Abstract
We present an analysis of the chicken (Gallus gallus) transcriptome based on the full insert sequences for 19,626 cDNAs, combined with 485,337 EST sequences. The cDNA data set has been functionally annotated and describes a minimum of 11,929 chicken coding genes, including the sequence for 2260 full-length cDNAs together with a collection of noncoding (nc) cDNAs that have been stringently filtered to remove untranslated regions of coding mRNAs. The combined collection of cDNAs and ESTs describe 62,546 clustered transcripts and provide transcriptional evidence for a total of 18,989 chicken genes, including 88% of the annotated Ensembl gene set. Analysis of the ncRNAs reveals a set that is highly conserved in chickens and mammals, including sequences for 14 pri-miRNAs encoding 23 different miRNAs. The data sets described here provide a transcriptome toolkit linked to physical clones for bioinformaticians and experimental biologists who wish to use chicken systems as a low-cost, accessible alternative to mammals for the analysis of vertebrate development, immunology, and cell biology.
Birds have played a central role in the history of biology, particularly in evolution (Darwin 1859) and ethology (Tinbergen 1953; Lorenz 1981). Since providing the first evidence of genetic linkage and the applicability of Mendels laws to vertebrates (Bateson and Punnett 1905–1908), chicken genetics has been largely restricted to practical problems of meat and egg production and to the analysis of disease resistance. The ensuing improvements in productivity have led to the chicken Gallus gallus becoming an agricultural animal of worldwide importance. Furthermore, the advent of tools for investigating gene function together with the accessibility of the chicken embryo suggest that it will remain an important and versatile experimental system for the foreseeable future (Brown et al. 2003).
Full-length cDNA clones are both essential for gene identification and annotation of complex genomes and provide a valuable resource for experimentalists interested in gene function. Consequently, there has been a large effort to produce annotated EST and full-length cDNA collections for mammals and plants (Okazaki et al. 2002; Seki et al. 2002; Imanishi et al. 2004; Ota et al. 2004). Earlier, we described an extensive chicken EST resource (Boardman et al. 2002), and here we extend that work and describe a collection of 19,626 full insert cDNA sequences. Our work is based on a set of cDNA libraries established from 22 different tissues of interest particularly to developmental biologists. A complementary project has been undertaken using tissue of immunological importance (Caldwell et al. 2004). In combination, these projects describe >4000 novel full-length coding cDNAs together with a collection of noncoding cDNAs. The finished cDNA sequences combined with ESTs generated previously have been of central importance in the chicken genome project (International Chicken Genome Sequencing Consortium [ICGSC] 2004) where they have provided direct experimental evidence for most of the Ensembl annotated genes. In the future these finished cDNA resources are likely to be widely exploited by experimentalists in the analysis of vertebrate gene function. The cDNA data and analysis presented here are supported by a Web-based resource page (www.chick.umist.ac.uk). This site provides a comprehensive set of interrogation tools to mine these data sets and should be of general utility to the growing community using chick-based systems.
Results
Clone set selection
The cDNA clones used for full insert sequencing were chosen from two collections that have been used previously to generate EST data, the BBSRC collection (23,115 clones) (Boardman et al. 2002) and the University of Delaware collection (368 clones) (www.chickest.udel.edu). The sequences generated from these clones provide transcriptome information for many of the tissues used by experimental biologists working with chicken systems, with the principal exception of Bursal cells, which have been analyzed separately (Caldwell et al. 2004). The cDNA libraries used in this project were constructed using total rather than cytoplasmic mRNA, and although this has the disadvantage that cDNAs with retained introns may have been selected for sequencing, an analysis of the BBSRC EST data set indicates that <1% of ESTs represent incompletely spliced cDNAs (www.chick.umist.ac.uk). The considerable advantage of using total mRNA is that the libraries contain cDNAs for nuclear noncoding RNAs.
We set out to generate a collection of finished cDNA sequences (defined as the complete consensus sequence from both strands of the cDNA insert) that provides the broadest possible coverage of the chicken transcriptome and maximizes the number of full-length cDNAs sequenced (defined as containing the 5′ and 3′ untranslated regions [UTRs] and coding sequence). Analyzing the 85,486 EST contigs (ECs) in the BBSRC EST collection, we identified protein coding ECs that had a BLASTX hit (E-value < 10-4) to the SWISS-PROT/TrEMBL database. From this set, we subtracted 1845 ECs corresponding to an existing EMBL database entry for a chicken cDNA together with a set of 2750 ECs whose 5′ reads matched the set planned to be sequenced in the Bursal full-length cDNA project, although only 2300 of these sequences have currently been completed (Caldwell et al. 2004). The remaining set were screened for those clones that had a BLASTX hit which mapped within 10 amino acids of the N terminus of an orthologous protein. This identified a set of 12,018 ECs, and the cDNA clone from each EC with the most 5′ sequence was selected for sequencing, some of which were expected to be full-length cDNAs whereas others might only contain a full-length coding sequence (cds). Clones that lacked up to the first 10 amino acids of the cds were also included in this set, since the missing region could in many cases be predicted from the genome sequence when completed and then a full coding sequence could be experimentally reconstituted using the polymerase chain reaction (PCR). A further set of 3914 clones were selected for full insert sequencing from ECs that had a possible open reading frame (ORF) >80 amino acids and a codon usage bias similar to that for other chicken genes, but lacked a reasonable BLASTX hit (E-value < 10-3) to SWISS-PROT/TrEMBL. A set of 368 cDNA clones from the University of Delaware collection, not represented in the BBSRC protein coding set, were selected for full insert sequencing on the basis that they had a 5′ EST read which matched the N terminus of an orthologous protein using the same BLAST criteria.
Putative noncoding cDNA clones were selected for sequencing using three strategies. Firstly, we identified 2220 ECs that had a BLASTN hit with an EST from another organism in dbEST but lacked an ORF >80 amino acids. The second set comprised 4447 ECs lacking an ORF >80 amino acids and were represented by five or more ESTs. Finally, a further 413 ECs were identified with BLASTN hits (>80% identity over 40 bp) to the RIKEN collection of putative mouse noncoding RNAs (Okazaki et al. 2002). Following the removal of redundancies between the three noncoding EC sets, a total of 6307 clones were selected for sequencing. For each EC, the clone that gave rise to the most 5′ EST sequence was chosen for sequencing. The overall statistics for the cDNA clones sequenced can be found in Table 1.
Table 1.
