathology of Parkinson’s disease (PD) and related neurodegenerative diseases, including dementia with Lewy bodies.[6] α-Synuclein amyloid fibrils are the main component of Lewy bodies, protein-rich inclusions found in the cytoplasm of dopaminergic neurons in the brains of patients suffering from these disorders.[7] It has been shown in vitro that AS, like other amyloidogenic peptides and proteins, forms ß-sheet rich oligomers and prefibrillar aggregates prior to the assembly of mature amyloid fibrils .[8] These intermediate species are now widely thought to act as the primary pathogenic agents in PD and related diseases,[9] for example, disrupting membrane homeostasis[4, 10] or impairing protein degradation pathways.[1, 11]
We report herein that amyloid growth is accompanied by the formation of energy states which lead to the development of intrinsic fluorescence in non-labelled, aggregated amyloidogenic proteins, such as AS. These corresponding energy states exhibit spectral overlap with the emission spectrum of a yellow fluorescent protein variant (termed Venus; but here referred to as YFP),[12] which we fused to the C-terminus of AS to form AS–YFP. FRET thus takes place between YFP and the growing AS oligomers with an efficiency that increases with oligomer size. FRET between YFP (donor) and the forming aggregate (acceptor) is conveniently quantified by measuring the excited state lifetime of YFP using time correlated single photon counting (TCSPC) fluorescence lifetime microscopy. The sensor was applied to monitor aggregation in vitro, that is, of the purified protein in solution, in cells in culture, and also in vivo in the nematode worm Caenorhabditis elegans (C. elegans). We found that AS–YFP is more likely to aggregate into fibrillar species in conditions representative of neuronal rather than non-neuronal environments and that AS–YFP-mediated toxicity is directly correlated with a lifetime signature of intermediate, pre-fibrillar species in a living organism. This method therefore permits a detailed characterisation of the species associated with neurodegenerative disorders in situ and in a non-invasive manner.
2. Results
2.1. Fluorescence Lifetime is a Sensitive Readout for Amyloid-Related Aggregation of AS
During the course of experimenting with amyloidogenic proteins in vitro we noticed that AS develops an intrinsic fluorescence, which is concomitant with its conversion into aggregates. The spectrum and the lifetime of the developing intrinsic fluorescence in amyloid fibrils both shift as the aggregates grow in size due to the increase in β-sheet content, which appear to promote electron delocalisation.[13] This intrinsic fluorescence can be excited in the broad range of 450 to 550 nm and emits in the 500 to 700 nm range with corresponding lifetimes ranging from 1000 to 3000 ps as the aggregates form (Figure 1A).[13] Due to the low quantum yield of this intrinsic fluorescence, it is not readily exploitable for in vivo imaging of amyloid formation, but instead it is possible to construct a FRET sensor by conjugation of a suitable fluorophore exhibiting spectral overlap with the aggregate-specific excitation spectrum. We have developed such a sensor by fusing YFP to AS, enabling YFP to act as a donor during FRET with the aggregated portion of the protein. As a convenient and accurate measure of FRET efficiency, we monitored the lifetime changes of the YFP donor fluorescence on a TCSPC confocal microscope setup.[14, 15]
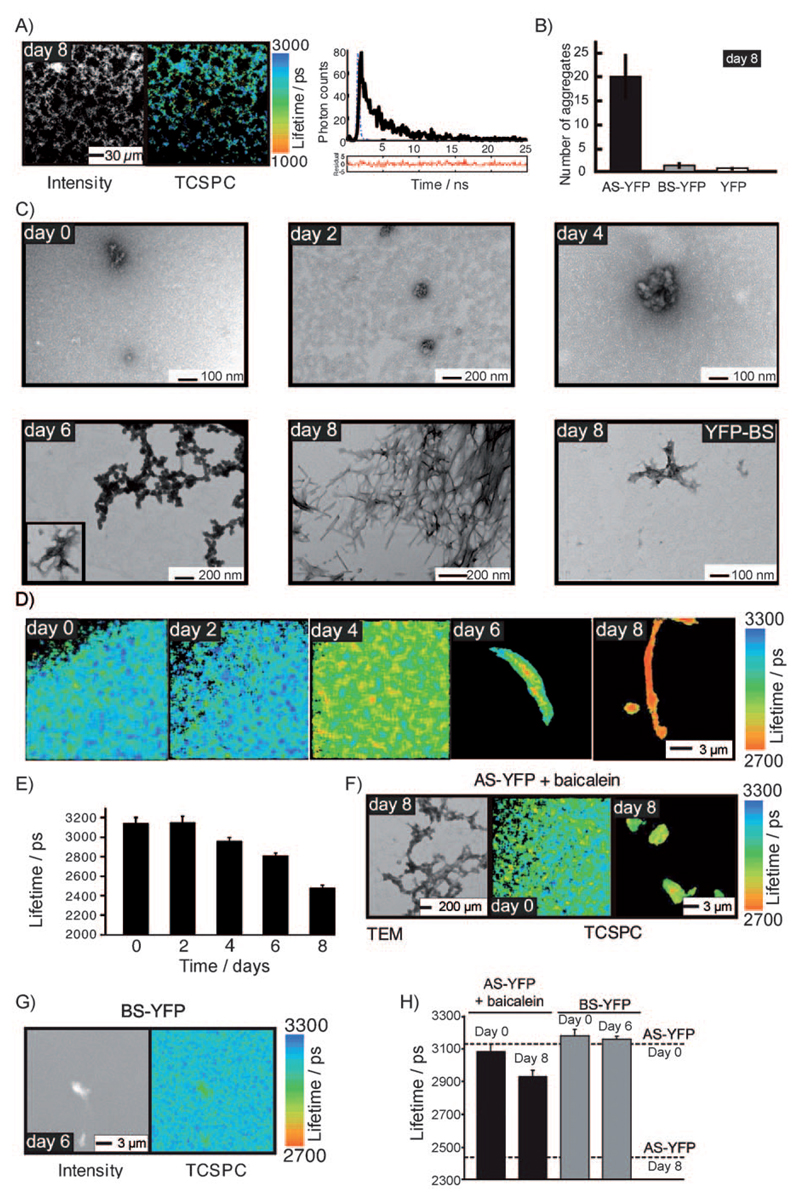
Figure 1. TCSPC lifetime imaging distinguishes different molecular species of AS–YFP formed during the aggregation process in vitro.
