Abstract
Most genetic association signals for type 2 diabetes risk are located in non-coding regions of the genome, hindering translation into molecular mechanisms. Physiological studies have shown a majority of disease-associated variants to exert their effects through pancreatic islet dysfunction. Systematically characterizing the role of regional transcripts in β-cell function could identify the underlying disease-causing genes, but large-scale studies in human cellular models have previously been impractical. We developed a robust and scalable strategy based on arrayed gene silencing in the human β-cell line EndoC-βH1. In a screen of 300 positional candidates selected from 75 type 2 diabetes regions, each gene was assayed for effects on multiple disease-relevant phenotypes, including insulin secretion and cellular proliferation. We identified a total of 45 genes involved in β-cell function, pointing to possible causal mechanisms at 37 disease-associated loci. The results showed a strong enrichment for genes implicated in monogenic diabetes. Selected effects were validated in a follow-up study, including several genes (ARL15, ZMIZ1 and THADA) with previously unknown or poorly described roles in β-cell biology. We have demonstrated the feasibility of systematic functional screening in a human β-cell model, and successfully prioritized plausible disease-causing genes at more than half of the regions investigated.
Type 2 diabetes risk is determined by a complex interplay between environmental and genetic factors, with heritability estimates ranging from 20-80% (1). Over the past decade, genome-wide association studies (GWAS) of ever-increasing size have discovered more than a hundred regions of the genome (loci) associated with type 2 diabetes risk (2). Studies in non-diabetic individuals have demonstrated that a large number of these association signals exert their effects on disease susceptibility through pancreatic islet dysfunction (3).
Despite these advances, progress in translating genetic findings into disease biology has been relatively slow. The majority of risk variants are located in non-coding regions of the genome, and pinpointing the underlying causal genes or “effector transcripts” has proved challenging (4). Recent efforts have focused on identifying structural or functional links between association signals and regional genes (5, 6). A complementary strategy uses candidate-gene biology to prioritize genes located near association signals. High-throughput screening (HTS) could facilitate the identification of genes implicated in β-cell function, and thereby highlight potential effector transcripts at type 2 diabetes GWAS loci. To date, such approaches have been limited by inadequacies of available human cellular models and the high cost of insulin immunoassays (~$2 per data point), the gold standard for measuring insulin. To circumvent these issues, previous studies have relied on rodent β-cell models and either used reporter assays as a proxy for insulin measurements, or focused on cellular proliferation (7–11).
Recently, the first glucose-responsive human β-cell line, EndoC-βH1, was generated (12, 13). The line is derived from fetal pancreatic buds matured in vivo, and displays modest but robust induction of insulin secretion in response to glucose and secretagogous. Detailed characterizations have shown the cell line to be an authentic model system for studying stimulus-coupled secretion (14–16).
To accelerate the discovery of causal genes for type 2 diabetes, the present study performed and validated a genetic screen in the EndoC-βH1 cell line. We identified genes at half of the type 2 diabetes-associated loci studied (37/75) where siRNA-mediated silencing resulted in β-cell dysfunction. This demonstrates the feasibility of performing systematic screening for insulin secretion in a human β-cell model, with implications for both high-throughput genetic and chemical compound screening. Our results can be integrated with existing lines of evidence to prioritize effector transcripts at GWAS loci, and highlight potential roles for ARL15, ZMIZ1 and THADA in the regulation of insulin secretion.
Research Design and Methods
RNA-seq
The EndoC-βH1 cell line was cultured as previously described and grown to near confluency (12). RNA was then TRIzol-extracted, and sequenced at the Oxford Genomics Centre (Wellcome Trust Centre for Human Genetics, University of Oxford) (see Fig. S4 for details). The raw sequencing data have been deposited at the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under accession number XXXXXXXXXXX.
Cellular assays
Cellular phenotypes were adapted for automated screening on a Perkin Elmer Janus liquid handling workstation based on previously described assays (Fig. S1A) (17). Briefly, 20,000 cells/well were reverse transfected in 96-well format at final siRNA concentrations of 25 nmol/L pre-incubated with 0.2 µL RNAiMAX in Opti-MEM. Custom libraries of siRNAs (ON-TARGETplus SMARTpools [Dharmacon] for the primary screen and Silencer Select [Thermo Fisher Scientific] for follow-up validation) were designed based on criteria described in table S2. In each case, non-targeting sequences based on the same chemistries were used as negative controls. Three days after transfection, cells were starved overnight in complete media containing 2.8 mmol/L glucose followed by 1 h starvation in 0 mmol/L media. Static insulin secretion assays were then performed for 1 h in complete media under the indicated conditions, after which cells were counted as described below.