Summary of chicken cDNA clone statistics
| EC contigs selected for sequencing | 22,607 |
| Coding | 16,300 |
| Non-coding | 6307 |
| Clones attempteda | 23,115 |
| Clones accepted by EMBL database | 19,626 (85%) |
| Average length | 1006 bp |
| Number >1 Kb | 7967 |
| Number >2 Kb | 354 |
| Longest clone (ChEST76n19) | 5339 bp |
| Clones annotated at 07/07/04 | 19,626 |
| Accepted putative coding cDNAs | 14,735 |
| That are spliced | 10,901 |
| Accepted putative noncoding cDNAs | 4891 |
| That are spliced | 1064 |
| Clones mapped to genomeb | 17,896 |
| Clones mapped to genomec | 18,519 |
| Clones mapped to Ensembl transcript | 12,211 |
| Putative antisense clonesd | 289 |
| Putative noncoding RNAse | 367 |
| Conserved noncoding clonesf | 43 |
Multiple clones for some ECs were attempted when the most 5′ clone failed
Ninety-eight percent identity over minimum of 100 bp via BLASTN
Exonerate matches with a rawscore > 1000
cDNAs found antisense to ensembl genes with at least five component ESTs in corresponding EST contig, and passing reverse clone quality control tests
Clones mapped to assembled genome, but not found to be within 5 Kb of an Ensembl chicken gene
Clones found in set e that which have a sequence that is also conserved in the human, mouse, and rat genomes
Chicken transcription units
We collated all the chicken sequence data from the EST and cDNA collections that are publicly available to estimate the total number of different transcripts produced from the chicken genome using two different protocols. Transcripts that mapped to the same segment of the genome but were alternatively spliced or antisense to another gene were separated into different groups. Protocol 1 mapped all publicly available G. gallus EST and cDNA resources to the predicted chicken transcripts from the annotation of the genome via Ensembl (Birney et al. 2004). Ensembl uses all available transcriptional evidence (including the majority of the cDNA set presented here) as well as protein similarity to build gene models. From this, 24,207 (85%) of the 28,416 Ensembl transcripts were matched, corresponding to 14,685 (83%) of the 17,709 Ensembl genes. The degree of transcriptome/genome coverage based on these figures is comparable to that seen for the RIKEN representative transcript and protein set and EST reads, in which 19,370 (86%) of the 22,444 annotated mouse Ensembl genes were covered (Okazaki et al. 2002).
Protocol 2 estimated the number of genes and transcripts from an assembly of all chicken EST and cDNA data, generated by clustering related ESTs into 101,244 gene bins, prior to assembly using PHRAP, which produced 141,532 ECs. To estimate the number of chicken genes and transcripts represented in this set, ECs were mapped to the Ensembl genes via BLASTX (E-value < 10-20, >95% identity) to collapse 49,221 ECs onto 14,856 Ensembl genes. Of the remaining 92,311 orphan ECs, a further 7549 were mapped with BLASTX (E-value < 10-30) to known proteins from UniProt (Apweiler et al. 2004), corresponding to a further 3414 genes. The remaining ECs may represent UTRs or additional chicken transcripts. A further 52,220 ECs could be assigned within 5 kb of 8421 Ensembl genes via BLASTN using their component ESTs to map them to the genome. A subset of 26,581 of these 52,220 ECs mapped between the predicted start and stop codon of an Ensembl gene, suggesting they represent novel transcripts. Together with the 7549 from the UniProt pipeline, this represents 34,130 potential novel transcripts not yet predicted or as yet unsequenced in the chicken genome, over and above the 28,416 transcripts identified by the Ensembl team (ICGSC 2004). This provides an estimate from this data on the number of chicken genes of 21,123 (17,709 Ensembl + 3414 novel) producing a total of 62,546 (28,416 Ensembl + 34,130 novel) protein coding transcripts. Indeed, this may well be an underestimate since transcripts assigned to genes that lie nearby, but not within, Ensembl genes may represent genuinely novel genes rather than UTRs.
Removing redundancy from the contig-based and EST-based pipelines, transcriptional evidence is available for 15,575 (88%) of the Ensembl genes, and hence transcriptional evidence in the form of a cDNA clone is available for 18,989 (15,575 + 3414) chicken genes. These chicken genes do not include the 19 pri-miRNAs identified in this study, antisense transcripts, or other ncRNAs whose function is currently unknown, which are considered separately in this work.
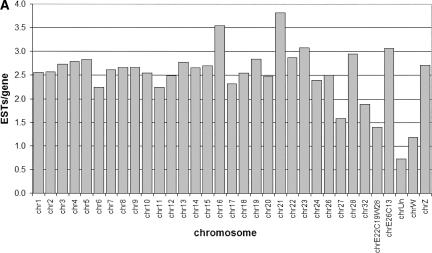
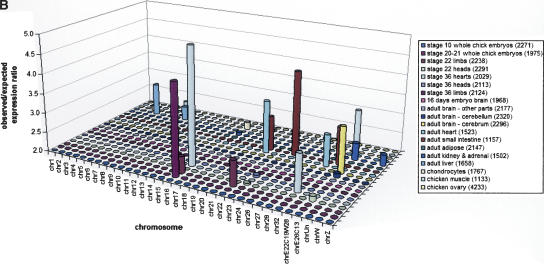
In light of apparent differences in gene density and conservation observed between macro- (chromosomes 1–5) and microchromosomes (ICGSC 2004), we examined the level of expression as measured by the number of ESTs per gene assigned to each chromosome. This analysis did not reveal any clear biases in expression from micro- versus macrochromosomes (Fig. 1A). We examined tissue-specific expression of ESTs across the genome, restricting the analysis to ESTs from unnormalized libraries from 19 different tissues. Figure 1B shows overrepresented transcripts on each chromosome for each tissue, plotting the ratio of observed to expected ESTs for each tissue–chromosome combination. Values greater than one indicate higher-than-expected expression from a given chromosome in a particular tissue, assuming a uniform level of expression across all chromosomes for each tissue and taking into account the number of available genes on each chromosome. For clarity, only those tissue-chromosome pairs showing a greater than twofold overrepresentation are shown. All but three tissues exhibit chromosomally specific EST expression with a P-value <0.001 from standard χ2 tests comparing observed to expected expression over all chromosomes for each tissue. Striking examples include several embryonic tissues (stage-36 hearts, stage-20 to -21 whole embryos, and stage-22 limbs), which are overrepresented by ESTs that map to chromosome 16. However, it should be noted the chromosome 16 is relatively poorly assembled (ICGSC 2004). The data do not show, however, any unequivocal macro- versus microchromosomal expression patterns for tissues, although it is interesting to note that liver and muscle appear to be overrepresented on a few macrochromosomes.