A) Left: Grey scale intensity image of unlabelled (wild type) AS at day 8 of the aggregation assay. Middle: corresponding TCSPC image. Aggregates of unlabelled AS exhibit a characteristic fluorescence life-time. Right: Representative fluorescence lifetime decay corresponding to one pixel in the TCSPC image. Blue line: instrument response curve; black solid line: measured lifetime decay; black dashed line: fitted exponential curve; red line: residuals of fitted data. B) Number of fluorescent aggregates of YFP variants per picolitre quantified in a blind fashion on a confocal microscope. C) TEM images showing the morphological changes of AS–YFP aggregates as a function of time. The last image of the series illustrates a rare aggregate in the BS–YFP control sample at day 8 of the aggregation assay. D) TCSPC confocal images of AS–YFP in different aggregation states during the time course of the aggregation assay. E) Bar plot of the average fluorescence lifetime of samples of AS–YFP recorded at days 0, 2, 4, 6, and 8 of the aggregation assay. Note how the drop in fluorescence lifetime correlates with the appearance of amyloid fibrils. F) Left: TEM image of AS–YFP treated with the generic amyloid inhibitor baicalein showing non-fibrillar amorphous aggregates. Right: TCSPC confocal image showing amorphous aggregates of baicalein-treated AS–YFP at days 0 and 8. G) TCSPC lifetime and intensity confocal images of BS–YFP incubated in vitro for 6 days at 37°C. Occasionally, small amorphous aggregates can be detected in BS–YFP samples at day 6. These aggregates do not, however, display a similar reduction in the fluorescence lifetime similar to that of the AS–YFP samples and do not show the typical amyloid morphology by TEM. H) Plot of average lifetime for AS–YFP samples treated with baicalein at days 0 and 8 and BS–YFP samples at days 0 and 6. Lifetimes for AS–YFP at days 0 and 8 are indicated for comparison. In all cases the mean lifetime ±SEM is plotted (n > 5).
In previous papers we have described comprehensive biophysical and physiological studies, which show that the AS–YFP fusion protein serves as a valid model for the aggregation of wild type (wt) AS both in vitro and in vivo.[16, 17] Standard in vitro aggregation assays have confirmed that AS–YFP forms fluorescent aggregates, which retain the amyloid character of the species formed by wt AS when subjected to standard in vitro aggregation assays (Figure 1C, day 8). Conversely YFP itself, and also a YFP fusion with ß-synuclein (BS–YFP), the latter a homologue of AS which has a much lower aggregation propensity than AS and which is not linked to neurotoxicity associated with disease,[18] generate negligible quantities of aggregates under the conditions prevailing here (Figure 1B and Figure 1C, BS–YFP).
The power of the FRET assay was established by measuring the fluorescence lifetime of purified AS–YFP throughout the time-course of its aggregation reaction in vitro, and then correlating the data with TEM images of the same samples taken at corresponding time points. The data show that during the initial phase of the aggregation processes only a small number of aggregates could be detected by TEM, and no measurable change is observed in the fluorescence lifetime (3140±60 ps on average) throughout this phase (Figures 1D,E, days 0 and 2). From day 4 onwards, however, TEM images show an increasing number of approximately spherical species, with diameters of up to about 60 nm (Figure 1C, day 4), that are typical of oligomeric precursors of fibrillar assemblies;[8] concomitantly with the appearance of these species, the average lifetime of the AS–YFP fluorescence is shortened by about 180 ps to 2960±40 ps (Figures 1D,E, day 4). The reduction in the fluorescence lifetime of AS–YFP is even more pronounced after longer periods of aggregation (Figures 1D,E, days 6 and 8), when the TEM data show that the oligomeric precursors are replaced by proto-fibrillar species (Figure 1C, day 6), and later by mature amyloid fibrils (Figure 1C, day 8) with widths of 16±2 nm typical of such species,[8] and the fluorescence lifetime decreases to 2480±30 ps (Figures 1D,E, day 8). Moreover, an analysis of lifetime histograms permits the nature of the aggregated species to be discerned (Supporting Information, Figure 1A).
In order to explore further the nature of the change in fluorescent properties of AS–YFP, control experiments were carried out under conditions where the propensity to aggregate is much reduced. No change in lifetime indicative of amyloid formation could be detected when AS–YFP was placed under aggregation conditions in the presence of baicalein, a flavonoid that strongly inhibits protein fibril formation (Figures 1F,H).[19] Furthermore, no decrease in lifetime was observed for BS–YFP when placed under conditions analogous to AS–YFP (Figure 1G and Figure 1H), consistent with the much lower aggregation propensity of BS compared to AS.
2.2. Fluorescence Lifetime Microscopy can Detect AS Aggregates within Living Cells and Define their Nature
We next evaluated the capability of the FRET sensor to probe the nature of the species resulting from aggregation of AS–YFP in a cellular environment. We introduced purified monomeric AS–YFP into cultured human neuroblastoma cells (SH-SY5Y) by means of electroporation, a powerful method for the rapid and precise delivery of biomolecules into cells through the induction of transient permeability within cellular membranes[20] (Figure 2A). Lifetime images were then recorded, starting from 3 h after electroporation and continuing for 4 days. AS–YFP remaining outside the cell, that is, recombinant AS–YFP that had not diffused into cells upon electroporation (referred to as ex cell in Figure 2B), yielded lifetimes comparable to monomeric AS–YFP measured in solution (see Figure 1E and Figure 2B), indicating that the electroporation process itself has no effect on the aggregation state of the protein.