Sample analysis
Following secretion assays, supernatants were analyzed for insulin content using AlphaLISA Human Insulin Immunoassays (Perkin Elmer) on a PHERAstar FS plate-reader (BMG). Supernatants (50-250 nL) and beads pre-diluted in water (500 nL) were dispensed into 384-shallow well microplates with an Echo 550 (Labcyte) acoustic liquid handler, before manual addition of immunoassay buffer to a final volume of 5 µL. Cell counts were measured using the CyQUANT Direct Cell Proliferation kit (Thermo Fisher) on an EnVision plate-reader (Perkin Elmer). All responses were normalized as indicated (see relevant figure legends), and expressed as a percentage of non-targeting (NT) control for each phenotype. Effect sizes are given as the percentage difference from NT (ResponseGene - ResponseNT), and the absolute values hereof (|ResponseGene - ResponseNT|).
Statistical analysis
Data analysis was performed using R 3.0.2. To identify significant responses, cell counts and normalized insulin secretion measurements for each gene were compared to NT control using Student’s two-sample t-test. The false-discovery rate (FDR) was controlled at 5% by applying the Benjamini-Hochberg procedure to produce adjusted p-values (q-values) for each phenotype. The Z-factor measuring the control response for each phenotype was calculated as
Results
We first developed an automated assay for disease-relevant phenotypes in the human β-cell line EndoC-βH1 (Fig. S1A). Selected targets were silenced in a parallel format using RNA interference (RNAi). Cells were then assessed for effects on cell number and insulin secretion under four different conditions: low glucose (1 mM), high glucose (20 mM), and high glucose with the sulphonlyurea tolbutamide (100 µM) or with the phosphodiesterase inhibitor IBMX (100 µM). Low and high glucose conditions were included to provide information on the effect of gene silencing under conditions representing the fasted and fed states in vivo. Tolbutamide and IBMX act on the depolarizing and the potentiating pathways of insulin secretion, respectively, and were included to provide additional mechanistic insights through modulation (e.g. synergy or pharmacologic rescue) of any primary defects observed in low or high glucose.
To reduce the cost of sample analysis, we made use of acoustic liquid handling to miniaturize insulin immunoassays. This generalizable method enabled us to maintain high sensitivity for insulin measurements (coefficient of variation [CV] < 3%; Fig. S2A-B), while obtaining a ten-fold reduction in the cost of sample analysis ($0.20 per data-point). Using the insulin gene (INS) as a positive control, we confirmed that we were able to robustly detect effects of gene silencing on the phenotypes of our assay (mean Z’ = 0.6 across conditions; Fig. S3).
Based on this combined analysis and assay pipeline, we designed a primary screen to assess the role of positional candidate genes for type 2 diabetes GWAS loci in β-cell function (Fig. S1B). For target selection, we considered all protein-coding genes located within 1 Mb of a type 2 diabetes association signal. To exclude genes not expressed in our cellular model, we performed whole-genome RNA sequencing of the EndoC-βH1. Our expression data strongly correlated with published sequencing data for enriched primary β-cells (ρ = 0.78; Fig. S4) and showed robust expression of key β-cell genes (12, 18) (Table S1). We included only genes expressed in both EndoC-βH1 and primary β-cells (Table S2), resulting in inclusion of 300 positional candidates from 75 type 2 diabetes GWAS loci.
We next performed our primary screen in triplicate, and derived standardized scores for each phenotype. Knockdown was visibly confirmed using PLK1, an essential gene, which caused extensive cell death across all conditions. In a representative subset of 16 genes we assessed knockdown efficiency at the transcript level and found the median residual expression to be 43% (Fig. S5), roughly equivalent to monoallelic loss-of-function. To account for differences in plating efficiency and proliferation, cell counts were used to normalize insulin secretion data on a per-well basis. Two criteria were then applied to identify robust effects (“hits”): (1) an FDR-adjusted q-value < 0.05; and (2) an absolute effect size among the top 5% (Fig. S6). This identified a total of 67 hits (15 for cell count and 52 for insulin secretion phenotypes) between 45 genes at 37 loci (Table 1).
Table 1. Effects of significant hits identified in a primary screen for β-cell dysfunction.
