Abstract
The CreBC two-component system (TCS) is a conserved regulatory system found in Escherichia coli, Aeromonas spp., Pseudomonas aeruginosa, and Stenotrophomonas maltophilia. In this study, we determined how CreBC TCS regulates secreted protease activities and swimming motility using creB, creC, and creBC in-frame deletion mutants (KJΔCreB, KJΔCreC, and KJΔBC) of S. maltophilia KJ. Compared to wild-type KJ, KJΔCreB had a comparable secreted protease activity; however, the secreted protease activities were obviously reduced in KJΔCreC and KJΔBC, suggesting that CreC works together with another unidentified response regulator (not CreB) to regulate secreted protease activity. Single gene inactivation of creB or creC resulted in mutants with an enhanced swimming motility, and this phenotype was exacerbated in a double mutant KJΔBC. To elucidate the underlying mechanism responsible for the ΔcreBC-mediated swimming enhancement, flagella morphology observation, RNA-seq based transcriptome assay, qRT-PCR, and membrane integrity and potential assessment were performed. Flagella morphological observation ruled out the possibility that swimming enhancement was due to altered flagella morphology. CreBC inactivation upregulated the expression of creD and flagella-associated genes encoding the basal body- and motor-associated proteins. Furthermore, KJΔBC had an increased membrane susceptibility to Triton X-100 and CreD upregulation in KJΔBC partially alleviated the compromise of membrane integrity. The impact of creBC TCS on bacterial membrane potential was assessed by carbonyl cyanide m-chlorophenyl hydrazine (CCCP50) concentration at which 50% of bacterial swimming is inhibited. CCCP50 of wild-type KJ increased when creBC was deleted, indicating an association between the higher membrane potential of KJΔBC cells and enhanced motility. Upregulation of the basal body- and motor-associated genes of flagella in KJΔBC cells may explain the increased membrane potential. Collectively, inactivation of creBC increased swimming motility through membrane potential increase and creD upregulation in S. maltophilia. The increased membrane potential may supply more energy for flagella propelling and CreD upregulation supports membrane stability, providing a strong membrane for flagellum function.
Introduction
Stenotrophomonas maltophilia is both a commensal microbe and opportunistic human pathogen that occurs naturally in a variety of habitats [1]. The ubiquitous nature of this microorganism stems mostly from its capacity to survive a variety of environmental conditions with the aid of its stress defense mechanisms. Two-component regulatory systems (TCSs) constitute a critical set of regulators that sense environmental signals and respond by coordinating the expression of an array of genes [2]. TCSs are composed of an inner membrane sensor kinase (SK), acting as a signal sensor, and a cognate response regulator (RR), which works as a transcription factor to activate or repress the expression of a variety of genes of the TCS regulon [3]. The genome of S. maltophilia K279a is equipped with at least 43 sets of TCSs [4], but only a few have been characterized, including SmeSR, SmeRySy, BfmAK, and CreBC. The SmeSR and SmeRySy systems are involved in the regulation of RND-type efflux pumps, as well as in multidrug resistance [5–6]. The BfmAK system controls biofilm development [7].
CreBC/BlrAB is a conserved TCS in many gram-negative bacteria such as Escherichia coli, Aeromonas spp. (named as BlrAB), Pseudomonas aeruginosa, and S. maltophilia. CreC/BlrB functions as an SK and CreB/BlrA as an RR. The functions of CreBC/BlrAB TCSs in E. coli, Aeromonas spp., and P. aeruginosa can be discerned when these systems are activated [8–10]. The CreBC TCS of E. coli is responsive to carbon sources and oxygen availability, and its activation is beneficial, as it mediates growth adaption to anaerobic environments [8]. The BlrAB TCS of Aeromonas spp., which acts as a regulator for β-lactamase expression, is activated by β-lactam challenge or by the functional loss of penicillin-binding protein 4 (PBP4) [10–12]. In response to PBP4 inactivation, the activated CreBC TCS of P. aeruginosa plays a major role in fitness, biofilm growth, and global regulation [13]. In addition, components of the creBC/blrAB regulon in E. coli, Aeromonas spp., and P. aeruginosa have also reported. Among these, a tightly controlled cre regulon gene was reported, namely creD/blrD, which is located downstream of the creBC/blrAB operon and is highly conserved in CreBC/BlrAB-harboring microorganisms. CreD expression is upregulated by activation of CreBC in E. coli, Aeromonas spp., and P. aeruginosa [8–9]; therefore, creD upregulation is considered an indicator of creBC TCS activation in systems of these bacterial species. Compared to those of E. coli, Aeromonas spp. and P. aeruginosa, the CreBC TCS of S. maltophilia has some unique features. The creBC operon of S. maltophilia is constitutively transcribed under laboratory culture conditions [14], although the extracellular stimulating signals remains unknown. There may be signals that further stimulate the creBC system. Furthermore, in contrast to E. coli, Aeromonas spp., and P. aeruginosa, creD of S. maltophilia is expressed separately from the adjacent creBC operon and has its own promoter. The promoter of creD (PcreD) is negatively regulated by creBC and positively regulated by bacterial culture density. Factors compromising bacterial growth such as plasmid carriage or antibiotics attenuate the promoter activity of PcreD [14]. CreD of S. maltophilia is responsible for cell division and cell envelope integrity [14].
Bacteria have developed different motility systems to move, ensuring a survival advantage under a wide variety of environments. Flagella-based swimming motility is a major mode of locomotion for bacterial movement through liquids. The bacterial flagellum is composed of approximately 20 proteins, with approximately 30 additional proteins required for its regulation and assembly [15]. The flagellum is usually described in three parts, specifically the basal body, the hook, and the helical filament [16]. The basal body is embedded within the cell membrane and is composed of a rotor, stator, and periplasmic rod. The rotor includes the cytoplasmic membrane MS ring (FliN protein) and the cytoplasmic C ring (FliG, FliM, and FliN proteins), which acts as a switch to determine the rotation of flagellum. MotA and MotB proteins are the main components of the stator. The proton flow from the periplasm to the cytoplasm mediated by the stator complex MotA/MotB is coupled to the rotation of the flagellum and thus drives swimming [17]. The periplasmic rod (FliE, FlgB, FlgC, FlgF, and FlgG proteins) runs between the hook and basal body, passing through the peptidoglycan layer-P ring (FlgI protein) and outer membrane-L ring (FlgH protein). The hook, connecting the basal body to the filament, consists of FlgE, FlgK, and FlgL. The helical filament, which is composed of flagellin (FliC) and the distal FilD cap, functions as a propeller [16].
Little is known about the function of CreBC in S. maltophilia, except that inactivation of MltD1 (a lytic transglycosylase) elicits a CreBC-mediated elevation in β-lactamase activity in the absence of β-lactam [18]. Since the CreBC TCS is active in laboratory culture conditions, without stress challenge, this prompted us to consider the involvement of the CreBC TCS of S. maltophilia in bacterial physiology. In this study, we sought to define the function of CreBC in bacterial growth, morphology, secreted protease activity, and swimming motility. Based on phenotypic and genetic studies of creB, creC, and creBC mutants, we demonstrated that CreC likely modulates swimming motility via the response regulator CreB, whereas CreC might be involved in cross talk with another unidentified response regulator, to modulate secreted protease activity. Furthermore, inactivation of creBC increases inner membrane potential and upregulates creD, which can contribute to enhanced swimming motility.