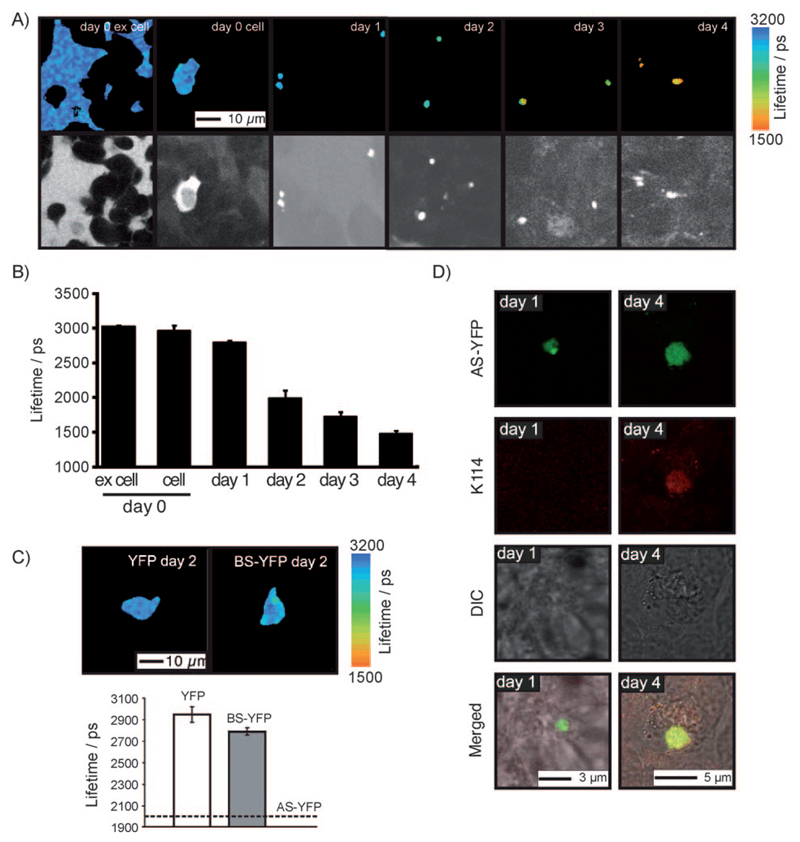
Figure 2. AS–YFP aggregation kinetics in a human SH-SY5Y neuroblastoma cell line are revealed by confocal and TCSPC fluorescence imaging.
A) Colour pictures: TCSPC images of SH-SY5Y cells after electroporation with AS–YFP: (ex cell) referring to extracellular fluorescence of AS–YFP measured 3 h after electroporation; (cell) referring to the homogeneous cytosolic distribution of AS–YFP at day 0. Day 1–4 images show the monotonic decrease in the fluorescence lifetime of cytosolic aggregates over four days in culture. Black and white pictures: Intensity data of corresponding TCSPC images. B) Average fluorescence lifetime of AS–YFP in SH-SY5Y cells in culture, determined for days 0 to 4 after electroporation. C) Average fluorescence lifetime for YFP and BS–YFP measured 2 days after electroporation into SH-SY5Y cells. The corresponding lifetime of AS–YFP is shown for comparison. D) K114 staining of 1- and 4-day-old AS–YFP aggregates in electroporated SH-SY5Y cells. Note that only AS–YFP aggregates observed at day 4 are K114 positive, indicating the amyloid nature of these deposits. In all cases the mean lifetime ±SEM is plotted (n > 5).
The cell culture medium was subsequently replaced with fresh medium, and from this point onwards we analysed as a function of time the location and characteristics of the AS–YFP species inside the cells by TCSPC microscopy. Using pixel-based analysis of the images, we measured an average lifetime of 3020±20 ps for the extracellular recombinant AS–YFP, and 2970±70 ps for the homogenously distributed cytoplasmic AS–YFP at the starting point of the experiment (Figures 2A,B) falling to 1480±40 ps after 4 days (Figures 2A,B, day 4). Measurements at subsequent times revealed that the lifetime of AS–YFP changed significantly over the time course of these experiments (Figures 2A,B). Importantly, no time-dependent changes in lifetime were observed in control cells electroporated with YFP alone or with the less amyloidogenic protein species BS–YFP (Figure 2C).
In order to correlate the observed decrease in the fluorescence lifetime of AS–YFP in SH-SY5Y cells with the nature of the aggregates, we counterstained the cells with K114, a fluorescent dye that shows enhanced fluorescence upon binding to amyloid species.[21] As shown in Figure 2D, K114-positive AS–YFP-aggregated species were found in cells at day 4 in culture, whereas no K114-positive aggregates could be detected at day 1 in culture. This finding clearly demonstrates that the shorter lifetimes observed with cells in culture at the longer incubation times are indicative of the appearance of deposits of amyloid AS–YFP fibrils.
2.3. The Nature and Kinetics of AS Aggregation in a Live Animal can be Probed by Fluorescence Lifetime Imaging
The capacity of the FRET sensor to identify different aggregated forms of AS–YFP within living cells in culture prompted us to monitor protein aggregation in vivo within a multicellular organism, namely the transparent nematode worm C. elegans. This model organism has been employed extensively for the study of protein aggregation, particularly in the context of Huntington’s, Alzheimer’s and Parkinson’s diseases.[16, 22–24] Moreover, the short lifespan of the worm makes it an ideal model organism for studying protein aggregation in the context of neuro-degeneration and ageing.[22] In a previous study, we established a C. elegans model where overexpression of the AS–YFP transgene in muscle cells resulted in fluorescent aggregates.[16]
Here, we apply the sensor in this system to explore its value in revealing amyloid aggregation directly in a higher-order organism. Images of the whole anterior regions of intact C. elegans were taken at various time points during the lifespan of AS–YFP transgenic worms, and the average fluorescence lifetime determined in each case. Again, the fluorescence lifetime of AS–YFP decreases (by ca. 130 ps) during the aggregation process (Figure 3A, second image in panel, and Figure 3B, black bars). These data indicate that TCSPC imaging is indeed able to probe directly the kinetics of aggregation of AS in a multicellular living organism. Importantly, no such reduction in lifetime is evident for the control animals containing a YFP only transgene, coincident with the absence of protein aggregates in this control (Figure 3A, first image of panel, and Figure 3A, white bars). Moreover, an analysis of lifetime histograms permits the nature of the aggregated species to be discerned in living C. elegans (Supporting Information, Figure 1B).