The table lists effect sizes (% deviation from NT control) for each gene with a least one significant effect across the five phenotypes measured. All insulin secretion measurements were normalized on a per-well basis to cell-counts, and the mean percentage deviations from NT control were then calculated for each condition. For cell counts, values were median-normalized for interplate differences, and the mean percentage deviations from NT control were calculated across conditions. *q < 0.05 by Student’s t-test (FDR-adjusted).
| Gene | Locus | Low glucose | High glucose | IBMX | Tolbutamide | Cell count |
|---|---|---|---|---|---|---|
| ABCC8 | KCNJ11 | 48.2* | 24 | 26.7* | 1.8 | -1.4 |
| ADAMTS9 | ADAMTS9 | 6.3 | -4.8 | 2.8 | -8 | 12.2* |
| ADIPOQ | ST64GAL1 | 87* | 23.2 | 23.1 | 8.8 | -7.3 |
| ARL15 | ARL15 | -5.5 | -25.9* | -2.1 | -15.5 | -1.5 |
| BCAR1 | BCAR1 | 5.9 | 25.2 | 28.5* | 9.5 | -7 |
| BCL6 | LPP | -20.7* | -8.9 | -1 | -12 | 5.6 |
| BMP8B | MACF1 | 7.8 | 16.5 | 9.5 | 26.9* | -1 |
| CCNT2 | TMEM163 | -32.6* | 1.8 | 4.7 | 5 | -2.8 |
| CDKAL1 | CDKAL1 | 2.4 | 1.7 | -10.6 | -23.5* | 7.5 |
| DGKQ | MAEA | 3.3 | 17.5 | 20.4 | 33.6* | -9.8 |
| DMRTA2 | FAF1 | 32.3* | 24.5 | 13.1 | 22.7 | -0.3 |
| ELAVL4 | FAF1 | -3.6 | 9.1 | 21.2 | 22.8 | -11.4* |
| ETV5 | IGF2BP2 | -12.4 | -25.6* | -10.9 | -12.6 | -2.5 |
| FAH | ZFAND6 | -23.1* | -20.6* | -16.9 | -27.2 | -2.3 |
| FBXW7 | TMEM154 | 45.2* | 9.6 | 8.7 | 11.7 | 9.9* |
| GINS4 | ANK1 | -16.1 | -14.2 | -9.5 | -17 | 8.7* |
| GLIS3 | GLIS3 | -13 | -10.6 | 6.4 | -9.3 | -10.3* |
| HEYL | MACF1 | 29.2 | 29.2* | 0.4 | 20 | -7.2 |
| HMGA2 | HMGA2 | 16.5 | 4.1 | 14.2 | 24.1* | -1.2 |
| HNF1A | HNF1A | 21.7 | 38.3* | 23.1 | 25.5* | 5.2 |
| HNF4A | HNF4A | 36.7* | 66.9* | 74.9 | 92.9* | -8.8 |
| IGF2 | DUSP8 | -26.3* | -10.4 | 1.2 | 0.1 | -1.3 |
| INS | DUSP8 | -53.5* | -44.8* | -48.7 | -38.7* | 0.9 |
| KCNK17 | KCNK16 | -9.4 | -17.3 | -2.5 | -1.2 | 9.8* |
| KCTD15 | PEPD | 21.7 | 9.4 | 12.5 | 0.8 | -10.9* |
| KIF11 | HHEX/IDE | 45.4* | 35* | 26.9 | 55.4* | -40.1* |
| LINGO1 | HMG20A | 0 | 19.1 | -12.4 | -14.9 | 7.9* |
| MFGE8 | AP3S2 | 30* | 3.4 | 2.8 | -1.7 | -5.7 |
| MIER3 | ANKRD55 | 5.9 | 36.5* | 5.8 | 18.2 | 1.9 |
| NDUFS4 | ARL15 | 1.6 | -4.9 | 3.9 | -1.7 | 10.8* |
| PABPC1L | HNF4A | -9.7 | -12.3 | -10.4 | -28.8* | -1.2 |
| PHF23 | SLC16A11 | -25.6* | -1.7 | -5.9 | 3.5 | -0.9 |
| PLA2R1 | RBMS1 | 8.6 | 1.6 | 10.8 | 1.6 | 9.1* |
