Systemic acquired resistance (SAR) is a form of broad-range disease resistance in plants that develops after exposure to certain avirulent necrotizing pathogens. Induction of SAR is dependent on the accumulation of the endogenous signaling molecule salicylic acid (SA) and the transmission of the SA signal via the activity of the key regulatory protein NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1). Resistance is conferred in large part by the SA- and NPR1-dependent activation of PR genes and the accumulation of PR proteins, some of which have antimicrobial activity. Although a number of key players in the development of SAR have been identified, the fundamental mechanism(s) of SA signal transduction remains unknown.
The basic model of SA action in SAR states that SA accumulation causes the translocation of NPR1 into the nucleus, where it interacts with members of the TGA family of transcription factors and enhances the binding of these factors to SA response elements in the promoters of PR genes, thus ultimately affecting the transcription of numerous genes in the SAR pathway (Després et al., 2000; Kinkema et al., 2000; Zhou et al., 2000; Subramaniam et al., 2001; Fan and Dong, 2002). The mechanism by which the nuclear translocation of NPR1 is effected is unclear. Furthermore, the in vivo interaction of NPR1 with TGA proteins is dependent on induction by SA, even though TGA proteins are expressed constitutively in the nucleus and some NPR1 protein is localized to the nucleus as well as the cytoplasm of unstimulated tissue. In other words, SA is thought to stimulate the enhanced nuclear translocation of NPR1 and to activate the protein (or its interacting TGA partner) to stimulate the interaction with TGA factors.
In this issue of The Plant Cell, Després et al., (pages 2181–2191) show that redox changes influenced by SA control the interaction of NPR1 and TGA1, thus enhancing the DNA binding activity of TGA1. The authors show that SA modulates the redox state of TGA1 in vivo, via the reduction of two Cys residues that form a disulfide bridge in the absence of SA, and that NPR1 interacts specifically with the reduced form of TGA1 (Figure 1). They further show that DNA binding activity per se of TGA1 is not affected by the redox status; rather, the redox-regulated interaction with NPR1 enhances the DNA binding of TGA1. These results demonstrate that TGA1 and TGA4 are likely to be functional interacting partners of NPR1 in the development of SAR and that the redox regulation of these factors confers an additional level of control over their interaction.
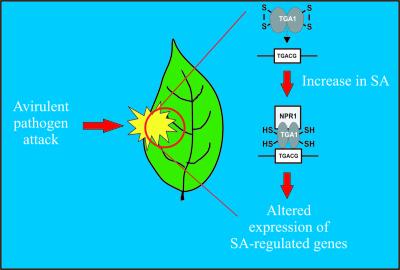
Figure 1.
SA-Mediated Redox Control of TGA1.
Després et al. show that TGA1 is present in an oxidized state in Arabidopsis leaves in the absence of SA, as manifested by the formation of a disulfide bridge between two conserved Cys residues. SA accumulation causes the reduction of these Cys residues, and the reduced form of TGA1 is capable of interacting with the regulatory protein NPR1. The interaction with NPR1 enhances the DNA binding of reduced TGA1, which, like other members of the TGA family, binds specifically to SA-regulatory sequences found in the promoters of numerous PR genes in an NPR1-dependent manner. TGA4 contains conserved Cys residues similar to TGA1 and also is likely to be redox regulated in this manner.
NPR1 was identified previously as a key factor in SA-regulated PR gene expression and the development of SAR because disease resistance and the SA-induced expression of PR genes are compromised severely in npr1 mutant plants (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). NPR1 contains a nuclear localization domain at the C terminus and two known protein–protein interaction domains: a BTB/POZ domain at the N terminus and an ankyrin repeat domain near the center of the protein sequence (Cao et al., 1997; Ryals et al., 1997). The presence of protein–protein interaction domains and the absence of a DNA binding domain, together with the mutant phenotype, suggest that NPR1 functions as a regulatory protein that interacts with and influences the activity of SA-responsive transcription factors.
Yeast two-hybrid experiments have shown that NPR1 interacts specifically with members of the TGA family of transcription factors (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000), and analysis of these factors has further shown that they are involved in the regulation of PR gene expression and the development of SAR (Pontier et al., 2001; Fan and Dong, 2002). The TGA gene family in Arabidopsis includes at least 10 members, 7 of which (TGA1 to TGA7) have been cloned (Xiang et al., 1997; Chuang et al., 1999; Niggeweg et al., 2000b). Of these seven, TGA1 and TGA4 fail to interact with NPR1 in the yeast two-hybrid system (Després et al., 2000; Zhou et al., 2000). Thus, TGA1 and TGA4 previously have been considered as unlikely candidates for important functional NPR1-interacting partners, and more attention has been focused on other TGA family members, such as TGA2.