Figure 1.
Distribution of ESTs across the chicken genome. (A) The subset of 39,362 ESTs from non-normalized cDNA libraries mapped to each individual assembled chromosome is shown, normalized by the number of genes on each chromosome, plotting the number of EST hits per gene. (B) Tissue-specific expression patterns for ESTs mapped to the genome. Only ESTs from nonnormalized cDNA libraries were used, and the ratio of actual to expected ESTs mapping from each tissue onto each chromosome is plotted, showing only those cases where the ratio exceeds two. Expected expression levels were calculated as the total number of ESTs expressed by each tissue multiplied by the fraction of all genes located on each given chromosome. Bars shown indicate tissues showing above-expected expression of ESTs on a particular chromosome, where all values are significant at P < 0.001. The legend shows the number of ESTs from each tissue mapped to the genome in brackets after each library name.
We have assessed the level of splicing of the transcripts in the finished cDNA sequences by mapping individual cDNAs back to the genome, using Exonerate (G. Slater and E. Birney, in prep.). This analysis shows that 10,901 (74%) of the predicted 14,735 coding cDNAs are spliced, whereas only 1064 (22%) of the 4891 noncoding cDNAs are spliced (Table 1). These frequencies of splicing are similar to those seen in the FANTOM 2 mouse cDNA set, where 82% of coding transcripts and 29% of noncoding RNAs were spliced (Okazaki et al. 2002). The surprisingly high level of nonspliced coding cDNAs may in part be due to some cDNAs not being full length and may represent single exons of larger spliced genes. Consistent with this, we find that only 169 (8%) of the 2260 full-length cDNAs are unspliced.
Functional annotation
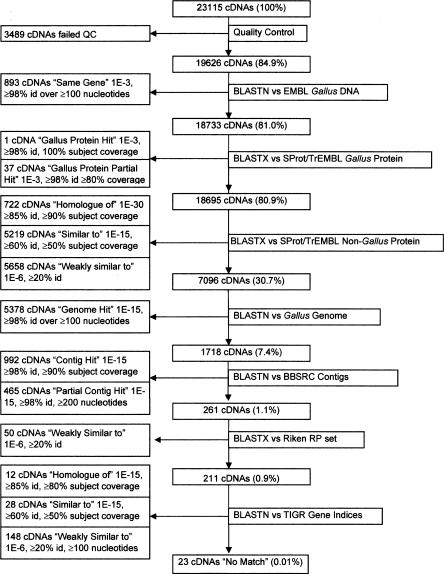
Annotation of the 19,626 cDNA sequences was BLAST-based and assigned a match to publicly available sequence data for 99.9% of the cDNA sequences (Fig. 2). The pipeline indicates matches to either SWISS-PROT/TrEMBL or Gallus EMBL entries for 12,530 (64%) of the cDNAs, providing a basic annotation for the majority (85%) of the 14,735 predicted coding cDNAs accepted by EMBL/GenBank. Of the remaining 7096 cDNAs, 97% match to the chicken genome or to the BBSRC contigs using the BLAST criteria defined in Figure 2. From the remaining 261 cDNAs, 91% match with expressed transcripts in other organisms, contained in the representative protein set from the Riken mouse cDNA project (Okazaki et al. 2002) and the TIGR gene indices (Quackenbush et al. 2001). The 23 cDNAs annotated as “no match” were further searched against satellite, repeats, and transposable element sequences using BLASTN, which did not reveal any matches with ≥95% identity and ≥90% coverage. All 23 no match cDNAs do align with BBSRC contigs with ≥96% identity over ≥60 nucleotides.
Figure 2.
cDNA Annotation pipeline. The diagram illustrates the pipeline that was used to annotate 19,626 finished cDNA sequences. The criteria for inclusion in any category are shown within the box for that category. Annotations are assigned in a top-down approach as indicated, leaving a final count of 23 unmatched cDNAs.
Gene Ontology (GO) terms (Harris et al. 2004) and InterPro assignments (Mulder et al. 2003) were assigned to the cDNA collection as well as the ESTs and are available from our Web site (www.chick.umist.ac.uk).
Full-length cDNAs
To estimate the total number of full-length cDNAs in our collection, we searched all the cDNA sequences (including the 4891 putative noncoding clones from the selection stage) against SWALL (Apweiler et al. 2004), RefSeq (Pruitt and Maglott 2001), and EMBL (Kulikova et al. 2004). In total, 2260 full-length cDNAs were identified that contained 5′ UTR sequence, the complete cds, and 3′ UTR sequence, taken as the superset from 1233, 1537, and 1538 matches to SWALL, RefSeq, and EMBL. Since we specifically subtracted clones that were being sequenced in the Bursal full-length cDNA project (Caldwell et al. 2004) the combination of these two projects has produced 4560 novel full-length coding cDNAs.
A “virtual” full-length cDNA sequence set has also been constructed using a BLAST based protocol, incorporating the EC sequence information. This set represents the longest electronically available sequence associated with a transcript, produced by extending cDNA sequences in silico using additional EST sequence from ECs. Using this set we are able to define an additional 1064 virtual full-length cDNAs with matches to the SWALL, RefSeq, and EMBL complete cds databases.
One thousand five hundred and fifty of the coding cDNA clones that we selected for sequencing as potentially being full length contained the 5′ UTR and start codon but were missing the 3′ end of the gene. Consistent with this result, the average size for the cDNA sequences obtained in this project was only 1.0 kb (Table 1) compared with CAP-trapped cDNA libraries in the mouse FANTOM 2 set, which have an average insert size of 2.35 kb and yield a high proportion of full-length cDNAs (Okazaki et al. 2002). This was unexpected since the cDNAs were generated with oligodT priming and may reflect inadequate protection of internal restriction sites during second-strand cDNA synthesis, physical fragmentation of cDNAs, or excessive poly(A) trimming. Internal priming at poly(A) sequences seems unlikely, however, since inspection of the genome corresponding to the 50 bp downstream of the 3′ ends of the 19,626 cDNAs reveals only 153 (0.8%) that contain internal poly(A) sites of 15 consecutive “A” bases.