| PRDX3 | GRK5 | 24 | 31.7* | 23.4 | 9.6 | 16.6* |
| PTHLH | KLHDC5 | -2.8 | -6.5 | -0.5 | -25* | -5.9 |
| RND3 | RND3 | -8.7 | -6.1 | 0 | -3 | -14.9* |
| SLC2A4 | SLC16A11 | 14.5 | 0 | 27.2* | 18.2 | -1.6 |
| SOCS7 | HNF1B | 3.9 | -18.5* | 11.2 | -14.7 | -1.6 |
| SPPL3 | HNF1A | -11.9 | -21.9* | -6.1 | -10 | -10.8* |
| STK38L | KLHDC5 | 15.2 | 40.9* | 4.7 | 25.2* | -3 |
| THADA | THADA | -1.9 | 6.5 | 27.5 | 24.8* | -10.3 |
| TLE1 | TLE1 | 4.3 | -5 | -23* | 16.2 | 4.6 |
| TM6SF2 | CILP2 | -22.6* | -8.3 | -0.8 | -12 | -8.7 |
| UPF2 | CDC123 | 7.5 | -12.5 | 4.7 | -24.9* | -3.2 |
| ZMIZ1 | ZMIZ1 | -29.5* | -21.4* | -19.8 | -16.8 | -15.2* |
For cell numbers, effect sizes for each gene were estimated based on 12 independently plated replicates (four conditions in triplicate), and therefore likely represent true differences in cellular proliferation and/or viability rather than random plating effects. Aside from KIF11, a gene with a known role in cell division, the largest effect sizes compared with NT control (CV = 4% for cell numbers) were observed for ZMIZ1 (-15.2%; q = 6.5x10-5) and PRDX3 (+16.5%; q = 9.2x10-5).
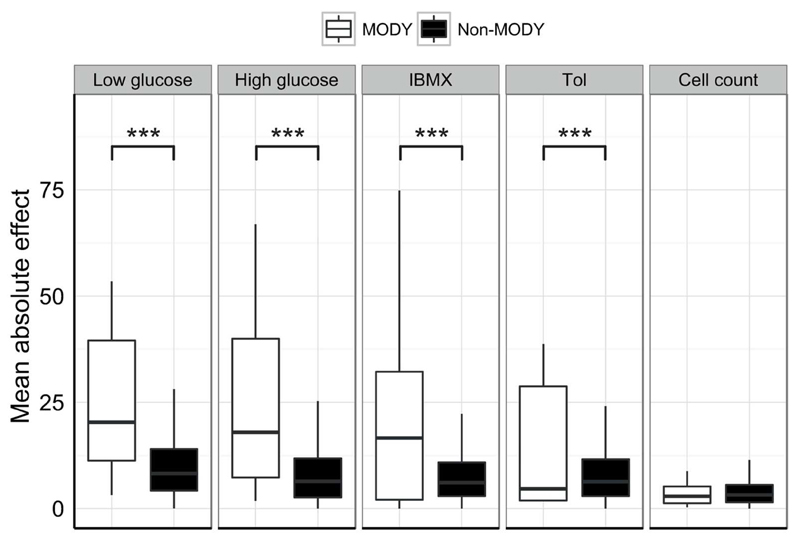
For the insulin secretion data, we first performed an enrichment analysis for genes implicated in maturity-onset diabetes of the young (MODY). MODY describes a collection of monogenic subtypes of diabetes, characterized by insufficient release or production of insulin. As would be expected for a set of bona fide regulators of β-cell function, we observed a strong enrichment of MODY genes among the significant hits (Fisher’s exact test, p = 5.5x10-9). Aggregating absolute effect sizes for MODY and non-MODY genes revealed this enrichment to be driven by altered insulin secretion, and not through effects on cell numbers (Fig. 1).
Figure 1. Comparing mean absolute effect sizes for MODY and non-MODY genes.
Boxplots of mean absolute effect sizes for MODY genes (white) and non-MODY genes (excluding controls, black) across the five phenotypes measured. Effect sizes were calculated as described for table 1, and the absolute values were then averaged for the two categories of genes. Among 14 identified MODY genes, eight fulfilled criteria for inclusion in the screen: HNF4A, GCK, HNF1A, HNF1B, PAX4, INS, ABCC8 and KCNJ11. Boxplots show median and interquartile ranges for groups of n = 8 and 292 data-points. ***q-value < 0.001 by Student’s t-test (FDR-adjusted). Tol = Tolbutamide.