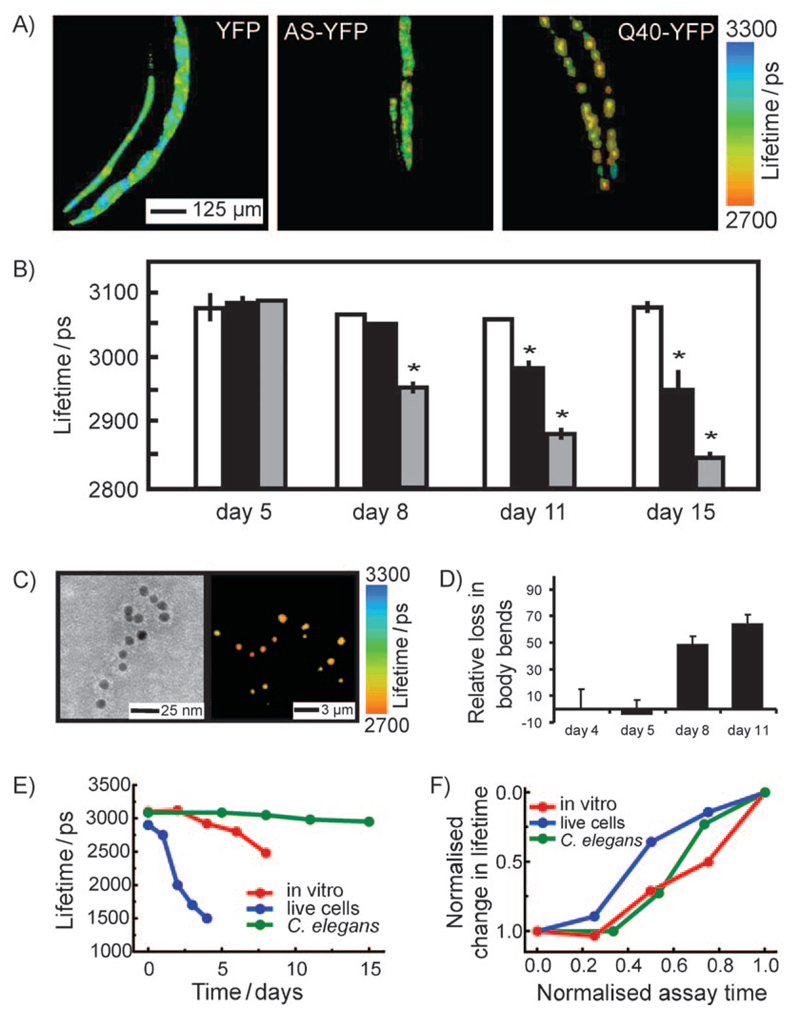
Figure 3. TCSPC lifetime imaging distinguishes the nature and kinetics of the aggregation of amyloidogenic YFP fusion protein variants during ageing in a living animal.
A) Left: TCSPC image of YFP transgenic C. elegans at day 11; middle: TCSPC image of AS–YFP transgenic C. elegans at day 11; right: TCSPC image of Q40-YFP transgenic C. elegans at day 11. Note how the fluorescent aggregates display a strong reduction in the fluorescence lifetime (red colour) with time. B) Mean fluorescence lifetimes of the whole anterior part of transgenic worms expressing YFP (white), AS–YFP (black), and Q40-YFP (grey) during ageing of the animals, determined at various days during the assay (mean lifetime ±SEM, ANOVA with Scheffes’ post hoc test, p<0.05). C) Ex situ studies of in vivo aggregated AS–YFP. Left: anti-AS immunogold-labeled TEM image of cell extracts from aged AS–YFP transgenic C. elegans (day 15). Only pre-fibrillar aggregates of AS–YFP are observed in these samples. Right: TCSPC image corresponding to the same sample as that shown on the right. D) AS–YFP-dependent toxicity during ageing in C. elegans. The plot shows the ratio (%) of number of bends/minute for the AS–YFP transgene versus the YFP only transgene. Because of the selective expression of the transgene in muscle cells, the toxicity phenotype is a perturbation of the motility of the worm and the toxicity readout is a reduction in the number of bends per minute. E) Time-dependence of the decrease of fluorescence lifetime for AS–YFP in the three systems studied herein (in vitro, red; living SH-SY5Y cells, blue; living C. elegans, green). F) Normalised kinetics of AS–YFP aggregation in vitro and in vivo determined by the decrease in fluorescence lifetime. The colour code is the same as in (E).
In order to explore the wider applicability of this methodology, we carried out an additional series of experiments to probe the aggregation of Q40-YFP, a 40-residue polyglutamine sequence (Q40) attached to YFP, again using C. elegans as a model organism. The Q40 construct forms intramuscular inclusions, and acts as a model for polyglutamine expansion disorders,[22] including Huntington’s disease and various types of spinocerebellar ataxia.[1] TCSPC confocal imaging of the Q40-YFP species in C. elegans shows a pronounced decrease in the lifetime of the fluorophore inside the protein-rich inclusions relative to the soluble monomeric form, as was found for AS–YFP. Indeed, the Q40-YFP construct shows a particularly large change in the mean fluorescence lifetime as a function of time, with the system displaying a shift of around 260 ps by the end of the assay (Figure 3A, third image of panel, and Figure 3B, grey bars), a result attributable to the much greater (almost 3 orders of magnitude) rate of aggregation of Q40 relative to AS in vitro.[25]
To investigate further the nature of the aggregated material in C. elegans, we isolated the aggregated AS–YFP species from the worms at different time points and subjected them to parallel TCSPC and immuno-TEM analysis. This analysis revealed the presence of AS immuno-positive, non-fibrillar aggregates with lifetimes of around 2700 ps (Figure 3C). This value is close to that observed for pre-fibrillar species observed in vitro, indicating that the aggregates formed in the C. elegans muscle tissue are less ordered species than more mature amyloid fibrils observed in vitro and in mammalian neurons.