Noncoding RNAs
Since noncoding clones picked for sequencing were chosen prior to chicken genome sequencing, we anticipated that they would contain a significant number of both 5′ and 3′ UTR sequences. To eliminate these, we mapped all the putative noncoding cDNAs to the chicken genome using Exonerate and removed clones that overlapped or lay within 5 kb of an Ensembl annotated gene. Antisense transcripts that lie within Ensembl genes were identified separately. The 5-kb cut-off perimeter around an Ensembl gene left a high-quality noncoding (HQNC) RNA set of 757 sequences that should be largely free of UTR sequences for known genes at the expense of excluding some genuine ncRNAs that are close to coding genes. A similar 5-kb cut-off was used recently to define noncoding human genes (Imanishi et al. 2004), and since the chicken genome is more compact than the human, we may have filtered even more stringently for chicken ncRNAs. Three hundred and sixty-eight sequences from the HQNC set could be mapped to an assembled part of the chicken genome, and of these, only 8% showed evidence for splicing. A single noncoding cDNA, which corresponded to one of the pri-miRNAs corresponding to clone CR390304, was subtracted from the HQNC set. These results and others are summarized in Table 2. Of the HQNC set, 294 sequences are represented by contigs containing two or more ESTs and 186 had five or more, indicating that these sequences are unlikely to represent genomic DNA contamination.
Table 2.
Noncoding cDNA statistics
| cDNA set | Total | Likely polyA signal | Upstream CpG island | Both poly A and CpG |
|---|---|---|---|---|
| Putative noncoding | 4891 | 4401 (90%) | 1012 (21%) | 855 (18%) |
| HQNC set | 757 | 583 (77%) | 124 (16%) | 93 (12%) |
| HQNC set mapped to assembled chromosomes | 367 | 339 (92%) | 57 (16%) | 50 (14%) |
| Conserved in one of human/mouse/rat | 67 | 61 (91%) | 13 (19%) | 12 (18%) |
| Conserved in all three of human/mouse/rat | 43 | 41 (95%) | 11 (26%) | 10 (23%) |
We used BLASTN to identify members of the HQNC set that were conserved in other organisms (BLASTN, E-value < 10-30, pairwise identity > 80%). This resulted in the identification of 43 sequences that were conserved in human, mouse, and rat genomes. Those conserved in the human genome all mapped >5 kb from a human Ensembl annotated gene, providing further evidence that they are unlikely to correspond to the 5′ or 3′ UTR for a known vertebrate gene. We performed in situ hybridizations on stage-20 to -21 chick embryos with three EST clones from the HQNC set, which came from embryonic cDNA libraries and mapped >5 kb from a known gene in human, mouse, or chicken genomes, but were unable to detect expression. This indicates that either the expression levels for these RNAs were low or that in vivo they are normally processed into smaller RNA molecules that fail to hybridize efficiently with the probes in the chick embryo.
The HQNC set was also subjected to analysis using ddbRNA (di Bernardo et al. 2003), which attempts to classify a candidate nucleotide sequence as either “RNA” or “other,” based on analysis of sequence alignments. In total, 45 of the 367 HQNC set were predicted to be noncoding RNA based on pairwise WU-BLASTN alignments with ESTs from vertebrate organisms in the TIGR Gene Indices (Quackenbush et al. 2001). A subset of 18 of the 45 are also conserved in the human, mouse, and rat genomes. For each of the HQNC set, the 5′ flanking genomic sequence was searched for CpG islands using CpG island searcher (Takai and Jones 2002). This analysis identified 56 with CpG islands, which are characteristic of transcriptional regulatory regions, and this frequency of CpG islands is similar to that found in mammalian coding genes. We also searched the 3′ flanking genomic region of the HQNC cDNAs for potential polyadenylation sites. In total, 87% of the HQNC set contained polyadenylation signals, defined as either “AAUAAA” or “AUUAAA,” within 1500 bases of the 3′ end of the transcript. This is higher than would be expected by chance for “random” DNA (52% to 72% for %GC 40% to 45%). Mouse noncoding cDNAs show a similar linkage with poly(A) signals (Numata et al. 2003).
In addition to nc RNA analyses, the level of antisense transcripts present in the collection was examined. Using Exonerate, we established that there are 786 cDNAs that are exclusively antisense to a single Ensembl gene (within 5 kb or overlapping), and of these, 467 directly overlap a gene. Five hundred and fifty-three of the antisense cDNAs come from multicomponent ECs, including 325 whose ECs had five or more components, indicating that they are unlikely to have arisen from genomic contamination of the cDNA libraries. We rejected 36 sequences from multicomponent ECs since they were represented by a single EST antisense to the rest in an EC and may correspond to reversed clones. This left a set of 289 high-quality candidate antisense sequences. An analysis of the GO terms for the coding genes associated with these antisense transcripts did not reveal any clear trends or biases, consistent with a recent analysis of ∼1600 sense/antisense transcript pairs from the human genome (Yelin et al. 2003).
MicroRNAs
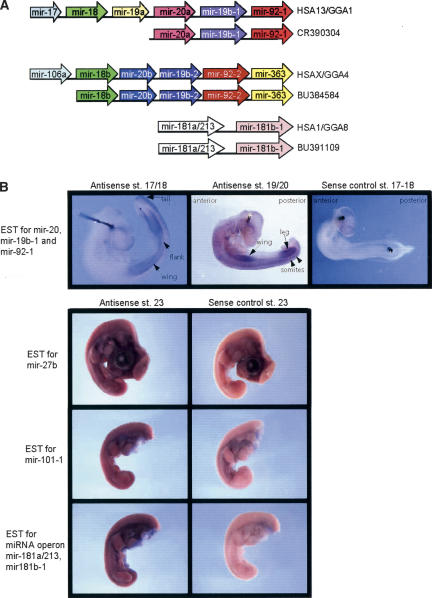
To identify conserved miRNA sequences within the EST data set, we performed a BLASTN search with the miRNA database (Griffiths-Jones 2004; http://www.sanger.ac.uk/Software/Rfam/mirna/search.shtml). This led to the identification of the pri-miRNAs for 23 miRNAs shown in Table 3 together with the cDNA libraries in which they are found. We identified three pri-miRNA operons that are highly conserved in humans (Fig. 3A). The pri-miRNA operon identified in BU384584, which maps to chicken chromosome 4, encoded three miRNAs that were readily identifiable as mir-18b, mir-19b-2, and mir-92-2. When we aligned the BU384584 sequence with the human genome, it matched an miRNA cluster on the X chromosome. The alignment revealed two more highly conserved regions that could be folded into stem-loop precursors within BU384584. One of these was closely related to mir-20a and we named it mir-20b, and the second is the predicted chicken ortholog of the recently identified human mir-363 (S. Pfeffer and T. Tuschl, in prep.). Within the human X-chromosome cluster, there was an additional 5′ sequence which encodes mir-106a, and this sequence was also found in the chicken chromosome 4 cluster, indicating that the full-length pri-miRNA operon for this cluster would encode mir-106a, mir-18b, mir-20b, mir-19b-2, mir-92-2, and mir-363. The pri-miRNA operon found in CR390304 encoded mir-20a, mir-19b-1, and mir-92-1, mapping to chicken chromosome 1. The flanking genome sequence contains three additional miRNA predictions, mir-17, mir-18, and mir-19a. All six miRNAs are conserved in order and orientation in an orthologous cluster on human chromosome 13. This suggests that the CR390304 cDNA may not be full length. mir-17 and mir-106a are closely related by sequence, and the data clearly show that the chicken miRNA clusters contained in BU384584 and CR390304 are paralogous, as recently discussed (Tanzer and Stadler 2004).