Results
Growth characteristics and morphology of the creBC mutant
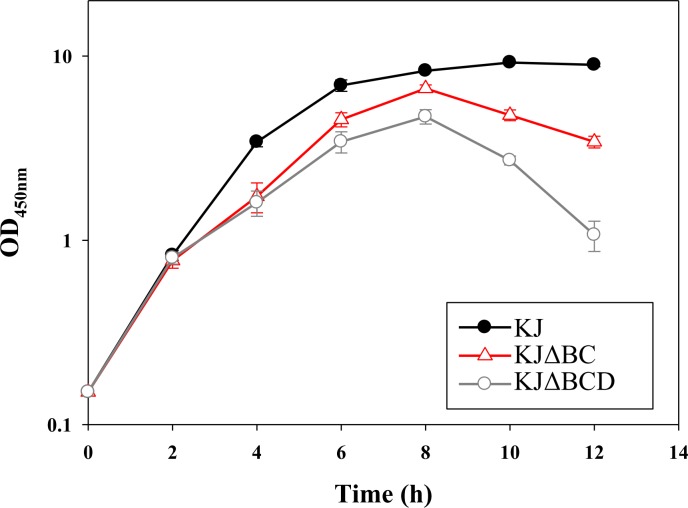
KJΔBC, an isogenic creBC in-frame deletion mutant, was constructed in our recent study [14]. The growth of KJ and KJΔBC was assessed by monitoring the OD450nm every 3 h. These strains exhibited indistinguishable growth patterns at 37°C (data not shown).
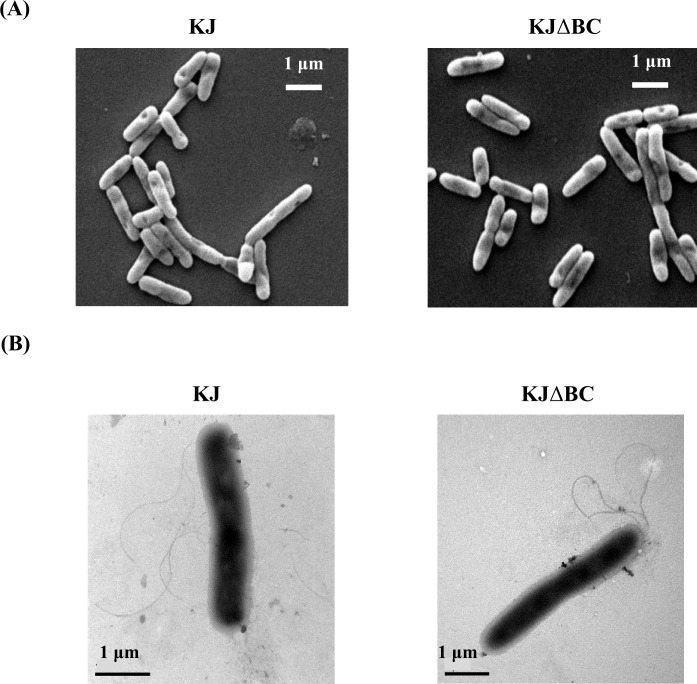
The impact of creBC inactivation on bacterial morphology was assessed by SEM. No observable morphological aberrations were noticed when comparing KJ and KJΔBC strains (Fig 1A).
Fig 1. Role of CreBC TCS in bacterial and flagella morphologies.
The overnight-cultured bacteria were adjusted to an initial OD450 of 0.15. After a 5-h incubation at 37°C, the logarithmic-phase cells were harvested for morphology observation. (A) Bacterial morphology. The samples for bacterial morphology observation were processed by glutaraldehyde-osmium tetraoxide (OsO4)-ethanol method and examined suing SEM. (B) Flagella morphology. The flagella were negatively strained with 1% phosphotungstic acid (pH 7.4) and observed by TEM.
CreC and creBC mutants, but not a creB mutant, display decreased secreted protease activity
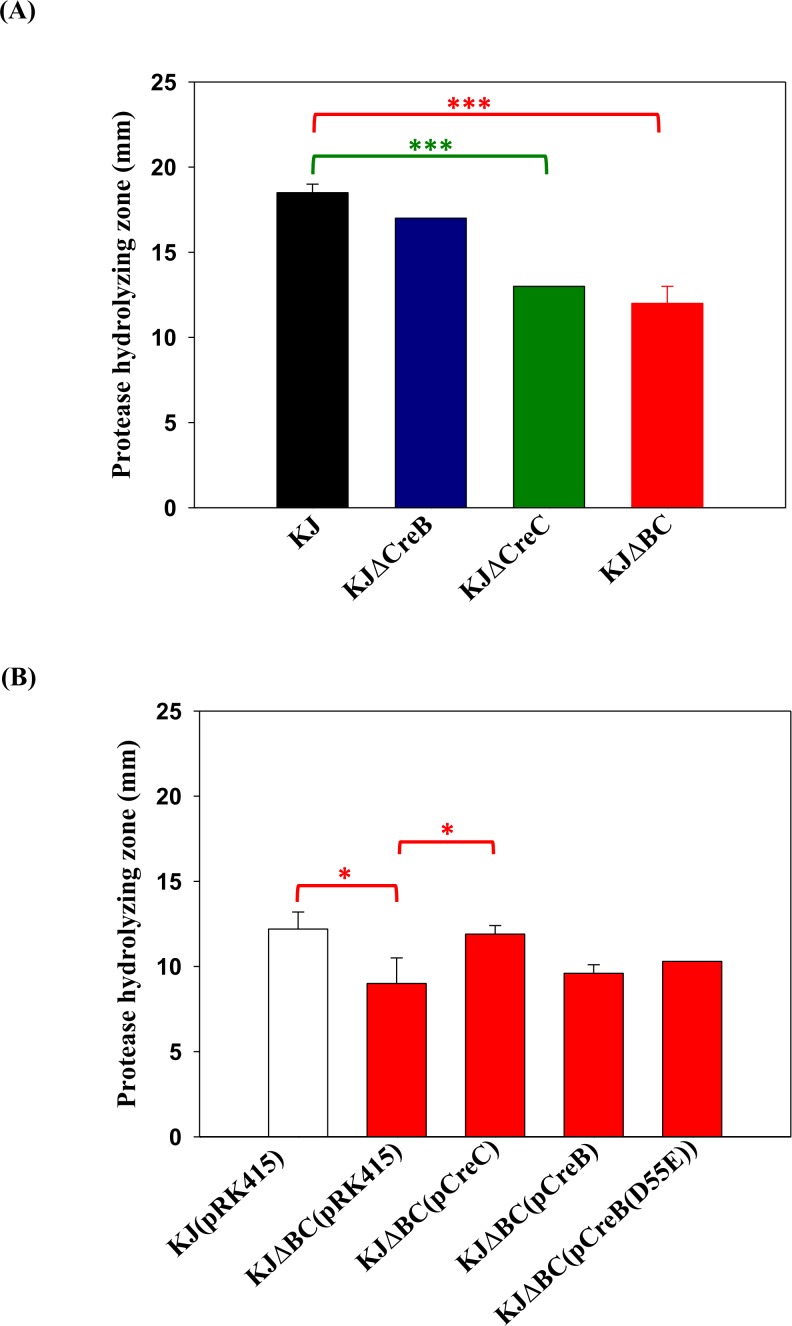
The activity of protease secreted from the bacteria was assayed using LB agar containing 1% skim milk. The protease hydrolyzing zones of KJΔCreC and KJΔBC were smaller than that of wild-type KJ; whereas, KJΔCreB displayed secreted protease activity that was similar to that of wild-type KJ (Fig 2A). This observation prompted us to consider the possibility that CreC is cognate with an unidentified response regulator, and governs secreted protease activity. To test this possibility, creC complementation assay was performed. We noticed that the hydrolysing zones observed in plasmid-carrying strains were generally smaller than those of their deletion-mutant counterparts [for example KJ vs. KJ(pRK415) and KJΔBC vs. KJΔBC(pRK415)] (Fig 2B). This might have resulted from the addition of tetracycline for plasmid maintenance. The protease hydrolysing zones of KJΔBC was reverted to the wild-type level when intact creC was complemented (KJΔBC(pCreC) in Fig 2B). To further verify the irrelevance of CreB in secreted protease activity, a complementation assay was performed. We used two different strategies to mimic constitutive activation of CreB to enable analysis of the corresponding CreB signalling pathway; one involved overexpression of CreB, while the other was overexpression of CreB(D55E) [14], in which amino acid 55 in CreB was converted from aspartate to glutamate. It is widely accepted that a mutation converting the conserved aspartate to glutamate at the site of phosphorylation constitutively activates the response regulator, acting as a phosphor-mimic variant of the response regulator. The plasmids pCreB and pCreB(D55E) did not rescue the effect of creBC deletion on secreted protease activity [KJΔBC(pRK415) vs. KJΔBC(pCreB) and KJΔBC(pCreB(D55E)] (Fig 2B), further confirming that CreB is not involved in secreted protease activity. These results supported the possibility that the CreC SK works together with another unidentified response regulator (not CreB) to regulate secreted protease activity.