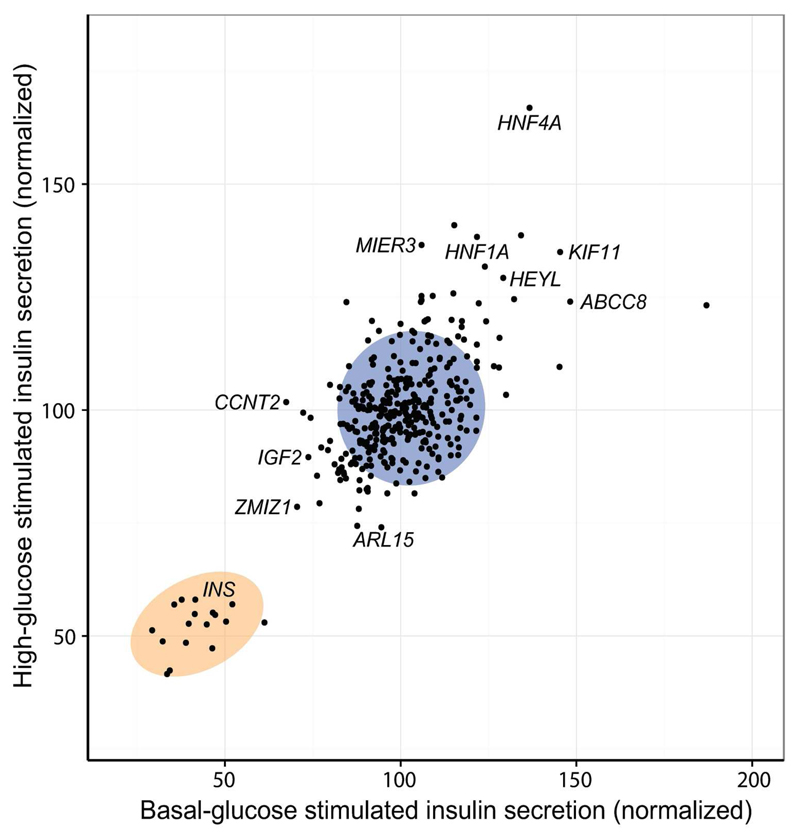
Further validating our secretion data, we observed strong positive correlations between the normalized responses across conditions (p-values < 2.2x10-16; Fig. 2), and found 10 of 35 genes to cause significant effects under two or more conditions. This included four known MODY genes and ZMIZ1, which, independently of the effect on cell numbers, was one of the strongest hits for reduced insulin secretion (q < 0.01 for low and high glucose). Knockdown of the ABCC8 gene, which encodes a subunit of the ATP-sensitive potassium channel, was found to significantly increase insulin secretion under low glucose and IBMX stimulation. As expected, the depolarization caused by this was masked under high glucose (as cells are already partially depolarized), and fully rescued by tolbutamide (due to pharmacological depolarization of the cells). The pattern of modulation by secretion conditions can thus be used to pinpoint specific biological pathways affected by gene silencing. To explore the relationship between conditions in greater detail, we performed clustering analysis on Z-scores derived from the normalized secretion values. This revealed high glucose and tolbutamide to be most similar in terms of modulating knockdown effects, with low glucose and tolbutamide being most dissimilar (Fig. S7).
Figure 2. Comparison of insulin secretion data for high and low glucose.
Normalized insulin secretion responses under high glucose versus low glucose, with selected hits annotated. The blue circle indicates the 95 % confidence contour for NT control, and the orange circle indicates the 95 % confidence contour for insulin (INS) positive controls. All measurements were normalized on a per-well basis to cell-counts, and averages for each condition were then subsequently normalized to the mean of NT control. Data points are mean of n = 3 and shown as percentage of NT control.