This result reveals the power of the TCSPC method to identify the nature of the aggregates within an organism rather than simply their presence. In this context, it is particularly interesting that we observed a strong correlation between the decrease in fluorescence lifetime and the number of body bends performed by AS–YFP expressing worms compared to control worms expressing YFP only (Figure 3D). As the reduction in the frequency of body bends is associated with cellular damage, the value of the fluorescence lifetime observed for AS-YFP in this study strongly supports the view that it is the pre-fibrillar species rather than mature fibrils that give rise to the high toxicity associated with neurodegenerative disorders.
Moreover, this work has enabled a sensor to be developed that permits the processes of polypeptide aggregation to be studied under laboratory conditions, in cells in culture and in live animals so that they can be directly compared (Figure 3E and Figure 3F). In particular, in all three aggregation reactions a lag phase could be identified that lasts for about 30% of the time of the assay, after which aggregation proceeds with comparable rates in each case (Figure 3F).
3. Discussion
We describe here the development of a novel FRET assay that is able to reveal the nature and the kinetics of self-association reactions taking place in vitro and in vivo for amyloidogenic polypeptides using confocal fluorescence lifetime imaging microscopy. Our method makes use of a YFP fusion with the polypeptide of interest in which fluorescence emitted by the YFP moiety is quenched as a result of FRET between the YFP moiety and the amyloid structure. Moreover, the occurrence of FRET reports not only on the conversion of the polypeptide species into aggregates, but also on the nature of the aggregates. These effects are conveniently and accurately determined by TCSPC measurements of the excited-state lifetime of the donor fluorophore (YFP) using confocal microscopy.
Herein, we focused particularly on AS because of its connection with PD. Species formed during self association reactions of AS–YFP in vitro were analysed by EM and fluorescence spectroscopy, and a distinct correlation is observed between the fluorescence emission lifetime and aggregate size. Following self-association and aggregation, proteins such as AS and other amyloidogenic polypeptides form highly organised supramolecular ß-sheet structures particularly rich in hydrogen bonds.[26] It has been suggested that stacking of ß-sheet structures can lead to electron delocalisation[27] and that this effect could be the origin of changes in the molecular properties of amyloidogenic polypeptides and polymer systems.[13, 28] Here, we exploited the fact that this phenomenon can lead to the formation of intrinsic energy states in the growing aggregates with excitation and emission spectra in the visible range. The growing amyloid structures can thus act as acceptors of energy from suitable donor fluorophore-labels such as YFP. HeteroFRET occurring between donors and acceptors provides an explanation for the observed lifetime reductions in AS–YFP upon aggregation and forms the basis of our aggregate sensor. Of particular importance is that the FRET efficiency, and thus the YFP donor lifetime, is strongly dependent on the size and structure of the aggregates. Note that a complementary energy transfer mechanism between YFP molecules can also occur during aggregation of AS–YFP, termed homoFRET.[17, 29] HomoFRET between YFP cannot, however, account for the observed lifetime reductions, because in homoFRET donors and acceptors have indistinguishable fluorescence properties, preventing a net lifetime reduction to be observed.[29] Indeed, in a previous study we have verified that homoFRET occurs in AS–YFP aggregates and that its efficiency decreases when the ratio of labelled to unlabelled protein is reduced.[17] In contrast, the lifetime reductions observed with the present technique in growing aggregates remain intact and independent of the fluorophore labelling density. Most importantly, the lifetime reductions are aggregate size specific because the probability for energy transfer increases as the protein structures grow, akin to a situation where the acceptor density is increased. Although homo-FRET is very sensitive and useful to detect the onset and presence of aggregates and changes in monomer/aggregate ratio, it does not provide size-specific information beyond cluster sizes of N~4. For the latter situation, fluorescence signals become almost completely depolarised and thus homoFRET is indistinguishable from different clusters.[29, 30]
The lifetime of AS–YFP within cells was found to decrease monotonically with time, indicative of aggregation and similar to the changes observed in the in vitro assay. Interestingly though, the lifetime reduction in cells in culture is more pronounced, and occurs more rapidly than that in vitro, with lifetimes as short as 1480±40 ps observed by the fourth day of culture, compared to that for the fibrils formed in vitro. The molecular environment in the cytoplasm, particularly the levels of dopamine present in SH-SY5Y cells, is known to increase the rate of AS aggregation[31] as are other factors, such as salt and intracellular calcium concentration.[31, 32] Moreover, AS has been shown under certain conditions to become localised and degraded in lysosomes and autophagosomes,[33] both of which are highly acidic compartments of the cells. Since a lower pH also enhances AS aggregation and modifies the morphology of the resulting fibrils[34] there are likely to be differences between the lifetime changes in vivo and in vitro. Further experiments to define these details should therefore increase the value of this approach, and enable the development of live cell screens with which to investigate, for example, the effects of mutations of variants or potential inhibitors of aggregation.
The total decrease in fluorescence lifetime observed for AS–YFP in C. elegans is less pronounced than that observed in vitro or in neuronal cells in culture (Figure 3B versus Figure 1E and Figure 2B). An ex vivo analysis of the aggregated material from C. elegans showed, however, that the major species present in aged worms are non-fibrillar AS–YFP aggregates, characterised by a longer fluorescence lifetime than the mature aggregates formed in human neuroblastoma cells. In accord with this finding, the aggregates are observed to have the high toxicity associated with pre-fibrillar amyloid species .[3, 35]
More generally, the ability to carry out quantitative measurements of protein aggregation kinetics in a living multicellular organism, demonstrated herein, opens the door for increasingly detailed studies of the structures and pathologies of aggregation-mediated processes, for example as a function of time in ageing animals. In addition to enhancing a general understanding of protein misfolding disorders, the sensor has potential to be developed into high throughput platforms to screen for modifiers of amyloid protein aggregation in vivo, with evident applications in the search for, and validation of, possible therapeutic agents.