Fig 2. The role of CreBC in the secreted protease activity.
Forty microliters of bacterial cell suspension was dipped onto LB agar containing 1% skim milk. After incubation at 37°C for 72 hour, the proteolytic activity of bacteria was assessed by measuring the transparent zones around the bacteria. Data represent the means from 3 independent experiments. Error bars represent the standard deviations for three triplicate samples. *, p < 0.05; ***, p < 0.001. (A) The secreted protease activities of creB, creC, and creBC mutants. (B) The secreted protease activities of creBC mutant and its derived complementation strains. Tetracycline (30 μg/ml) was added for the plasmid maintenance.
The creBC mutant displays enhanced swimming motility
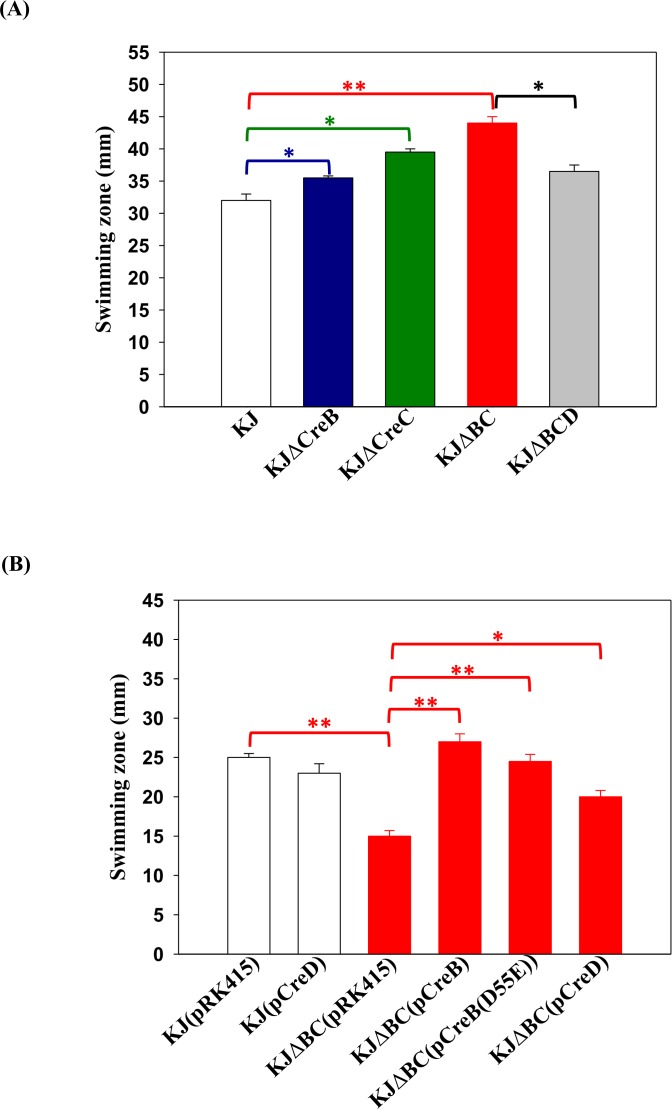
The swimming motilities of KJΔCreB, KJΔCreC, KJΔBC, and wild-type KJ were examined by assessing their migration through semi-solid agar (0.15% agar). Compared to wild-type KJ, KJΔCreB and KJΔCreC displayed enhanced swimming motility, and this phenotype was exacerbated through simultaneously deletion of CreB and CreC, namely KJΔBC (Fig 3A). Next, the complementation assay was performed to further confirm the involvement of the CreBC TCS in swimming motility. The empty vector pRK415 was introduced into KJ and KJΔBC as a control. To our surprise, the swimming zone of KJΔBC(pRK415) was smaller than that of KJ(pRK415) (Fig 3B), opposite to the results observed for KJΔBC and KJ strains (Fig 3A). The phenotypic deviation mediated by plasmid introduction indicates that the maintenance of plasmid pRK415 in KJ strain may alter the expressions of creBC-regulated genes, which are responsible for the swimming phenotype. This is reminiscent of creD, whose expression is negatively regulated by CreBC TCS and positively regulated by the bacterial culture density [14]. Factors that decrease the bacterial culture density, such as plasmid carrying and tetracycline addition for plasmid maintenance, may attenuate the creD expression [14]. A speculated model was thus proposed herein that CreD upregulation in KJΔBC mutant may contribute to the enhanced swimming motility in KJΔBC, as observed for KJ and KJΔBC (Fig 3A). However, CreD upregulation in KJΔBC was counteracted by the introduction of plasmid pRK415 and the addition of tetracycline [14], which may account for the swimming motility observations in KJ(pRK415) and KJΔBC(pRK415) (Fig 3B). To assess this, we first determined the creD transcript levels in KJ and KJΔBC, as well as in KJ(pRK415) and KJΔBC(pRK415). Consistent with previous results [14], the creD transcript showed a 2.67±1.02-fold increase in KJΔBC compared to that in wild-type KJ. However, the creD transcript was decreased in plasmid-carriage strains (KJ(pRK415) and KJΔBC(pRK415)) (S1 Fig.), signifying that plasmid introduction and tetracycline addition attenuate the promoter activity of PcreD., consistent with our previous finding [14] Next, three strategies were adopted to link CreD to the swimming phenotype: (i) a ΔcreD allele was introduced into KJΔBC, yielding KJΔBCD, (ii) KJΔBC was complemented with creB- and creB(D55E)-containing plasmids, yielding KJΔBC(pCreB) and KJΔBC(pCreB(D55E)) respectively, and (iii) a creD-containing plasmid was introduced into KJΔBC and wild-type KJ, generating KJΔBC(pCreD) and KJ(pCreD) respectively. The swimming zone of KJΔBCD was smaller than that of KJΔBC, but not as small as that of wild-type KJ (Fig 3A), indicating that creD upregulation in the ΔcreBC background partially contributed to enhanced swimming. In the plasmid-harbouring counterpart, swimming motility alterations caused by creBC inactivation were reverted to wild-type levels when either CreB or CreB(D55E) was complemented (Fig 3B). Furthermore, the swimming zone of KJΔBC(pCreD) was larger than that of KJΔBC(pRK415), but not as large as that of KJ(pRK415) (Fig 3B). However, overexpression of CreD in the wild-type KJ had only a minor effect on swimming motility (Fig 3B, KJ(pRK415) vs. KJ(pCreD)). Therefore, CreD overexpression is not the sole parameter contributing to enhanced swimming of KJΔBC. Some factors in ΔcreBC background, but not in the wild-type background, are involved swimming enhancement. Taken together, these data support that (i) swimming motility in S. maltophilia is negatively regulated by CreBC TCS; (ii) ΔcreBC-mediated CreD upregulation partially contributes to increased swimming motility in KJΔBC; and (iii) in addition to creD, other genes regulated by CreBC TCS contribute to enhanced motility in KJΔBC.