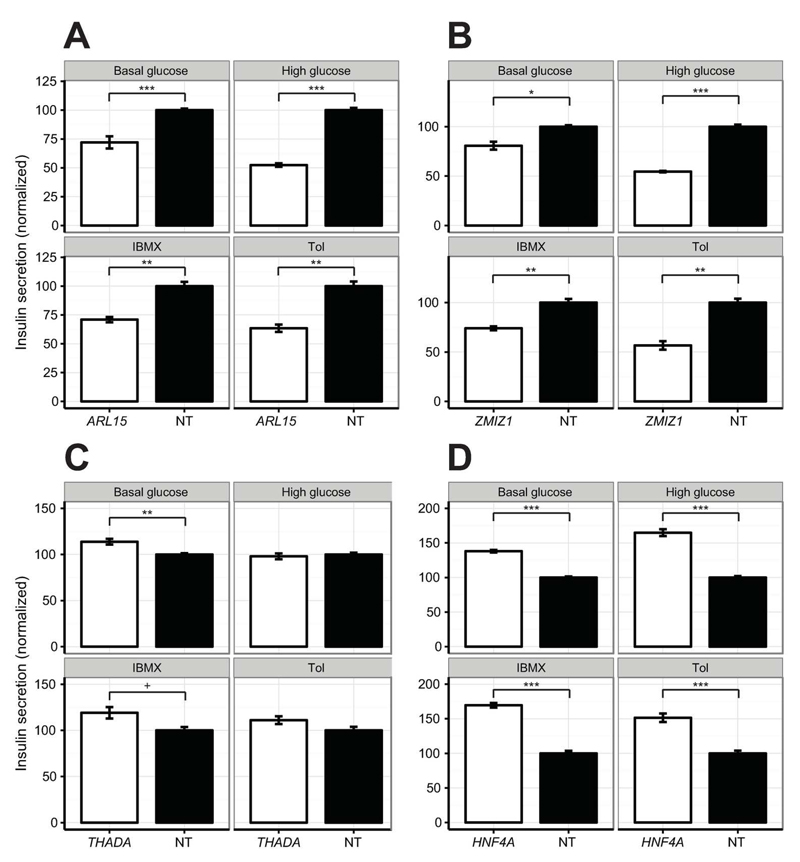
Finally, we assessed the contribution of off-target effects by performing a small-scale validation experiment using siRNAs designed with an alternate algorithm. The sequences were confirmed to be different from those of the primary screen, and could thus be used to establish the biological relevance of positive hits. We selected eight target genes, representing hits for both positive and negative defects across the four conditions, and confirmed that knockdown efficiency was satisfactory (median residual expression = 19.3 %; Fig. S8). Compared with insulin secretion results from the primary screen, we observed an excellent linear correlation (ρ = 0.85, p = 6.7x10-10, Fig. S9) and 88% directional consistency in normalized responses. The validated hits included several genes with limited prior evidence of a role in the regulation of β-cell function, including; ARL15 and ZMIZ1, which were found to significantly reduce insulin secretion across conditions (q-values < 0.05; Fig. 3A-B), and THADA, which modestly elevated insulin secretion across three conditions, though the effect under low glucose was not observed in the primary screen (q = 5.6x10-3; Fig. 3C). Interestingly, gene silencing of the known MODY gene HNF4A was confirmed to cause a paradoxical increase in insulin secretion across all four conditions tested (q-values < 0.001, Fig. 3D).
Figure 3. Insulin secretion data for selected genes in a follow-up validation experiment.
Insulin secretion for (A) ARL15, (B) ZMIZ1, (C) THADA, and (D) HNF4A (white) versus non-targeting (NT; black) negative control under the indicated conditions. Measurements were processed as described for Fig 2, and shown as percentage of NT control. Data points are the mean of n = 6 for NT and n = 3 for other genes, and error bars are SEM. +q-value < 0.1, *q-value < 0.05, **q-value < 0.01, ***q-value < 0.001 by Student’s t-test (FDR-adjusted). Tol = Tolbutamide.
Discussion
High-throughput screens for β-cell dysfunction offer the opportunity to systematically characterize the role of genes in a disease-relevant tissue for type 2 diabetes. Previous efforts have focused on non-human model systems (7–10), reporter-based proxy measurements for insulin (7, 8), and/or phenotypes not directly related to insulin production and secretion (10, 11). Here, we report a genetic screening strategy for the interrogation of multiple disease-relevant phenotypes in the human β-cell line EndoC-βH1. In a primary screen of 300 positional candidates, we successfully identified 15 genes regulating cell number (proliferation and/or viability), and 35 genes regulating insulin secretion. This is, to our knowledge, the first systematic, large-scale effort to identify genes involved in insulin secretion. Importantly, the identified hits can be used to prioritize novel effector transcripts for type 2 diabetes GWAS loci, and may shed further light on mechanisms underlying genes previously implicated in β-cell dysfunction.