Experimental Section
Expression and Purification of Proteins: For recombinant expression in E. coli, the cDNA of AS–YFP or BS–YFP was amplified from the C. elegans expression plasmid pENG001 and pENG002[16] and cloned into the vector PET41C (Novagen, Merck, Nottingham, UK).[17] This vector adds an octorepeat of histidine residues at the C-terminus of the exogenous gene under the control of the T7 promoter. α- and β- Synuclein-YFP proteins were expressed in E. coli (BL21) and purified by nickel affinity chromatography and size exclusion chromatography .[17] Purified YFP was a gift from Dr. Sophie Jackson, University of Cambridge.
Amyloid Formation Assays: Aggregation assays for YFP, AS–YFP and BS–YFP were performed in Eppendorf tubes at 37°C with gentle agitation (220 rpm) in a total volume of 250 to 500 μL. Samples contained 50 μm protein in 25 mm Tris HCl pH 7.4, 100 mm NaCl, 0.01% NaN3. Aliquots were taken every 24 h and stored at 4 °C until measurements were made (typically within 4 to 48 h).
Transmission Electron Microscopy and Immunogold Labelling: For TEM analysis of in vitro aggregated proteins, samples were diluted 1:10 in water and 5 μL aliquots of the solutions were applied to Formvar/carbon-coated 400-mesh copper grids (Agar Scientific Ltd., Stansted, UK). Samples were stained with 2% (w/v) uranyl acetate. Images were obtained at various magnifications using a Phillips CEM100 transmission electron microscope. For immunogold labelling experiments, extracts of C. elegans were diluted 1:5 in ddH2O and adsorbed on to glow discharged, carbon coated, nickel/Formvar TEM grids for 30 seconds. They were rinsed three times on drops of phosphate buffered saline (PBS) containing 0.1% bovine serum albumin (BSA) and incubated on drops of mouse anti-AS antibodies (LB509, Zymed, Invitrogen, UK) diluted 1:50 in PBS–BSA for 30 min. They were rinsed on 6 drops of PBS-BSA and incubated on drops of goat anti-mouse IgG conjugated to 10 nm colloidal gold for 30 min at a dilution of 1:100 in PBS-BSA. They were then rinsed 3 times on PBS–BSA and 3 times on ddH2O. They were finally negatively stained on drops of 2% uranyl acetate in ddH2O for 30 seconds, blotted dry and viewed in a FEI Technai G20 operated at 120 Kv. Images were captured with a AMT XR60B digital camera running Deben image capture software.
Cell Cultures and Electroporation: Human neuroblastoma cells (SH-SY5Y) were grown in 1:1 minimal essential medium (MEM) and nutrient mixture F-12 Ham (Sigma, Gillingham, UK) with sodium bicarbonate including 15% heat inactivated foetal bovine serum, 1% MEM non-essential amino acids, 2 mm N-glutamine (Sigma, Gillingham, UK) and 1% penicillin-streptomycin (10,000 U ml K1) and 0.1% fungizone (amphotericin B, 250 mg mL K1) (Invitrogen, Paisley, UK). Before inserting the proteins (AS–YFP, BS–YFP, and YFP) via nucleofection (Amaxa nucleofector, Lonza, Cologne, Germany), the cells were harvested using trypsin (0.5 gL−1 of trypsin) with EDTA (0.2 gL−1 of EDTA 4Na) in Hanks’ Balanced Salt Solution (Invitrogen, Paisley, UK), rinsed with growth medium and divided into several 1 × 106 cell batches. Cells were subsequently transfected using the Amaxa nucleofector with 4 and 8 μg protein in nucleofection buffer (Lonza, Cologne, Germany). Three hours after electroporation the cells were imaged on a confocal laser scanning microscope (Olympus, see below for more details) before the cell culture medium was changed to MEM free of phenol red but containing sodium bicarbonate plus 30 mm HEPES, 2 mm glutamine, 1% MEM non-essential amino acid solution and 1% penicillin-streptomycin (10 000 U ml K1) and 0.1% fungizone (amphotericin B, 250 mg mLK1), and 15% heat inactivated foetal bovine serum. The cells were incubated in total for 4 days and imaged on a daily basis. All cell culture experiments described in the manuscript were repeated at least 3 times and 10-15 cells were analysed.
K114 Staining and Imaging: K114 was dissolved in 10 mg mL−1 DMSO and further diluted to a final concentration of 50 μm using PBS. Cells that had been electroporated (see above) with or without AS–YFP for one or 4 days were analysed. The medium of the cells was removed before incubating the cells with 50 μm K114 for 35 min at 37°C. After 35 min the staining solution was removed and fresh PBS was added to the cells before imaging them on a Leica SP5 confocal microscope using a 60 × oil immersion lens. Images were taken in sequential mode using the 458 nm laser line to excite K114 and the 514 nm laser line to excite YFP. Emission filters for K114 were set at 465–500 nm and for YFP at 550–600 nm to avoid cross excitation.