Fig 3. The role of CreBC TCS in the swimming motility.
Two microliters of bacterial cell suspension was inoculated onto the swimming agar (1% tryptone, 0.5% NaCl, and 0.15% agar). Results were expressed as diameters (millimetres) of swimming zones after 48 h of incubation at 37°C. Data represent the means from 3 independent experiments. Error bars represent the standard deviations for three triplicate samples. *, p < 0.01; **, p < 0.001. (A) The swimming motility of creB, creC, creBC, and creBCD mutants. (B) The swimming motility of creBC mutant and its derived complementation strains. Tetracycline of 30 μg/ml was added for the plasmid maintenance.
Inactivation of creBC upregulates the expression of flagella-associated genes encoding the basal body and motor-associated proteins
The flagellum is a critical organelle for bacterial swimming motility. Flagella morphologies in KJ and KJΔBC cells were further assessed to elucidate the possible link between flagella morphology and motility. Most KJΔBC cells maintained similar flagella numbers and morphologies compared to those of KJ cells (Fig 1B), although we observed a few instances in which KJΔBC cells had more flagella. The minority of hyper-flagella KJΔBC cells was likely not the main cause of enhanced swimming in KJΔBC cells.
To further clarify the underlying mechanism of ΔcreBC-mediated swimming enhancement, transcriptome sequencing (RNA-seq) was performed to examine gene expression in KJ and KJΔBC cells. For analysis, we defined differentially expressed genes as those with an absolute fold change greater than 3. The transcriptome data revealed that 723 genes were differentially regulated between KJ and KJΔBC (S1 Table). Of these, 673 (93%) and 50 (7%) were upregulated and downregulated, respectively, in response to creBC inactivation, indicating that the CreBC TCS mainly acts as gene repressor in wild-type KJ.
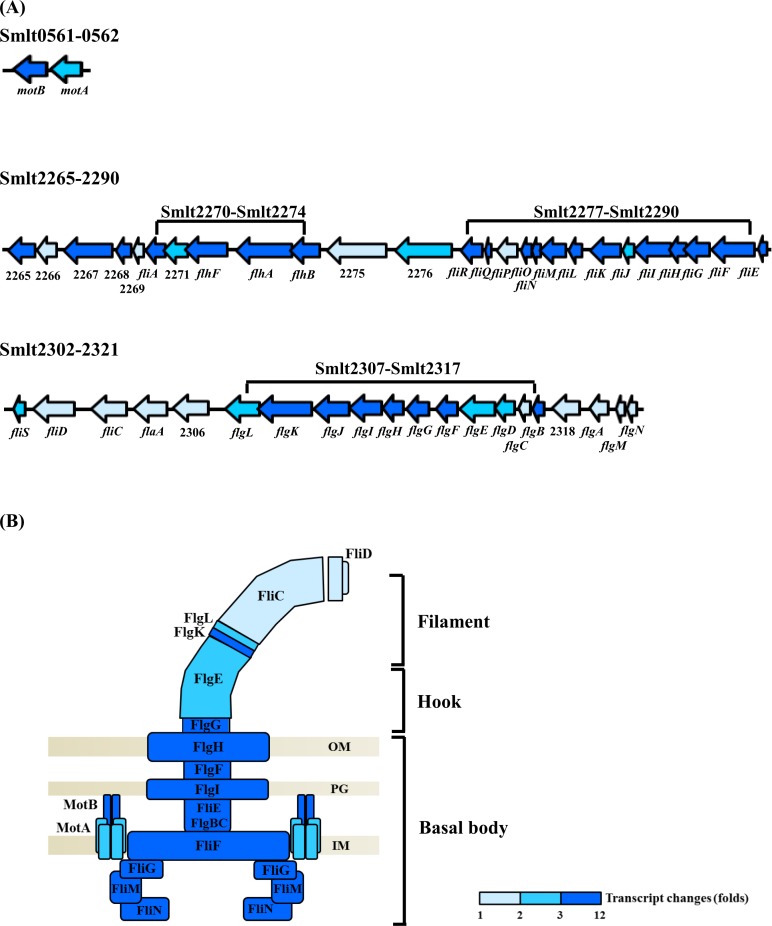
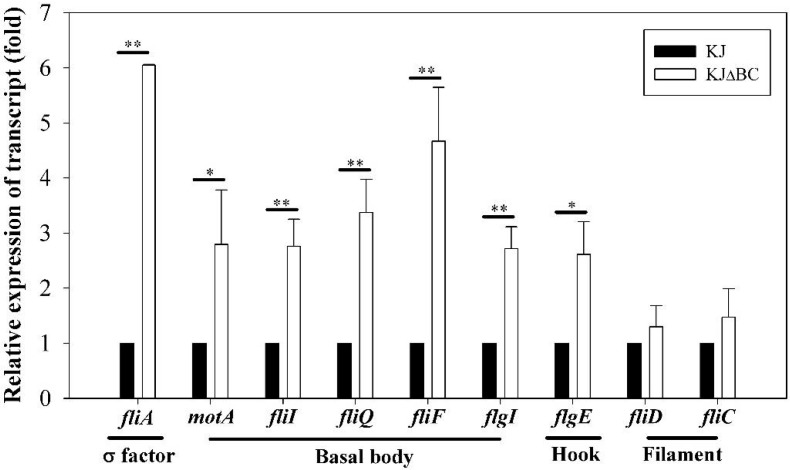
From the transcriptome results, we noticed that a cohort of putative flagella-related genes was significantly upregulated with creBC inactivation. These genes are located in three clusters of the S. maltophilia genome, Smlt0561–Smlt0562, Smlt2265–Smlt2290, and Smlt2302–2321 (Fig 4A). After further classifying upregulated genes, we noticed that proteins encoded by the Smlt0561–0562, Smlt2277–2290, and Smlt2307–2317 clusters comprised the stator, C-ring/MS-ring/motor switch, and P-ring/L-ring/rod/hook, respectively (Fig 4B). Of note, the expression of Smlt2270 (fliA), encoding the putative RNA polymerase sigma factor FliA for flagella regulon, and Smlt2272 (flhF), encoding the putative flagella biosynthesis regulator FlhF, were elevated by 5.33-fold and 9.42-fold, respectively, in KJΔBC cells compared to that in KJ cells. In addition, whereas creBC was inactivated, the upregulation of genes encoding rotor proteins (FliF, FliG, FliM, and FliN) and stator proteins (MotA and MotB) ranged from 2.64- to 7.37-fold (Table 1). Nevertheless, it was noticed that the expression of fliC and fliD, which encode filament and filament cap proteins, respectively, was not significantly altered when CreBC was inactivated (Table 1 and Fig 4).