In a series of elegant experiments, Després et al. showed that NPR1 does interact with TGA1 (and likely also with TGA4) in a SA-dependent manner in planta. First, the authors used a transient transfection assay in Arabidopsis leaves, similar to the yeast two-hybrid assay, to show that the interaction of TGA1 and NPR1 in planta is weak in the absence of SA but strong 24 h after SA treatment. Next, they constructed multiple chimeric proteins using segments of TGA1 and TGA2 and tested them in the yeast two-hybrid system to determine which regions of the proteins were critical for the interaction. These tests identified a 30–amino acid region of TGA2 that conferred upon TGA1 the ability to interact with NPR1 when swapped with the homologous region of TGA1. A comparison of the protein sequences among all seven Arabidopsis TGA proteins revealed that TGA1 and TGA4 contain two Cys residues in this region that are lacking in all other TGA proteins, and subsequent mutation of these Cys residues in either TGA1 or TGA4 produced a mutant protein capable of interacting with NPR1 in the yeast two-hybrid assay. In further experiments, Després et al. confirmed that NPR1 interacts specifically with the reduced form of TGA1, that TGA1 can form an intramolecular disulfide bridge, and that SA treatment reduces the Cys residues in the protein. They conducted additional experiments to show that the reduction of TGA1 does not regulate the DNA binding activity of the protein directly; rather, NPR1 interaction with reduced TGA1 enhances TGA1 DNA binding activity, as it does for other TGA factors.
The work of Després et al. broadens the scope of NPR1-interacting factors. The precise function of the various TGA factors is unknown. There is evidence that at least some of them function as positive regulators of PR gene expression, because numerous PR genes are induced in a SA- and NPR1-dependent manner. Fan and Dong (2002) inhibited TGA2 function in Arabidopsis by overexpression of a dominant-negative TGA2 mutant protein. TGA factors are basic Leu zipper (bZIP) proteins capable of forming homodimers or heterodimers (Lam and Lam, 1995). Fan and Dong (2002) used a dominant-negative construct containing only the C-terminal end of TGA2 to produce a protein capable of interacting with NPR1 but lacking both DNA binding and dimerization activity. This mutant TGA2 could inhibit other TGA factors by competition for binding with NPR1, because all TGA factors require interaction with NPR1 to enhance DNA binding. However, the inhibition of other TGA factors was expected to be less severe than with a mutant TGA2 lacking only the DNA binding domain but retaining the Leu zipper dimerization domain, because the latter protein could inhibit the DNA binding of multiple TGA factors via the formation of heterodimers. These authors found that expression of this dominant-negative TGA2 in a wild-type background produced an npr1-like phenotype of reduced induction of PR genes after treatment with a SA analog and enhanced disease symptoms after infection with an avirulent bacterial pathogen, suggesting that TGA2 functions as a positive regulator of SA-induced genes. Transformation of plants using a chimeric reporter system further demonstrated that Arabidopsis TGA2 is a transcriptional activator.
Interestingly, experiments with a dominant-negative TGA construct in tobacco designed to knock out the activity of all members of the TGA family strongly suggested that some of the TGA factors likely function as negative regulators of gene expression (Pontier et al., 2001). This mutant tobacco TGA2 construct was shown to function as a trans-dominant suppressor of DNA binding activity by suppressing the DNA binding activity of numerous TGA family proteins in addition to TGA2. Expression of the mutant TGA2 in tobacco plants resulted in the inhibition of SA-induced expression of two glutathione S-transferase genes that contain TGA binding sites in their promoters but, surprisingly, enhanced SA-induced expression of PR genes and concomitant enhanced SAR. These results suggested that the family of TGA factors have diverse functions and that at least some of them function as repressors (Pontier et al., 2001). However, the expression of a similar dominant-negative TGA2 mutant in tobacco by Niggeweg et al. (2000a) resulted in decreased SA induction of several potential TGA target genes. These studies highlight the complex nature of NPR1–TGA factor interactions.
Although the main characteristics of TGA DNA binding sequences are known, it is not known which cis sequences function predominantly in vivo and which might confer negative as opposed to positive regulatory function. Some of the next steps will be to determine which TGA factors function as negative regulators and which function as positive regulators (or indeed if some of them function as both) and which cis-acting sequences are the main targets of the different TGA factors in vivo. These are difficult questions to answer, in part because knockout experiments suggest that members of the TGA family have overlapping or redundant functions, and work in Arabidopsis and tobacco suggests that there may be differences between plant species. Furthermore, TGA factors might function as homodimers or heterodimers, greatly increasing the potential for functionally distinct interactions with NPR1. In addition, there may be other as yet unidentified NPR1-interacting partners. The possibility of redox control associated with some TGA factors adds yet another twist to the complicated tale of plant disease resistance.
References
- Cao, H., Bowling, S.A., Gordon, S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Chuang, C.F., Running, M.P., Williams, R.W., and Meyerowitz, E.M. (1999). The PERIANTHA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13, 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., and Fobert, P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., and Lam, Y.K. (1995). Binding site requirements and differential representation of TGF factors in nuclear ASF-1 activity. Nucleic Acids Res. 23, 3778–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000. a). Tobacco transcription factor TGA2.2 is the main component of as-1 binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000. b). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788. [DOI] [PubMed] [Google Scholar]
- Pontier, D., Miao, Z.-H., and Lam, E. (2001). Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 27, 529–538. [DOI] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S.W., and Brisson, N. (2001). Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19, 769–772. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Miao, Z., and Lam, E. (1997). DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 34, 403–415. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. 13, 191–202. [DOI] [PubMed] [Google Scholar]