The known MODY gene HNF4A was unexpectedly observed to cause a consistent increase (> 40%) in insulin secretion across all conditions. HNF4A encodes the transcription factor Hepatocyte nuclear factor 4 alpha (HNF4α), and is mutated in about 10% of all MODY cases (19). HNF4A loss-of-function mutations that cause monogenic diabetes later in life have also been associated with increased birth-weight (indicative of increased fetal insulin secretion) and congenital hyperinsulinism in early infancy (20). The underlying reason for this switch from elevated to reduced insulin secretion is unknown, but it has been speculated that gradual β-cell exhaustion or, alternatively, a shift in the modulating co-factors of HNF4α may underlie this phenomenon (21, 22).
Among the hits with limited prior evidence of a role in β-cell function, we independently validated ZMIZ1, ARL15, and THADA. Overexpression and knockdown of ZMIZ1, encoding Zinc Finger, MIZ-Type Containing 1 (ZMIZ1), has recently been shown to negatively impact on insulin secretion in primary human islets (6). Moreover, a nearby type 2 diabetes association signal overlaps a cis-eQTL for the gene, supporting its candidacy as the regional effector transcript (6). ARL15 encodes ADP-Ribosylation Factor-Like 15, a relatively uncharacterized member of the ARF-family of proteins involved in regulation of vesicle trafficking and biogenesis. The gene is highly expressed in β-cells and located downstream of an islet-active enhancer bound by key β-cell transcription factors (18, 23) (Fig. S10A). THADA encodes the protein Thyroid Adenoma-Associated (THADA), and contains a coding disease-association signal that has also been associated with reduced β-cell function (24) (Fig. S10C). Consistent with the directionality of our findings, expression profiling has shown the gene to be more highly expressed in patients with type 2 diabetes compared with controls (25). All three genes thus emerge as strong candidates for future studies.
While successfully enabling unbiased functional characterization, our current screening strategy has a number of limitations. False negatives (i.e. true causal genes not identified as hits) could arise as a result of primary effects of the causal gene on non-beta cell tissues, or through effects on genes expressed at different developmental stages. Likewise, overexpression or greater knockdown efficiency may in some cases be required to expose a disease-relevant phenotype. Among the targets analyzed for silencing efficiency, a variable range of knockdown was observed (34% - 88%), and some genes might remain undetected due to insufficient silencing. Conversely, false-positive effects (i.e. non-beta cell regulators identified as hits) also cannot be excluded, and unexpected findings should be further functionally validated (e.g. SLC2A4 effect on IBMX-stimulated insulin secretion). Though the EndoC-βH1 cell line has been found to recapitulate many aspects of β-cell function, it remains a possibility that some findings would not translate directly into human physiology. Finally, a subset of the identified hits may represent true β-cell regulators that are independent of any disease risk variants and, though still of biological importance, not genuine effector transcripts for type 2 diabetes. In addition to the possibility of more than a single effector transcript per locus, this phenomenon likely also contributes to the relatively high proportion of multi-hit loci observed in the primary screen (8/37).
Despite these limitations, our screening strategy successfully replicated well-established biological mechanisms, and identified genes involved in β-cell function at half of the loci investigated. This demonstrates, for the first time, the feasibility of performing scalable screens for insulin secretory defects in human pancreatic β-cells, and opens up the possibility, not only for large-scale genetic manipulations, but also for compound HTS to therapeutically manipulate human β-cells. Insights from this and subsequent functional screens can be integrated with complementary lines of evidence from exome-wide association studies, chromatin conformation capture and cis-eQTL studies to prioritize genes for follow-up studies. Ultimately, this could accelerate the translation of genetic association signals into molecular mechanisms for β-cell dysfunction, insulin insufficiency, and type 2 diabetes.
Supplementary Material
Acknowledgments
This study was funded in Oxford by the Wellcome Trust (095101/Z/10/Z and 098381). SKT is a Radcliffe Department of Medicine scholar. MvdB is supported by a Novo Nordisk postdoctoral fellowship run in partnership with the University of Oxford. MIM is Wellcome Trust Senior Investigator. ALG is a Wellcome Trust Senior fellow in Basic Biomedical Science.
Footnotes
SKT, AC, DE, MIM, ALG conceived and designed the study. RS provided protocols. SKT, AC, CB and AB performed the experiments. SKT, MvdB, MIM and ALG analyzed and interpreted the data. SKT, MIM and ALG wrote the manuscript. SKT, AC, MvdB, CB, AB, RS, DE, MIM and ALG edited and approved the manuscript. ALG is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors declare that there is no conflict of interest.