Fig 4. The transcript changes (folds) of flagella-related genes between KJ and KJΔBC cells by transcriptome analysis.
Total mRNA was extracted from KJ and KJΔBC logarithmic-phase cultures. The ribosomal RNA (rRNA) depletion, adapter-ligated cDNA library construction and enrichment, and cDNA sequencing were performed as described in Materials and Methods. Transcript changes (folds) of a gene is expressed as the transcript in KJΔBC relative to the transcript in wild-type KJ (KJΔBC/KJ). Navy blue color indicates the transcript change of the gene is greater than or equal to 3. Blue color indicates the transcript change of the gene is less than 3 and greater than 2. Light blue color indicates the transcript change of the gene is less than 2 and greater than 1. (A) The genomic organizations of the flagella-related genes. Based on the transcriptome assay, the flagella-related genes, upregulated in case of creBC inactivation, are located in three clusters, Smlt0561-0562, Smlt2265-2290, and Smlt2302-2321. The orientation of gene is indicated by the arrow. (B) Schematic diagram of bacterial flagellum. The flagellum consists of the basal body, the hook, and the filament. The composition proteins are labelled. OM, outer membrane; PG, peptidoglycan layer; IM, inner membrane.
Table 1. Flagella-associated genes differently expressed in S. maltophilia KJ and KJΔBC cells.
| Locus | Normalized expression | Fold change RNAseq | Encoded protein | |
|---|---|---|---|---|
| KJ | KJΔBC | |||
| Smlt0561 | 12.47 | 49.33 | 3.95 | flagellar motor protein MotB |
| Smlt0562 | 40.50 | 108.31 | 2.64 | flagellar motor protein MotA |
| Smlt2265 | 10.09 | 39.25 | 3.88 | flagellar motor protein MotD |
| Smlt2266 | 24.16 | 45.68 | 1.89 | flagellar motor protein |
| Smlt2267 | 9.92 | 46.19 | 4.62 | two component sensor kinase |
| Smlt2268 | 16.94 | 64.45 | 3.80 | chemotaxis protein |
| Smlt2269 | 18.76 | 31.80 | 1.69 | two component response regulator |
| Smlt2270 | 8.14 | 43.39 | 5.33 | RNA polymerase sigma factor, FliA |
| Smlt2271 | 17.64 | 47.89 | 2.71 | ParA family ATPase flagella number regulator |
| Smlt2272 | 7.78 | 73.37 | 9.42 | flagellar biosynthesis regulator, FlhF |
| Smlt2273 | 4.93 | 32.43 | 6.57 | flagellar biosynthesis protein FlhA |
| Smlt2274 | 6.63 | 28.54 | 4.30 | flagellar biosynthesis protein FlhB |
| Smlt2275 | 40.32 | 67.55 | 1.67 | esterase/peptidase |
| Smlt2276 | 11.89 | 27.22 | 2.28 | transmembrane GGDEF EAL domain signaling protein |
| Smlt2277 | 3.74 | 22.42 | 5.99 | flagellar biosynthetic protein FliR |
| Smlt2278 | 1.95 | 21.86 | 11.20 | flagellar biosynthetic protein FliQ |
| Smlt2279 | 11.81 | 19.14 | 1.61 | flagellar biosynthesis protein FliP |
| Smlt2280 | 4.70 | 45.87 | 9.75 | flagellar protein FliO |
| Smlt2281 | 3.52 | 14.98 | 4.24 | flagellar rotor switch protein FliN |
| Smlt2282 | 6.15 | 21.93 | 3.56 | flagellar rotor switch protein FliM |
| Smlt2283 | 4.84 | 16.82 | 3.46 | flagellar basal body-associated protein FliL |
| Smlt2284 | 11.31 | 59.35 | 5.24 | flagellar hook-length control protein FliK |
| Smlt2285 | 11.25 | 27.44 | 2.43 | flagellar FliJ protein |
| Smlt2286 | 6.55 | 48.97 | 7.47 | flagellum-specific ATP synthase FliI |
| Smlt2287 | 5.53 | 27.03 | 4.88 | flagellar assembly protein FliH |
| Smlt2288 | 6.83 | 24.42 | 3.57 | flagellar rotor switch protein FliG |
| Smlt2289 | 5.43 | 29.72 | 5.46 | flagellar MS-ring protein FliF |
| Smlt2290 | 6.47 | 22.29 | 3.44 | flagellar hook-basal body complex protein FliE |
| Smlt2302 | 16.41 | 47.87 | 2.91 | flagellar protein FliS |
| Smlt2303 | 23.84 | 39.02 | 1.63 | flagellar hook-associated protein FliD |
| Smlt2304 | 75.80 | 77.76 | 1.02 | Flagellin FliC |
| Smlt2305 | 21.09 | 30.04 | 1.42 | flagellin FlaA |
| Smlt2306 | 32.67 | 59.85 | 1.83 | flagellin |
| Smlt2307 | 12.47 | 25.68 | 2.05 | flagellar hook-associated protein FlgL |
| Smlt2308 | 13.92 | 45.03 | 3.23 | flagellar hook-associated protein FlgK |
| Smlt2309 | 8.96 | 44.14 | 4.92 | flagellar rod assembly protein/muramidase FlgJ |
| Smlt2310 | 10.13 | 35.53 | 3.50 | flagellar basal body P-ring protein FlgI |
| Smlt2311 | 10.83 | 43.08 | 3.97 | flagellar basal body L-ring protein, FlgH |
| Smlt2312 | 11.89 | 36.44 | 3.06 | flagellar basal body rod protein FlgG |
| Smlt2313 | 8.07 | 24.53 | 3.03 | flagellar basal body rod protein FlgF |
| Smlt2314 | 15.49 | 39.00 | 2.51 | flagellar hook protein FlgE |
| Smlt2315 | 9.24 | 22.33 | 2.41 | flagellar basal body rod modification protein FlgD |
| Smlt2316 | 12.90 | 16.16 | 1.25 | flagellar basal body rod protein FlgC |
| Smlt2317 | 7.98 | 24.11 | 3.02 | flagellar basal body rod protein FlgB |
| Smlt2318 | 15.18 | 24.79 | 1.63 | two-component response regulator chemotaxis signal |
| Smlt2319 | 34.26 | 55.22 | 1.61 | flagellar basal body P-ring biosynthesis protein FlgA |
| Smlt2320 | 192.76 | 195.71 | 1.01 | FlgM |
| Smlt2321 | 57.48 | 94.73 | 1.64 | flagella protein FlgN |
A set of nine flagella-related genes was chosen for validation by qRT-PCR, including Smlt0562 (motA), Smlt2270 (fliA), Smlt2278 (fliQ), Smlt2286 (fliI), Smlt2289 (fliF), Smlt2303 (fliD), Smlt2304 (fliC), Smlt2310 (flgI), and Smlt2314 (flgE) (Fig 5). Overall, the results of qRT-PCR analysis were in good agreement with the transcriptome data (Table 1).
Fig 5. The transcript changes (folds) of selected flagella-related genes between KJ and KJΔBC cells by qRT-PCR.