Table 3.
miRNAs found in the BBSRC chicken EST/cDNA collections
| Libraries in which miRNA is found
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA | Pri-miRNA Genbank I.D. (coordinates of stem-loop precursor in brackets) | St. 10 whole embryo | St. 20–21 whole embryo | 16 day embryo brain | St. 22 limbs | St. 36 limbs | St. 36 trunk | St. 36 head | Chondrocytes | Muscle | Ovary | Adult kidney + adrenal | Adult adipose | Adult small intestine | Adult brain cerebellum | Adult brain other parts | Adult pancreas |
| let-7b | CR524335 (693–778) | • | • | • | • | ||||||||||||
| let-7d | BU121246 (70–172) | • | |||||||||||||||
| mir-9 | CR405877 (385–471) | • | • | ||||||||||||||
| mir-9* | CR405877 (385–471) | • | • | ||||||||||||||
| mir-18b | BU384584 (46–129) | • | |||||||||||||||
| mir-19b-1 | CR390304 (219–305) | • | • | • | |||||||||||||
| mir-19b-2 | BU384584 (359–443) | • | • | ||||||||||||||
| mir-20a | CR390304 (85–182) | • | |||||||||||||||
| mir-20b | BU384584 (224–319) | • | |||||||||||||||
| mir-22 | CR353219 (86–178) | • | |||||||||||||||
| mir-24 | BU434799 (515–582) | • | |||||||||||||||
| mir-27b | BU346082 (102–196) | • | |||||||||||||||
| mir-30b | BU436353 (418–495) | • | • | ||||||||||||||
| mir-92-1 | CR390304 (341–418) | • | • | • | |||||||||||||
| mir-92-2 | BU384584 (511–586) | • | • | ||||||||||||||
| mir-101-1 | CR406426 (617–728) | • | |||||||||||||||
| mir-181a | BU391109 (74–179) | • | |||||||||||||||
| mir-181b-1 | BU391109 (269–357) | • | • | • | • | • | • | ||||||||||
| mir-199a-1 | BU253899 (134–241) | • | |||||||||||||||
| mir-213 | BU391109 (74–179) | • | |||||||||||||||
| mir-221 | BU382625 (395–493) | • | |||||||||||||||
| mir-301 | BU260747 (455–547) | • | • | ||||||||||||||
| mir-363 | BU384584 (644–721) | • | • | ||||||||||||||
Figure 3.
Pri-miRNA expression patterns in the chick embryo. (A) Organization of the three miRNA operons found in human (HSA) and chick (GGA) genomes is shown together with the corresponding GenBank identification of the cDNA/EST that encodes the miRNAs. (B) In situ expression patterns for miRNAs in the chick embryo. In each case the results with the antisense probes that detect the pri-miRNAs are shown in the left panels. The sense control results are shown in the right panels. st. indicates Hamburger-Hamilton stage.
We carried out in-situ hybridization expression analysis in whole chick embryos for a total of seven single microRNAs—mir-9, mir-22, mir-24, mir-27b, mir-30b, mir-101-1, and mir-199a-1—and two miRNA operons encoded in the CR390304 and BU391109 clones. Antisense and control sense RNA probes were generated using the EST clones corresponding to the pri-miRNAs. For the single miRNA ESTs and the double miRNA operon (BU391109), we carried out analysis on stage-23 embryos (Hamburger and Hamilton 1951), with two embryos per antisense probe and two for the corresponding sense control. In the case of the six miRNA operon (CR390304), we analyzed stage-17/18 and stage-19/20 embryos. We were unable to detect any significant difference from the sense controls for mir-22, mir-30b, mir24, mir-9/9*, and mir-199a-1. This may be due to rapid processing of the pri-miRNAs by DROSHA and DICER producing mature miRNAs, which hybridize weakly to the probes. Alternatively these pri-miRNAs may be expressed at low levels in the embryos or at later stages in development. The single miRNAs that gave detectable expression were mir-27b and mir-101-1, which showed widespread expression in the embryo and no staining for the sense control RNA probes (Fig. 3B). Similarly for the miRNA operon, which encodes mir-181a and mir-213 from the opposite sides of the precursor hairpin together with mir-181b-1 from an adjacent hairpin, we see general expression throughout the embryo (Fig. 3B). For the larger miRNA operon encoding mir17,18,19a,20a,19b-1,92-1, in situ hybridization showed that this pri-miRNA was expressed in a tissue-restricted manner that appears to be developmentally regulated. In stage-17 to -18 embryos, expression is seen in the emerging wing bud and leg bud, as well as the interlimb (flank) region. By stage 19–20, high-level expression is maintained in the limb buds but is now lower in the flank region. Expression is also seen in the tailbud and in posterior somites at these stages.
Surprisingly, we found that mir-24 is found in a pri-miRNA (BU434799), which also contains a partial ORF for the protein coding Ensembl gene ENSGALG00000012615. The pri-miRNA shares the terminal three exons of this protein coding gene but uses an alternative 5′ and 3′ exon. Such chimeric transcripts have been reported previously for various pri-miRNAs (Smalheiser 2003). It is interesting to note that 27 of the predicted miRNAs in the chicken genome map to intronic regions of protein coding genes (ICGSC 2004). This may ensure coordinate regulation of pri-miRNAs with the corresponding mRNA if DROSHA uses the pre-mRNA or intron lariat as the substrate for processing to produce the miRNA. Alternatively, these embedded miRNAs may be expressed from independent promoters.
Since DROSHA (Lee et al. 2003) and DICER (Bernstein et al. 2001) are required for miRNA function, we examined chicken EST databases and the genome sequence for these two genes. For DROSHA we found the chicken gene on chromosome 2 and ESTs corresponding to chick DROSHA were found in eight different cDNA libraries from developmental stage 10 through to adult tissues, suggesting that the miRNA processing pathway is active even at early stages of development. Chicken DICER gene was found on chromosome 5 and in 11 different embryonic and adult cDNA libraries. Furthermore, in situ hybridization studies show that DICER transcripts are widespread in stage-20 to -24 chick embryos, suggesting extensive usage of miRNAs throughout development (data not shown). This is consistent with the observation of widespread expression for three pri-miRNAs in the chick embryo at these stages (Fig. 3B).