Experiments on C. elegans: For details on animal strains and culturing of worms refer to .[16, 23] Briefly, the worms were synchronised by selection of individuals in the fourth larval stage. The worms were grown on NGM plates seeded with OP50. One day after synchronisation, animals were placed on NGM plates containing 5-fluoro-2′deoxy-uridine (FUDR) to inhibit growth of offspring. C. elegans protein extracts were prepared from whole animal frozen pellets in PBS containing proteinase inhibitors (Roche, Indianapolis, USA) using Fastprep24 (MP, Solon, USA). For imaging, transgenic worms were mounted on 2% agarose pads containing 40 mm NaN3 as anaesthetic on glass microscope slides. The slides were then mounted onto an inverted confocal microscope attached to a time correlated single photon counting (TCSPC) device. For motility assays, at different ages, animals were placed in a drop of M9 and were allowed to recover for 1 min after which the numbers of body bends were counted for 1 min. At least 8 animals were analysed, unless stated otherwise. Statistical analyses were performed in Graphpad Prism (GraphPad Software, San Diego, CA, USA).[36]
TCSPC Imaging: All samples were assayed on a home-built confocal microscopy platform that permits excitation and emission spectra as well as lifetime and polarisation state to be recorded in every image pixel. The equipment is a modified version of the multiparametric imaging system detailed in,[14] which has been used extensively in functional studies of protein-protein interactions.[37] A brief description follows. A pulsed supercontinuum source (SC 450, Fianium Ltd., Southampton, UK) was used for excitation, emitting a train of sub 10 ps pulses at 40 MHz repetition rate over a spectrum extending from 430 nm to over 2000 nm. The output beam was collimated and passed through a hot mirror assembly to remove infrared components at wavelengths greater than 700 nm. The visible portion of the spectrum was passed through an acousto-optic tuneable filter (AA Opto-electronic AOTFnC-VIS) whose RF modulator was driven by software developed in house using Lab-View (National Instruments, TX, USA). Arbitrary excitation wave-lengths could be selected featuring a spectral bandwidth of around 1-2 nm with an output power of around 1 mW per nm obtainable over the entire visible supercontinuum spectrum. The light was passed into a modified confocal scan unit (Olympus Fluoview FV300), and reflected onto the sample with a 20/80 broad bandwidth coated beam splitter so that 20% of the excitation light passed on to the sample and 80% of the fluorescence signal reflected towards the confocal pinhole. The fluorescence light was band pass filtered and passed onto a fast photomultiplier tube (PMC-100, Becker & Hickl GmbH, Berlin, Germany). Lifetimes were recorded using time correlated single photon counting, TCSPC, circuitry (SPC-830, Becker & Hickl GmbH, Berlin, Germany). An excitation wavelength of 515 nm was used, with the signal emission bandpass extending from 525 nm to 560 nm. Photon count rates were kept below 1% of the laser repetition rate to prevent pulse pile-up. Images were acquired during 100 to 300 s, and photo-bleaching was verified to be negligible during these acquisition times. All TCSPC images were processed using SPCImage (Becker & Hickl GmbH, Berlin, Germany) and fitted with a monoexponential decay function. Pixel binning was increased until approximately 3500 to 5000 photons were obtained per pixel. Image processing and data analysis were carried out using various codes developed in-house using Matlab (The Mathworks Ltd., Cambridge, UK).[15, 38]
Supplementary Material
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cphc.201000996.
Acknowledgements
We would like to thank Drs. Elin Esbjçrner Winters, Claire Michel, and Thomas Jahn (Cambridge, UK) for critically reading this manuscript and for many valuable suggestions. This work is part of a Wellcome Trust/MRC Initiative for Neurodegenerative Disorders (CMD and CFK). GSSK is supported by grants from EPSRC and the Wellcome Trust. CWB held an EMBO long-term postdoctoral fellowship during the initial period of these studies and acknowledges current funding from a EU-FP7 Marie Curie Fellowship. CWB, SS and AE acknowledge funding from a CamBridge-Sense Innovation grant. AE is supported by the UK Engineering and Physical Sciences Research Council, EPSRC. EAAN acknowledges the ZonMW Research Institute of Diseases in the Elderly and De Nederlandse Hersenstichting for funding. CMD acknowledges funding by the BBSRC, the Wellcome Trust and the Leverhulme Trust, and CFK acknowledges funding from EPSRC, the BBSRC and the Wellcome Trust.
References
- [1].Dobson CM. Nature. 2003;426:884–90. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- [2].Selkoe DJ. Nat Cell Biol. 2004;6:1054–61. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- [3].Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Nature. 2008;451:720–25. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Caughey B, Lansbury PT. Annu Rev Neurosci. 2003;26:267–98. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- [5].a) Bacskai BJ, Skoch J, Kajdasz ST, Wang Y, Huang G-F, Mathis CA, Klunk WE, Hyman BT. Proc Natl Acad Sci USA. 2003;100:12462–67. doi: 10.1073/pnas.2034101100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]; c) Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. Nature. 2009;457:1159–62. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]; d) Dai X, Eccleston ME, Yue Z, Slater NKH, Kaminski CF. Polymer. 2006;47:2689–98. [Google Scholar]; e) Bacskai BJ, Skoch J, Hickey GA, Allen R, Hyman BT. J Biomed Opt. 2003;8:368–75. doi: 10.1117/1.1584442. [DOI] [PubMed] [Google Scholar]
- [6].Dawson TM, Dawson VL. Science. 2003;302:819–22. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- [7].Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. Proc Natl Acad Sci USA. 1998;95:6469–73. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Volles MJ, Lansbury PT. Biochemistry. 2003;42:7871–78. doi: 10.1021/bi030086j. [DOI] [PubMed] [Google Scholar]