Total mRNA was extracted from KJ and KJΔBC logarithmic-phase cultures. cDNA was prepared by RT-PCR and used as the template for qRT-PCR. The expression of target gene transcripts in qRT-PCR were normalized to the level of expression of the 16S rRNA gene by using the ΔΔCT method. Data are the means from three independent experiments. Error bars represent the standard deviations for three triplicate samples. *, p < 0.05; **, p < 0.005.
CreD upregulation in KJΔBC alleviates the ΔcreBC-mediated membrane integrity compromise
The observation that the deletion of the creD allele from KJΔBC compromised swimming motility (Fig 3A) supports the involvement of CreD in the ΔcreBC-mediated increase in swimming motility. CreD is an inner membrane protein with six transmembrane α-helix domains (http://www.cbs.dtu.dk/services/TMHMM/). In our recent study, we showed that the creD mutant, KJΔCreD, has cell division defects and aberrations in cell envelope integrity [14], strengthening the possibility that CreD acts in the architectural frame of the inner membrane and plays a critical role in the maintenance of membrane integrity. Intact membrane architecture is essential for the successful assembly and function of the flagellum. Therefore, we considered whether CreD upregulation in KJΔBC cells contributes to the maintenance of membrane integrity, inferring a relationship between CreD and swimming motility. Membrane susceptibility of KJ, KJΔBC, and KJΔBCD to the detergent triton X-100 was assessed. The growth of KJΔBC in the presence of triton X-100 was compromised compared to that of wild-type KJ, and this impairment was further exacerbated when the ΔcreD allele was introduced into the chromosome of KJΔBC (Fig 6). This observation supports the contention that CreD upregulation in KJΔBC alleviates the ΔcreBC-mediated membrane integrity compromise, which might benefit flagellum construction and swimming motility.
Fig 6. The roles of creBC and creD in membrane susceptibility to Triton X-100.
The overnight-cultured bacteria were inoculated into fresh LB broth containing Triton X-100 of 200 μg/ml at the initial OD450 of 0.15. The bacterial growth was monitored by recording the OD450nm. Data are the means from three independent experiments. Error bars indicate the standard deviations for three triplicate samples.
KJΔBC cells show elevated CCCP50 value compared to KJ cells
The inner membrane of bacterial cells harbors a membrane potential, which is formed by the differences in the concentrations of ions on opposite sides of an inner membrane. One of the functions of the membrane potential is to provide power to operate flagellum rotation and drive swimming [19]. Given the upregulation of genes encoding rotor and stator proteins in KJΔBC cells (Figs 4 and 5), we evaluated whether upregulation of stator proteins in KJΔBC cells would provide a higher membrane potential, contributing to enhanced swimming. Carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a chemical inhibitor of oxidative phosphorylation, causes uncoupling of the proton gradient and thus abolishes swimming motility [20]. Therefore, we determined the CCCP50 of KJ cells and KJΔBC cells to determine to what extent membrane potential contributes to swimming. CCCP50 is defined as the CCCP concentration at which 50% of bacterial swimming is inhibited. We found that inactivation of creBC increased the CCCP50 value by approximately 1.3-fold (Table 2). Next, a complementation assay was performed. As a control, we also introduced the empty pRK415 vector into KJ and KJΔBC. Surprisingly, empty vector introduction had an opposite effect on CCCP50 values; the CCCP50 value of KJΔBC(pRK415) was lower than that of KJ(pRK415) (Table 2). A similar observation for plasmid introduction-mediated phenotypic deviation was obtained in the aforementioned swimming phenotype (Fig 3B). The plasmid introduction-mediated phenotypic deviation will be discussed below. Complementation of KJΔBC with pCreB or pCreB(D55E) reverted the CCCP50 value to the wild-type level (Table 2), supporting that creB-mediated signalling affects the membrane potential. Furthermore, we found that the CCCP50 values of the strains assayed (Table 2) were positively correlated with swimming motility (Fig 3).
Table 2. The CCCP50 values of S. maltophilia KJ, its isogenic creBC mutant (KJΔBC), and the complementary strains.
| Strain | CCCP50a (μg/ml) |
|---|---|
| KJ | 33.4 ± 1.0 |
| KJΔBC | 43.2 ± 0.4 |
| KJ(pRK415) | 31.4 ± 1.1 |
| KJΔBC(pRK415) | 24.4 ± 0.8 |
| KJΔBC(pCreB) | 39.8 ± 8.5 |
| KJΔBC(pCreB(D55E)) | 31.9 ± 1.8 |
a CCCP50 is defined as the required CCCP concentration at which the fifty percent of bacterial swimming is inhibited.
Discussion
The CreBC/BlrAB TCS in E. coli, Aeromonas spp., and P. aeruginosa is considered a defense system, helping bacteria to alleviate stresses, since it is activated by certain pressures such as anaerobic environments or β-lactam challenge. However, the CreBC of S. maltophilia is constitutively active in laboratory LB-cultured conditions without foreign stresses, signifying its possible role in the maintenance of bacterial physiology not limited to stress defense. Our results in this study provide evidence that the CreBC TCS of S. maltophilia negatively modulates swimming motility.
Swimming motility is an important mechanism for bacterial survival, allowing bacteria to approach nutrient sources, invade host cells, and escape from attack [21]. Nevertheless, swimming motility is an energy-consuming process, and inefficiencies result in energy waste, which curtails utilizable energy for bacterial growth [22]. Therefore, exquisitely modulating swimming motility is critical for bacterial survival in different environmental niches. It has been reported that TCSs generally act as positive regulators of swimming motility in response to environmental stimuli. Examples of this include the BceSR TCS of Burkholderia cenocepacia, the RpfCG TCS of Xanthomonas albilineans, and the QseBC TCS of Aeromonas hydrophila [23–25]. In a recent study, Zheng et al. successfully constructed 51 histidine kinase (HK) mutants of S. maltophilia and swimming motility was assessed in these mutants. Of 51 HK mutants, six had deficiency in swimming motility (not caused by growth defects) and no mutations were found to enhance swimming motility [7]. Unfortunately, a creC mutant could not be successfully constructed and therefore was not included in their assays. However, their findings provided an indication that at least six constitutively active TCS systems positively regulate swimming motility in S. maltophilia in the laboratory-cultured conditions [7]. In this study, we successfully constructed the creBC mutant and verified the role of the CreBC TCS in the negative regulation of swimming motility.
Examples of TCS systems acting as negative modulators of swimming motility have seldom been reported, with the exception of the GacS-GacA systems of P. fluorescens F113 and P. chlororaphis O6 [26–27]. The underlying mechanisms of GacSA TCS regulation of swimming motility are attributed to increasing flagella elongation [26] or flagella numbers [27]. In this study, we provide another example of a TCS (specifically CreBC) negatively regulating swimming motility in S. maltophilia, as a mutation in the creBC genes resulted in increased motility (Fig 3A). Distinct from GacS-GacA in Pseudomonas spp., CreBC inactivation-mediated swimming motility in S. maltophilia might result from increased motor output of the flagellum, rather than alterations in flagella numbers or morphology. This inference was supported by the observations that the motor-associated genes (motA, motB, fliG, fliM, fliN, and fliQ) were highly upregulated and the helical filament-associated genes (fliC and fliD) were normally expressed in KJΔBC (Table 1, Figs 4 and 5). Furthermore, the membrane potential of KJΔBC cells was higher than that of wild-type KJ cells (Table 2).