Resource availability
The sequence data described in this article can be accessed via the chicken transcriptome Web site (www.chick.umist.ac.uk). The facilities available on this Web site include the following: (1) BLAST, keyword, and ID search of the cDNA/EST data; (2) a functionally annotated database of the cDNA data; (3) an in silico subtraction system that allows the identification of ESTS/cDNAs that are restricted to particular cDNA libraries; (4) a peptide database that can be used for protein identification by peptide mass fingerprinting; and (5) a database of single nucleotide polymorphisms for the chicken based on EST data (Wong et al. 2004).
All the cDNA clones described in this article are available from our distributors at either (http://www.ark-genomics.org/) or (http://www.hgmp.mrc.ac.uk/geneservice/index.shtml). The cDNA sequences generated in this project are available directly from the EMBL nucleotide database or may be downloaded from http://www.sanger.ac.uk/Users/mdr/chicken/accepted_chicken_cDNAs_16_06_04.fasta.gz or www.chick.umist.ac.uk. A list of the ENSEMBL genes for which an antisense transcript was identified can be downloaded from www.chick.umist.ac.uk.
Discussion
This project set out to produce a collection of full-length cDNA clones using existing cDNA libraries generated from a wide range of embryonic and adult tissues, which would be of particular value to developmental biologists, immunologists, and cell biologists. To this end, we have generated 2260 full-length coding cDNA sequences that have no overlap with the sequences from the Bursal full-length cDNA project (Caldwell et al. 2004). Consequently, the combined projects provide full-length coding cDNA sequences linked to physical clones for 26% of the 17,709 chicken genes predicted by Ensembl. It is worth noting that signal peptide sequences appear to be relatively poorly conserved between human and chicken, and a significant fraction of human genes may have erroneously annotated ATG starts, upstream and in-frame from the true ATG start (ICGSC 2004). As a consequence of this, the number of full-length cDNA sequences in these collections may be greater than calculated by our pipelines using current gene models in other organisms.
Frequently cDNAs are incomplete and are missing the 5′ end of the clone because oligodT primed reverse transcriptase does not extend to the 5′ end of the mRNA. Consequently, the 5′ end of the cDNA is traditionally the difficult part to obtain, and in these cases, 5′ RACE is generally used to amplify the missing region and then a complete cDNA is assembled. The 1550 incomplete cDNA clones that lack the 3′ end of the gene identified in this study could readily be assembled into full-length clones using 3′ RACE, to significantly expand the full-length cDNAs available and therefore still represent a useful resource. Alternatively, these sequences may be used as a guide together with the Ensembl annotated genes to design RT-PCR primers to obtain the full-length cDNA. The incomplete coding cDNAs also provide high-quality sequence that allows exon definition across significant parts of a gene and are currently being used in the design and construction of cDNA based microarrays.
A second aim of this project was to produce cDNA sequences for ncRNAs, which would include antisense transcripts, miRNAs, and other uncharacterized RNAs. Through the EST and cDNA sequencing efforts, we have identified transcripts that encode 23 different miRNAs that are conserved in other organisms from Caenorhabditis elegans to man. These miRNAs represent ∼25% of those predicted in the chicken genome (ICGSC 2004). There may be additional chicken-specific miRNAs contained in the cDNA collection. Indeed, a large number of ncRNAs have the potential to form miRNA-like stem loop precursors (data not shown), but experimental validation of these potential miRNAs is required.
The role of miRNAs in developmental regulation is well established in C. elegans, and increasingly tissue-specific and developmental regulation of miRNAs is being found in other organisms (Pasquinelli et al. 2000; Aravin et al. 2003; Krichevsky et al. 2003; Suh et al. 2004). One of the miRNA operons we analyzed showed both tissue-specific and developmental regulation, suggesting it plays a role in vertebrate development, and it will be interesting to identify the target mRNAs for this miRNA operon. One might expect that the various mRNA targets predicted for an miRNA operon correspond to genes with functions in a related pathway, given that the expression of the gene regulators (miRNAs) is so tightly coordinated in an operon configuration.
Together with pri-miRNAs, we sequenced many other cDNAs that were noncoding, including a significant fraction that represent antisense transcripts. It is becoming increasingly apparent that antisense transcripts form a significant part of the overall transcriptome for mammals. For example, a recent analysis of antisense transcripts in the human genome identified ≥1600 sense–antisense pairs, which corresponds to >8% of the 40,000 estimated human genes (Yelin et al. 2003). The specific functions of the antisense transcripts identified in this study remain to be determined, although they may regulate mRNA stability and translation or invoke RNA interference type responses.
Using a 5-kb perimeter from a known gene, a set of 296 ncRNAs, similar in size to the set identified here, has been described for human cDNAs (Imanishi et al. 2004). The small number of ncRNAs identified in these two projects following stringent filtering suggests that some of the recently identified ncRNAs in other cDNA sequencing projects may correspond to UTR sequences. It is also possible that some of the noncoding RNAs we and others have sequenced might correspond to UTRs from genes yet to be identified. Interestingly, genome annotators note that a significant fraction of conserved noncoding sequences between human and chicken are found far removed from annotated coding regions (ICGSC 2004). We also find that 43 of our HQNC set of ncRNAs are also conserved in human, rat, and mouse genomes, as well as being >5 kb from annotated genes, although in this case we provide additional evidence that these elements are transcribed.
The function of these ncRNAs remains elusive, although it is interesting to note that only 8% of the HQNC set are spliced compared with 75% of the coding cDNAs. The low level of splicing found in ncRNAs may arise for several reasons; a ncRNA that is spliced would be expected to recruit an exon-junction complex (Tange et al. 2004) and as such would probably be subject to non-sense–mediated decay if it were exported to the cytoplasm as it would not be translated. Therefore it may be the case that spliced ncRNAs have a nuclear function. Unspliced ncRNAs would fail to recruit an exon junction complex, bypass non-sense–mediated decay surveillance and would not be restricted to a nuclear function. Of course, unspliced, or spliced ncRNAs might also be processed in the nucleus to smaller RNAs, such as smRNAs (Kuwabara et al. 2004) in a manner analogous to pri-miRNAs (Lee et al. 2002) and could then be exported to the cytoplasm for further processing or direct use.