- [9].Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507–11. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- [10].Stefani M, Dobson CM. J Mol Med. 2003;81:678–99. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- [11].Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Science. 2004;305:1292–95. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- [12].Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- [13].Del Mercato LL, Pompa PP, Maruccio G, Della Torre A, Sabella S, Tamburro AM, Cingolani R, Rinaldi R. Proc Natl Acad Sci U S A. 2007;104:18019–24. doi: 10.1073/pnas.0702843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Frank JH, Elder AD, Swartling J, Venkitaraman AR, Jeyasekharan AD, Kaminski CF. J Microsc. 2007;227:203–15. doi: 10.1111/j.1365-2818.2007.01803.x. [DOI] [PubMed] [Google Scholar]
- [15].Elder AD, Domin A, Kaminski Schierle GS, Lindon C, Pines J, Esposito A, Kaminski CF. J R Soc Interface. 2009;6:S59–S81. [Google Scholar]
- [16].van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA. PLoS Genet. 2008;4:e1000027. doi: 10.1371/journal.pgen.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].van Ham TJ, Esposito A, Kumita JR, Hsu ST, Kaminski Schierle GS, Kaminski CF, Dobson CM, Nollen EA, Bertoncini CW. J Mol Biol. 2010;395:627–42. doi: 10.1016/j.jmb.2009.10.066. [DOI] [PubMed] [Google Scholar]
- [18].Hashimoto M, Rockenstein E, Mante M, Mallory M, Masliah E. Neuron. 2001;32:213–23. doi: 10.1016/s0896-6273(01)00462-7. [DOI] [PubMed] [Google Scholar]
- [19].Zhu M, Rajamani S, Kaylor J, Han S, Zhou F, Fink AL. J Biol Chem. 2004;279:26846–57. doi: 10.1074/jbc.M403129200. [DOI] [PubMed] [Google Scholar]
- [20].Deora AA, Diaz F, Schreiner R, Rodriguez-Boulan E. Traffic. 2007;8:1304–12. doi: 10.1111/j.1600-0854.2007.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Crystal AS, Giasson BI, Crowe A, Kung MP, Zhuang ZP, Trojanowski JQ, Lee VM. J Neurochem. 2003;86:1359–68. doi: 10.1046/j.1471-4159.2003.01949.x. [DOI] [PubMed] [Google Scholar]
- [22].Morley JF, Brignull HR, Weyers JJ, Morimoto RI. Proc Natl Acad Sci USA. 2002;99:10417–22. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH. Proc Natl Acad Sci USA. 2004;101:6403–08. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].a) Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Science. 2006;313:1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]; b) Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Proc Natl Acad Sci USA. 2008;105:728–33. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].a) Chen S, Berthelier V, Yang W, Wetzel R. J Mol Biol. 2001;311:173–82. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]; b) Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr Proc Natl Acad Sci USA. 2000;97:571–6. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].a) Nielsen JT, Bjerring M, Jeppesen MD, Pedersen RO, Pedersen JM, Hein KL, Vosegaard T, Skrydstrup T, Otzen DE, Nielsen NC. Angew Chem. 2009;121:2152–2155. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:2118–21. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]; b) Heise H. ChemBioChem. 2008;9:179–89. doi: 10.1002/cbic.200700630. [DOI] [PubMed] [Google Scholar]; Walsh P, Neudecker P, Sharpe S. J Am Chem Soc. 2010;132:7684–95. doi: 10.1021/ja100431q. [DOI] [PubMed] [Google Scholar]
- [27].Gazit E. FASEB J. 2002;16:77–83. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]
- [28].a) Lemmer U, Heun S, Mahrt RF, Scherf U, Hopmeier M, Siegner U, Gorbel EO, Mullen K, Bassler H. Chem Phys Lett. 1995;240:373–78. [Google Scholar]; b) Shukla A, Mukherjee S, Sharma S, Agrawal V, Radha Kishan KV, Guptasarma P. Arch Biochem Biophys. 2004;428:144–53. doi: 10.1016/j.abb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- [29].Chan FT, Kaminski CF, Kaminski Schierle GS. ChemPhysChem. 2011 doi: 10.1002/cphc.201000833. In press. [DOI] [PubMed] [Google Scholar]
- [30].Runnels LW, Scarlata SF. Biophys J. 1995;69:1569–83. doi: 10.1016/S0006-3495(95)80030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pham CL, Leong SL, Ali FE, Kenche VB, Hill AF, Gras SL, Barnham KJ, Cappai R. J Mol Biol. 2009;387:771–85. doi: 10.1016/j.jmb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- [32].Mosharov EV, Larsen KE, Kanter E, Phillips KA, Wilson K, Schmitz Y, Krantz DE, Kobayashi K, Edwards RH, Sulzer D. Neuron. 2009;62:218–29. doi: 10.1016/j.neuron.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].a) Pan T, Kondo S, Le W, Jankovic J. Brain. 2008;131:1969–78. doi: 10.1093/brain/awm318. [DOI] [PubMed] [Google Scholar]; b) Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, Di Monte DA. J Biol Chem. 2010;285:13621–9. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].a) Hoyer W, Antony T, Cherny D, Heim G, Jovin TM, Subramaniam V. J Mol Biol. 2002;322:383–93. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]; b) Hoyer W, Cherny D, Subramaniam V, Jovin TM. Biochemistry. 2004;43:16233–42. doi: 10.1021/bi048453u. [DOI] [PubMed] [Google Scholar]
- [35].Campioni S, Mannini B, Zampagni M, Pensalfini A, Parrini C, Evangelisti E, Relini A, Stefani M, Dobson CM, Cecchi C, Chiti F. Nat Chem Biol. 2010;6:140–7. doi: 10.1038/nchembio.283. [DOI] [PubMed] [Google Scholar]
- [36].Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. PLoS Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].a) Mauritz JM, Esposito A, Tiffert T, Skepper JN, Warley A, Yoon YZ, Cicuta P, Lew VL, Guck JR, Kaminski CF. Med Biol Eng Comput. 2010;48:1055–63. doi: 10.1007/s11517-010-0668-0. [DOI] [PubMed] [Google Scholar]; b) Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. J Virol. 2010;84:6782–98. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dai XW, Yue ZL, Eccleston ME, Swartling J, Slater NKH, Kaminski CF. Nanomedicine. 2008;4:49–56. doi: 10.1016/j.nano.2007.12.002. [DOI] [PubMed] [Google Scholar]; d) Esposito A, Tiffert T, Mauritz JM, Schlachter S, Bannister LH, Kaminski CF, Lew VL. PLoS One. 2008;3:e3780. doi: 10.1371/journal.pone.0003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].a) Esposito A, Choimet JB, Skepper JN, Mauritz JM, Lew VL, Kaminski CF, Tiffert T. Biophys J. 2010;99:953–60. doi: 10.1016/j.bpj.2010.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schlachter S, Schwedler S, Esposito A, Kaminski Schierle GS, Moggridge GD, Kaminski CF. Opt Express. 2009;17:22747–60. doi: 10.1364/OE.17.022747. [DOI] [PubMed] [Google Scholar]; c) Chakrabortee S, Meersman F, Kaminski Schierle GS, Bertoncini CW, McGee B, Kaminski CF, Tunnacliffe A. Proc Natl Acad Sci USA. 2010;107:16084–9. doi: 10.1073/pnas.1006276107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.