Interestingly, we found that some phenotypes of the KJΔBC mutant showed dramatically changes when an empty vector (pRK415) was introduced, including the swimming motility (Fig 3B) and CCCP50 value (Table 2). There are two possible explanations for this as follows: (i) plasmid introduction may affect the expression of some genes in the CreBC regulon, and these genes affect the phenotypes assayed. CreD, a member of the CreBC regulon, is such an example. (ii) Plasmid carriage is an energy-consuming process, which may redistribute the energy utilization in bacteria and thus alter some energy-dependent phenotypes. This may also explain why some complementation assays performed by ectopic expression of the mutated genes in this study could not fully restore the phenotypes of the mutant to the wild-type level.
We have previously indicated that creD expression is upregulated in response to CreBC inactivation [14]. In this study, we further demonstrated that CreD upregulation makes a significant contribution to cell membrane integrity in KJΔBC (Fig 6). The relevance of CreD upregulation and cell membrane integrity for enhanced swimming motility are highlighted by the fact that enhanced swimming motility and cell membrane integrity in KJΔBC are compromised by creD inactivation (Figs 3A and 6). According to these observations, we propose a model for the negative regulatory role of the CreBC TCS in swimming motility in S. maltophilia. The CreBC TCS is constitutively active, signifying its importance in bacterial physiology. Activated CreB maintains the expression of flagella-associated genes at an adequate level, especially the membrane proton flow related genes (motA and motB), preventing inefficient swimming and thus refining bacterial energy utilization in S. maltophilia. In the absence of a functional CreBC TCS, some physiological functions of S. maltophilia are compromised, such as secreted protease activity (Fig 2A) and oxidative stress tolerance (our unpublished data), threatening bacterial survival during stress challenge. Nevertheless, ΔcreBC-mediated swimming motility enhancement could provide a survival benefit for KJΔBC by effectively escaping from stresses. The underlying mechanisms for enhanced swimming motility in KJΔBC may involve the upregulation of creD and increased membrane potentials. Although the exact reason for the membrane potential elevation of KJΔBC cells remains unclear, upregulation of flagella-associated genes encoding basal body- and motor-associated proteins may be involved. Increased CreD enforces the membrane integrity, which is critical for flagellum assembly and motility.
Materials and methods
Bacterial strains and culture conditions
A complete list of strains, plasmids, and primers used in this study is shown in S2 Table. S. maltophilia KJ acts as the parental wild type strain [28]. Cells were grown at 37°C in Luria-Bertani (LB) broth.
Construction of deletion mutants KJΔCreB and KJΔCreC
The KJΔCreB and KJΔCreC in-frame deletion mutants were constructed by double-crossover homologous recombination between the wild-type KJ chromosome and plasmids pΔCreB and pΔCreC, respectively. The pΔCreB was prepared as follows: the intact creB gene was amplified from the wild-type KJ chromosome using the primers CreB-F and CreB-R (S2 Table) and cloned into pEX18Tc, yielding pEXCreB. Plasmid pEXCreB was digested by PstI and then self-ligated to generate pΔCreB, in which the internal 402-bp PstI-PstI fragment of creB was deleted. The pΔCreC was prepared as follows: two DNA fragments targeting the 5’ terminus and the 3’ terminus of the creC genes were obtained by PCR using the primer sets CreCN-F/CreCN-R and CreCC-F/CreCC-R (S2 Table). The PCR amplicons were digested and subsequently cloned into pEX18Tc. Plasmid mobilization, transconjugants selection, and mutant confirmation were performed as described previously [29]. The 44 to 177 amino acids of CreB and the 4 to 359 amino acids of CreC were thus deleted in the in-frame deletion mutants KJΔCreB and KJΔCreC respectively.
Plasmids construction
The plasmids pCreB and pCreC were constructed by cloning creB and creC amplified with primers CreB-F/CreB-R and CreCN-F/CreCC-R into the pRK415, respectively. The creB(D55E) allele was generated by site-directed mutagenesis using primer extension PCR as described previously [14] and cloned into pRK415 at HindIII/XbaI sites to generate pCreB(D55E). All constructs were verified by sequencing. All primers are listed in S2 Table in the supplemental materials.
Scanning electron microscope (SEM)
The bacterial cells for SEM observation were prepared as described previously14. Briefly, the bacterial cells (OD450nm of 1.0) were harvested by centrifugation and washed three times with PBS (pH7.4). The bacteria were pre-fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4), washed, post-fixed with 1% osmium tetraoxide (OsO4), and then dehydrated by ethanol. The high-resolution FEI Inspect S scanning electron microscope was used for the observation of bacterial cells.
Secreted protease activity assay
The secreted protease activity of bacteria was assayed using LB agar containing 1% skim milk. For the convenience of bacterial suspension loading, the skim milk agar was prepared with a 6-mm-diameter hole in the center. The overnight cultured bacteria were adjusted to an OD450 of 1.0 and 40 μl of the bacterial suspension was dripped onto the hole of the skim milk agar plates. After incubation at 37°C for 72 h, the secreted protease activity of bacteria was assessed by measuring the protease hydrolyzing zones around the bacteria.
Swimming assay
The bacterial strains tested were grown to an OD450 of 1.0, and 2 μl of bacterial suspension was inoculated at the swimming agar surface (1% tryptone, 0.5% NaCl, and 0.15% agar) [30]. The plates were incubated at 37°C for 48 h. Results are expressed as diameters (millimeters) of swimming zones. For the determination of CCCP50, swimming assay was performed using the swimming agar containing the CCCP of 0,10, 20, 30, 40, 50, and 60 μg/ml, respectively. CCCP50 is defined as the CCCP concentration at which 50% of bacterial swimming is inhibited.
Flagella staining
The bacterial strains tested were grown to an OD450 of 1.0. And the bacterial suspension was diluted twice with PBS (pH 7.4). The bacterial suspension was negatively stained with 1% phosphotungstic acid (pH 7.4) on Formvar-coated copper grids [29]. The presence of flagella was observed by transmission electron microscope (TEM) (Hitachi H-7650 microscope).
Transcriptome sample preparation and sequencing
Total RNA isolation, ribosomal RNA (rRNA) depletion, adapter-ligated cDNA library construction and enrichment, and cDNA sequencing were performed as described previously [6]. After trimming of low quality of bases (< Q30), the first 12 bases and adapters, the trimmed Reads were mapped to the Stenotrophomonas maltophilia K279a genome (GenBank acc. no. NC_010943.1) and run RNA-seq analysis by CLC Genomics Workbench v 6.0 (CLC Bio). RNA-seq data representing the alignment of sequences (short reads) to coding sequences (CDS) were quantified as reads per kilobase CDS length per million reads analyzed (RPKM). The sequence dataset was deposited in NCBI Sequence Read Archive (SRA) database under STUDY accession number SRP100809.
Quantitative real-time PCR (qRT-PCR)
Total cellular DNA-free RNA extraction, cDNA preparation, and the transcripts of the flagella-related genes determination were carried out as described previously [29]. A complete list of primers used for qRT-PCR is shown in S2 Table. The 16S rRNA gene was used as the normalizing gene. The relative expression of mRNA from each gene of interest was determined by the comparative cycle threshold (CT) method [31].
Triton X-100 susceptibility test
The envelope integrity of bacteria was assessed by evaluating the bacterial capability to protect the membranes against Triton X-100. Overnight cultures of the bacteria strains were diluted to an OD450 of 0.15 with LB broth containing 200 μg/ml Triton X-100. The OD450nm was read at an interval of 2 h.