The completion of the first draft of the chicken genome sequence this year represents a valuable resource for people interested in vertebrate gene function. However, because biologists are largely occupied with the expressed part of the genome, transcriptome resources are essential if the experimental advantages of the chicken are to be widely exploited. In this project we have produced a transcriptome resource, complementary to the genome sequence which we anticipate, together with the Bursal full-length cDNA collection, will greatly benefit the existing chicken scientific community. It is hoped that the easy identification of the chicken orthologs of other vertebrate genes and access to physical clones will encourage others to use chicken systems. The chicken can provide a low-cost, rapid alternative to experiments in other vertebrates, particularly with the advent of simple gain or loss of function technologies for the chick embryo (Brown et al. 2003; Katahira and Nakamura 2003).
Methods
Sequencing of cDNAs and quality control
DNA preparation and sequencing were carried out using standard methods, and the data generated were processed by a suite of in-house programs (http://www.sanger.ac.uk/software/sequencing/) prior to assembly with the PHRED (Ewing and Green 1998; Ewing et al. 1998) and PHRAP (http://phrap.org/) algorithms. The assembled data were subjected to two rounds of automated primer picking using a modified Primer3 code (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Sequences were subsequently assembled using the GAP4 program (Bonfield et al. 1995) to allow manual finishing. Finished cDNAs had both strands completely sequenced with every base having a quality of PHRED ≥30 and no ambiguities. Unfinished clones after two rounds were subjected to subsequent rounds of automated or manual primer picking until finished. An automated system (cDNA_DB) for quality control of finished cDNA sequences was developed, which confirmed that the cDNA sequence of the clones corresponded to that for the original EST read and checked the coding cDNA sequences for frameshifts. The software package (cDNA_DB) and documentation are freely available at http://www.sanger.ac.uk/Software/cdna_db/ under the terms of the Perl Artistic License.
Mapping ESTs and contigs to Ensembl genes
ESTs were mapped to the Ensembl final build gene set using National Center for Biotechnology Information (NCBI) BLASTN (version 2.2.3) against the Ensembl cDNAs (>98% identity over 100 bp), and then taking the top-scoring match to each EST to provide a unique gene assignment. Assembled contigs were compared to the Ensembl gene set via two protocols. The first used BLASTN with a 10-30 E-value cutoff, and percentage identity 95% over 50-bp cutoff. The second mapped the contigs to the genome via their component ESTs. Using start and end positions of ESTs mapped to the genome (described below), the full extent of the matching chromosomal region was defined, and a pairwise BLASTN was conducted between the contig and chromosomal segment ±5 kb. Chromosomal matches were then cross-referenced with Ensembl gene positions to classify the contigs as inside, within 5 kb, or nongenic.
Mapping ESTs and cDNAs to the genome
cDNAs were mapped to the genome using Exonerate 0.8.2 with the est2genome model with softmasking from the Ensembl repeatmasker run, reporting only the highest scoring alignment. The cDNA alignments were then compared to the Ensembl Gene models and classified as inside, nearby (within 5 kb), or nongenic. To map the ESTs to the genome, BLASTN searches with the ESTs were conducted against each of the assembled chromosomes without low-complexity sequence masking, reporting matches with a minimum 95% identity over 50 bp. Ambiguously mapped ESTs were excluded from further analysis by inter- and intrachromosomal paralogy filters. The interchromosomal paralog filter indicates potential paralogy when (1) the top two hits across all chromosomes are within ±5% identity, and (2), the E-value of the second top hit is less than the square root of the E-value of the top hit. The intrachromosomal paralogy filter excludes ESTs with two or more HSPs that have identities within 5% of one another and whose alignments overlap by at least 20 nucleotides.
Full-length cDNA assignment
The cDNA sequences were classified as full length by three alternative pipelines, using SWALL (Apweiler et al. 2004), RefSeq (Pruitt and Maglott 2001), and complete cds sequences from EMBL (Kulikova et al. 2004) as the reference databases. Individual cDNAs are annotated as full length if they have a contiguous BLASTX alignment (E-value cutoff 10-30) covering the complete sequence of the top scoring, SWALL entry, ignoring SWALL entries annotated as fragments. Full-length cDNAs are also annotated if the cDNA contains a complete ORF that covers both the SWALL entry's start and stop codons within the BLASTX alignment space.
The RefSeq and EMBL pipelines involved two stages, searching against either 76,467 vertebrate RefSeq sequences or 92,912 EMBL complete coding sequence (cds) vertebrate genes. In the first stage, BLASTN searches of the cDNAs against the database yielded a candidate hit list (E-value cutoff 10-20) of putative full-length sequences that either covered the start and stop codon of the subject sequence, or possessed sufficient sequence up/downstream of the match to contain putative start and stop signals. In the second stage, pairwise TBLASTX search of the subject sequence and candidate cDNA assigned full-length status to cDNAs with E-values better then 10-30 that covered the complete database sequence from start to stop codon. In a few instances, some cDNAs covered all but the start methione, and these were also included as full-length sequences.
Noncoding RNA searches
Putative noncoding status was assigned to all cDNAs not assigned as coding (by virtue of BLAST hits to known proteins, or presence of a predicted ORF using in-house software, EORF, I. Overton, C. Evans, A. Whetton, T. McLaughlin, S.A. Wilson, and S.J. Hubbard, in prep.). The members of this set of 4891 cDNAs were mapped via Exonerate (G. Slater and E. Birney, in prep.) onto the assembled chicken genome. Those found within 5 kb of an Ensembl gene were rejected as potential UTRs. Previously characterized pri-miRNAs found in this set (identified via BLASTN and manual inspection), were also removed.
Potential CpG islands were identified using CpG island searcher (Takai and Jones 2002) searching the corresponding genome segment, including 5 kb upstream of the 5′ end of the HQNC cDNA. Potential polyadenylation signals were found by searching for AATAAA and ATTAAA motifs at the genomic locus corresponding to the 3′ end of the cDNA. This analysis showed 87% of the HQNC set contained polyadenylation signals.
In situ hybridization
In situ hybridization on whole chick embryos was performed as previously described (Nieto et al. 1996). Briefly embryos were fixed in 4% paraformaldehyde overnight before being dehydrated in methanol and stored at -20°C. Embryos were rehydrated through methanol washes before being permeabilized and postfixed in 4% PFA. Embryos were prehybridized for at least 1 h at 65°C–70°C. Digoxigenin-labelled RNA probes were preheated to 65°C–70°C and embryos hybridized overnight. Color reaction was performed using standard techniques.