References
- [1].Ali O. Genetics of type 2 diabetes. World journal of diabetes. 2013;4:114–23. doi: 10.4239/wjd.v4.i4.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Prasad RB, Groop L. Genetics of Type 2 Diabetes-Pitfalls and Possibilities. Genes. 2015;6:87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dimas AS, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63:2158–71. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Thomsen SK, Gloyn AL. The pancreatic beta cell: recent insights from human genetics. Trends Endocrinol Metab. 2014;25:425–34. doi: 10.1016/j.tem.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fadista J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A. 2014;111:13924–9. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van de Bunt M, et al. Transcript Expression Data from Human Islets Links Regulatory Signals from Genome-Wide Association Studies for Type 2 Diabetes and Glycemic Traits to Their Downstream Effectors. PLoS Genet. 2015;11:e1005694–e1005694. doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burns SM, et al. High-Throughput Luminescent Reporter of Insulin Secretion for Discovering Regulators of Pancreatic Beta-Cell Function. Cell Metabolism. 2015;21:126–137. doi: 10.1016/j.cmet.2014.12.010. [DOI] [PubMed] [Google Scholar]
- [8].Ku GM, et al. An siRNA Screen in Pancreatic Beta Cells Reveals a Role for Gpr27 in Insulin Production. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu W, et al. Identification of glucose-dependant insulin secretion targets in pancreatic beta cells by combining defined-mechanism compound library screening and siRNA gene silencing. Journal of Biomolecular Screening. 2008;13:128–134. doi: 10.1177/1087057107313763. [DOI] [PubMed] [Google Scholar]
- [10].Lee JA, et al. Open Innovation for Phenotypic Drug Discovery: The PD2 Assay Panel. Journal of Biomolecular Screening. 2011;16:588–602. doi: 10.1177/1087057111405379. [DOI] [PubMed] [Google Scholar]
- [11].Walpita D, et al. A Human Islet Cell Culture System for High-Throughput Screening. Journal of Biomolecular Screening. 2012;17:509–518. doi: 10.1177/1087057111430253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ravassard P, et al. A genetically engineered human pancreatic beta cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121:3589–97. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Weir GC, Bonner-Weir S. Finally! A human pancreatic beta cell line. J Clin Invest. 2011;121:3395–7. doi: 10.1172/JCI58899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Andersson LE, et al. Characterization of stimulus-secretion coupling in the human pancreatic EndoC-betaH1 beta cell line. PLoS One. 2015;10:e0120879. doi: 10.1371/journal.pone.0120879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Krishnan K, et al. Calcium signaling in a genetically engineered human pancreatic beta-cell line. Pancreas. 2015;44:773–7. doi: 10.1097/MPA.0000000000000318. [DOI] [PubMed] [Google Scholar]
- [16].Gurgul-Convey E, et al. Physiological characterization of the human EndoC-betaH1 beta-cell line. Biochem Biophys Res Commun. 2015;464:13–9. doi: 10.1016/j.bbrc.2015.05.072. [DOI] [PubMed] [Google Scholar]
- [17].Pal A, et al. Loss-of-Function Mutations in the Cell-Cycle Control Gene CDKN2A Impact on Glucose Homeostasis in Humans. Diabetes. 2016;65:527–33. doi: 10.2337/db15-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nica AC, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–62. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shields BM, et al. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia. 2010;53:2504–2508. doi: 10.1007/s00125-010-1799-4. [DOI] [PubMed] [Google Scholar]
- [20].Pearson ER, et al. Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. Plos Medicine. 2007;4:760–769. doi: 10.1371/journal.pmed.0040118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Glaser B. Type 2 diabetes: Hypoinsulinism, hyperinsulinism, or both? Plos Medicine. 2007;4:619–620. doi: 10.1371/journal.pmed.0040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rahman SA, et al. Molecular mechanisms of congenital hyperinsulinism. Journal of Molecular Endocrinology. 2015;54:R119–R129. doi: 10.1530/JME-15-0016. [DOI] [PubMed] [Google Scholar]
- [23].Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–43. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Simonis-Bik AM, et al. Gene Variants in the Novel Type 2 Diabetes Loci CDC123/CAMK1D, THADA, ADAMTS9, BCL11A, and MTNR1B Affect Different Aspects of Pancreatic beta-Cell Function. Diabetes. 2010;59:293–301. doi: 10.2337/db09-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Parikh H, et al. Prioritizing genes for follow-up from genome wide association studies using information on gene expression in tissues relevant for type 2 diabetes mellitus. BMC Medical Genomics. 2009;2:72. doi: 10.1186/1755-8794-2-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.