Supporting information
(JPG)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by grant MOST 104-2320-B-010-023-MY3 from Ministry of Science and Technology of Taiwan and grant 40419001 from Professor Tsuei-Chu Mong Merit Scholarship. The authors acknowledge the High-throughput Genome and Big Data Analysis Core Facility of National Core Facility Program for Biotechnology, Taiwan (MOST 104-2319-B-010-001), for sequencing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grant MOST 104-2320-B-010-023-MY3 from Ministry of Science and Technology of Taiwan and grant 40419001 from Professor Tsuei-Chu Mong Merit Scholarship. The authors acknowledge the High-throughput Genome and Big Data Analysis Core Facility of National Core Facility Program for Biotechnology, Taiwan (MOST 104-2319-B-010-001), for sequencing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25: 2–41. doi: 10.1128/CMR.00019-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010;64: 539–559. doi: 10.1146/annurev.micro.112408.134054 [DOI] [PubMed] [Google Scholar]
- 3.Gao R, Stock AM. Biological insights from structures of two-component protein. Annu Rev Microbiol. 2009;63: 133–154. doi: 10.1146/annurev.micro.091208.073214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki AK, Sebaihia M, et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008;17: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li XZ., Zhang L, Poole K. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2002;46: 333–343. doi: 10.1128/AAC.46.2.333-343.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu CJ, Huang YW, Lin YT, Ning HC, Yang TC. Inactivation of SmeSyRy two-component regulatory system inversely regulates the expression of SmeYZ and SmeDEF efflux pumps in Stenotrophomonas maltophilia. PLoS One. 2016;11: e0160943 doi: 10.1371/journal.pone.0160943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng L, Wang FF, Ren BZ, Liu W, Liu Z, Qian W. Systematic mutational analysis of histidine kinase genes in the nosocomial pathogen Stenotrophomonas maltophilia identifies BfmAK system control of biofilm development. Appl Environ Microbiol. 2016;82: 2444–2456. doi: 10.1128/AEM.03951-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avison MB, Horton RE, Walsh TR, Bennett PM. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J Biol Chem. 2001;276: 26955–26961. doi: 10.1074/jbc.M011186200 [DOI] [PubMed] [Google Scholar]
- 9.Moya B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, et al. β-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 2009;5: e1000353 doi: 10.1371/journal.ppat.1000353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tayler AE, Ayala JA, Niumsup P, Westphal K, Baker JA, Zhang L, et al. Induction of β-lactamase production in Aeromonas hydrophilia is responsive to β-lactam-mediated changes in peptidoglycan composition. Microbiology 2010;156: 2327–2335. doi: 10.1099/mic.0.035220-0 [DOI] [PubMed] [Google Scholar]
- 11.Alksne LE, Rasmussen BA. Expression of the AsbA1, OXA-12, and AsbM1 β-lactamases in Aeromonas jandaei AER 14 is coordinated by a two-component regulon. J Bacteriol. 1996;179: 2006–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niumsup PA, Simm AM, Nurmahomed K, Walsh TR, Bennett PM, Avison MB. Genetic linkage of the penicillinase gene, amp, and blrAB, encoding the regulator of β-lactamase expression in Aeromonas spp. J Antimicrobl Chemother. 2003;51: 1351–1358. [DOI] [PubMed] [Google Scholar]
- 13.Zamorano L, Moya B, Juan C, Mulet X, Blazquez J, Oliver A. The Pseudomonas aeruginosa CreBC two-component system plays a major role in the response to β-lactam, fitness, biofilm growth, and global regulation. Antimicrob Agents Chemother. 2014;58: 5084–5095. doi: 10.1128/AAC.02556-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang HH, Lin YT, Chen WC, Huang YW, Chen SJ, Yang TC. Expression and functions of CreD, an inner membrane protein in Stenotrophomonas maltophilia. PLoS One. 2015;10: e0145009 doi: 10.1371/journal.pone.0145009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaban B, Hughes HV, Beebv M. The flagellum in bacterial pathogens: from motility and a whole lot more. Semin Cell Dev Biol. 2015;46: 91–103. doi: 10.1016/j.semcdb.2015.10.032 [DOI] [PubMed] [Google Scholar]
- 16.Morimoto YV, Minamino T. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 2014;4: 217–234. doi: 10.3390/biom4010217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toutain CM, Zegans ME, O’Toole GA. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol. 2005;187: 771–777. doi: 10.1128/JB.187.2.771-777.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang YW, Wu CJ, Hu RM, Lin YT, Yang TC. Interplay among membrane-bound lytic transglycosylase D1, the CreBC two-component regulatory system, the AmpNG-AmpDI-NagZ-AmpR regulatory circuit, and L1/L2 β-lactamase expression in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2015;59: 6866–6872. doi: 10.1128/AAC.05179-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terashima H, Kojima S, Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol. 2008;270: 39–85. doi: 10.1016/S1937-6448(08)01402-0 [DOI] [PubMed] [Google Scholar]
- 20.Kanda E, Tatsuta T, Suzuki T, Taguchi F, Naito K, Inagaki Y, et al. Two flagellar stators and their roles in motility and virulence in Pseudomonas syringae pv. tabaci 6605. Mol Genet Genomics. 2011;285: 163–174. doi: 10.1007/s00438-010-0594-8 [DOI] [PubMed] [Google Scholar]
- 21.Ottemann KM, Miller JF. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24: 1109–1117. [DOI] [PubMed] [Google Scholar]
- 22.Berg HC. The rotary motor of bacterial flagella. Biochemistry 2003;72:19–54. [DOI] [PubMed] [Google Scholar]
- 23.Khajanchi BK, Kozlova EV, Sha J, Popov VL, Chopra AK. The two-component QseBC signaling system regulates in vitro and in vivo virulence of Aeromonas hydrophilia. Microbiology 2012;158: 259–271. doi: 10.1099/mic.0.051805-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rott P, Fleites LA, Mensi I, Sheppard L, Daugrois JH, Dow JM, et al. The RpfCG two-component system negatively regulates the colonization of sugar can stalks by Xanthomonas albilineans. Microbiology 2013;159: 1149–1159. doi: 10.1099/mic.0.065748-0 [DOI] [PubMed] [Google Scholar]
- 25.Merry CR, Perkins M, Mu L, Peterson BK, Knackstedt RW, Weingart CL. Characterization of a novel two-component system in Burkholderia cenocepacia. Curr Microbiol. 2015;70: 556–561. doi: 10.1007/s00284-014-0744-z [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, Rivilla R, Martin M. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS One. 2012;7: e31765 doi: 10.1371/journal.pone.0031765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JS, Kim YH, Anderson AJ, Kim YC. The sensor kinase GacS negatively regulates flagellar formation and motility in a biocontrol bacterium, Pseudomonas chloroaphis O6. Plant Pathol J. 2014;30: 215–219. doi: 10.5423/PPJ.NT.11.2013.0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu RM, Huang KJ., Wu LT, Hsiao YJ, Yang TC. Induction of L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2008;52: 1198–1200. doi: 10.1128/AAC.00682-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang TC, Huang YW, Hu RM, Huang SC, Lin YT. AmpD1 is involved in expression of the chromosomal L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2009;53: 2902–2907. doi: 10.1128/AAC.01513-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray TS, Kazmiercrak BI. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol. 2006;188: 6995–7004. doi: 10.1128/JB.00790-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (delta deltaC(T)) method. Methods. 2001;25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPG)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.