Acknowledgments
We thank ARK Genomics for supplying cDNA clones for sequencing. I.M.O was supported by a BBSRC committee studentship, and K.B. and P.E.B. were supported by MRC studentships. C.T. is supported by the Royal Society. This work was supported by a grant from the BBSRC and work at the Sanger Institute is supported by the Wellcome Trust.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3011405. Article published online before print in December 2004.
References
- Apweiler, R., Bairoch, A., Wu, C.H., Barker, W.C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. 2004. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 32: D115-D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337-350. [DOI] [PubMed] [Google Scholar]
- Bateson, W. and Punnett, R.C. 1905. –1908. Experimental studies in the physiology of heredity. Reports to the Evolution Committee of the Royal Society: Reports 2–4.
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. [DOI] [PubMed] [Google Scholar]
- Birney, E., Andrews, D., Bevan, P., Caccamo, M., Cameron, G., Chen, Y., Clarke, L., Coates, G., Cox, T., Cuff, J., et al. 2004. Ensembl 2004. Nucleic Acids. Res. 32: D468-D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman, P.E., Sanz-Ezquerro, J., Overton, I.M., Burt, D.W., Bosch, E., Fong, W.T., Tickle, C., Brown, W.R.A., Wilson, S.A., and Hubbard, S.J. 2002. A comprehensive collection of chicken cDNAs. Curr. Biol. 12: 1965-1969. [DOI] [PubMed] [Google Scholar]
- Bonfield, J.K., Smith, K., and Staden, R. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23: 4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W.R.A., Hubbard, S.J., Tickle, C., and Wilson, S.A. 2003. The chicken as a model for large-scale analysis of vertebrate gene function. Nat. Rev. Genet. 4: 87-98. [DOI] [PubMed] [Google Scholar]
- Caldwell, R., Kierzek, A., Arakawa, H., Bezzubov, Y., Zaim, J., Fiedler, P., Kutter, S., Blagodatski, A., Kostavska, D., Koter, M., et al. 2004. Full-length cDNAs from bursal lymphocytes to facilitate gene function analysis. Genome Biol. (in press). [DOI] [PMC free article] [PubMed]
- Darwin, C. 1859. The origin of species (ed. G. Beer). Oxford World's Classic, Oxford Paperbacks, Oxford.
- di Bernardo, D., Down, T., and Hubbard, T. 2003. ddbRNA: Detection of conserved secondary structures in multiple alignments. Bioinformatics. 19: 1606-1611. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred, II: Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred, I: Accuracy assessment. [DOI] [PubMed]
- Griffiths-Jones, S. 2004. The microRNA registry. Nucleic Acids. Res. 32: D109-D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger, V. and Hamilton, H. 1951. A series of normal stages in the development of the chick embryo. J. Morphol. 88: 49-92. [PubMed] [Google Scholar]
- Harris, M.A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B., Mungall, C., et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32: D258-D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi, T., Itoh, T., Suzuki, Y., O'Donovan, C., Fukuchi, S., Koyanagi, K.O., Barrero, R.A., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. 2004. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLOS Biol. 2: 1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (ICGSC). 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature (in press). [DOI] [PubMed]
- Katahira, T. and Nakamura, H. 2003. Gene silencing in chick embryos with a vector-based small interfering RNA system. Develop. Growth Differ. 45: 361-367. [DOI] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., and Kosik, K.S. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikova, T., Aldebert, P., Althorpe, N., Baker, W., Bates, K., Browne, P., van den Broek, A., Cochrane, G., Duggan, K., Eberhardt, R., et al. 2004. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 32: D27-D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K., and Gage, F.H. 2004. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116: 779-793. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation:stepwise processing and subcellular localization. EMBO J. 21: 4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415-419. [DOI] [PubMed] [Google Scholar]
- Lorenz, K. 1981. The foundations of ethology. Springer-Verlag, Vienna.
- Mulder, N.J., Apweiler, R., Attwood, T.K., Bairoch, A., Barrell, D., Bateman, A., Binns, D., Biswas, M., Bradley, P., Bork, P., et al. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31: 315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto, A., Patel, K., and Wilkinson, D.G. 1996. In situ hybridisation analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 51: 219-235. [DOI] [PubMed] [Google Scholar]
- Numata, K., Kanai, A., Saito, R., Kondo, S., Adachi, J., Wilming, L.G., Hume, D.A., RIKEN GER Group, Arakawa, T., Carninci, P., et al. 2003. Identification of putative noncoding RNAs among the RIKEN Mouse Full-Length cDNA Collection. Genome Res. 13: 1301-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36: 40-45. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A.E., Reinhart, B.J., Slack, F., Martindale, M.Q., Kuroda, M.I., Maller, B., Hayward, D.C., Ball, E.E., Degnan, B., Muller, P., et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86-89. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. 2001. The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29: 159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y. et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141-145. [DOI] [PubMed] [Google Scholar]
- Smalheiser, N.R. 2003. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol. 4: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, M.R., Lee, Y., Kim, J.K., Kim, S.K., Moon, S.H., Lee, J.Y., Cha, K.Y., Chung, H.M., Yoon, H.S., Moon, S.Y., et al. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270: 488-498. [DOI] [PubMed] [Google Scholar]
- Takai, D. and Jones, P.A. 2002. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. 99: 3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange, T.O., Nott, A., and Moore, M.J. 2004. The ever-increasing complexities of the exon junction complex. Curr Opin. Cell Biol. 16: 279-284. [DOI] [PubMed] [Google Scholar]
- Tanzer, A. and Stadler, P.F. 2004. Molecular evolution of a microRNA cluster. J. Mol. Biol. 339: 327-335. [DOI] [PubMed] [Google Scholar]
- Tinbergen, N. 1953. The herring gull's world. Collins, London.
- Wong, G.K. and The International Chicken Polymorphism Map Consortium. 2004. A polymorphism map for chicken with 2.8 million SNPs. Nature (in press).
- Yelin, R., Dahary, D., Sorek, R., Levanon, E., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotech. 21: 379-386. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.ark-genomics.org/; cDNA clone distributor.
- http://www.chickest.udel.edu; University of Delaware collection.
- http://www.chick.umist.ac.uk; comprehensive set of interrogation tools.
- http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi/; Primer3 code.
- http://www.hgmp.mrc.ac.uk/geneservice/index.shtml; cDNA clone distributor.
- http://phrap.org/; PHRAP.
- http://www.sanger.ac.uk/Software/Rfam/mirna/search.shtml; BLASTN search.
- http://www.sanger.ac.uk/software/sequencing/; suite of in-house programs.
- http://www.sanger.ac.uk/Software/cdna_db/; software package.