Abstract
The calcium-dependent protein kinase (CDPK) is a ser/thr protein kinase that plays vital roles in plant growth, development, and responses to multiple stresses. Despite an important member of the stress responsive gene family, little is known about the evolutionary history and expression patterns of CDPK genes in melon. Herein, a total of 18 CDPK genes and 7 CDPK-related protein kinases (CRK) genes were identified in the melon genome via bioinformatic analysis, which were unevenly distributed across eleven chromosomes with an apparent exception for chromosome 3. Comparative syntenic analysis between Cucumis melo L. and Arabidopsis thaliana revealed that 13 CmCDPKs and 19 AtCPKs existed in 20 corresponding syntenic blocks. In addition, based on gene structure and phylogenetic analyses, all CmCDPKs were divided into four groups (CDPK I-IV) and CmCRKs clustered into one group (CRK I). Interestingly, group CDPK IV was clearly distinct from the other three CDPK groups, but clustered with CRK I on the phylogenetic tree, implying their origination from a common ancestor. Furthermore, CmCDPKand CmCRK genes were differentially expressed in response to various stimuli, such as biotic stress (Podosphaera xanthii), abiotic stress (salt and cold), and hormone (abscisic acid) treatment. To our knowledge, this is the first report on CDPK and CRK gene families in melon, which provides a basic foundation for functional characterizations of CmCDPK and CmCRK genes in the future.
Introduction
To survive frequently occurring environmental stresses during plant growth process, plants have evolved an effective defense mechanism comprised of sophisticated signal transduction pathways. Calcium (Ca2+), a universal second messenger, plays an important role in plant growth, development, and responses to various environmental stimuli [1–3]. During exposure of plants to stress, transient changes in Ca2+ concentrations in the cytoplasm can be sensed and decoded by an array of specific Ca2+ sensors and Ca2+-binding proteins, such as calmodulins (CaM), calmodulin-like proteins (CaML), calcineurin B-like proteins (CBL), and the calcium-dependent protein kinase (CDPK) [4–6]. CaM, CaML, and CBL serve as Ca2+ sensors that can combine and interact with target proteins to transfer signals to downstream pathways. However, CDPKs feature the unique activity of both Ca2+ sensing and response within a single protein that can directly translate Ca2+ signals into downstream phosphorylation signals. Thus, only CDPKs can function both as Ca2+ sensors and effectors [5,7–9].
Calcium-dependent protein kinases (CDPKs) are ser/thr protein kinases, also named CPKs, which have been identified throughout the plant kingdom as well as in several protozoa, but are absent in animals [10–12]. In different plant species, CDPKs have been identified and characterized via four typical domains, including a variable N-terminal domain (consisting of the myristoylation and palmitoylation sites), a catalytic ser/thr protein kinase domain, an auto-inhibitory domain (acting as a pseudosubstrate combined with kinase domain to inhibit activity), and a C-terminal regulatory calmodulin-like domain, containing EF-hand motifs for Ca2+ binding [13–15]. CDPK-related kinases (CRKs) are another type of protein kinase that share similar domain structures with CDPK, such as the ser/thr kinase domain. However, CRKs belong to an autocephalous gene family due to a lack of EF-hand domains [16,17].
To date, genome-wide analyses have identified 34 CDPK genes in Arabidopsis [13], 31 CDPK genes in rice [18,19], 20 CDPK genes in wheat [20], 30 CDPK genes in poplar [21], and 41 CDPK genes in cotton [22]. In recent years, an increasing number of CDPK genes have been identified in horticultural plants, such as 19 CDPK genes in cucumber [23], 29 CDPK genes in tomato [16] and 31 CDPK genes in pepper [17]. Compared to extensive studies of CDPK genes, only a few studies have been focused on the identification of CRK genes. By far, genome-wide analyses have identified eight CRKs in Arabidopsis [14], five CRKs in rice [19], nine CRKs in poplar [21], six CRKs in tomato [16] and five CRKs in pepper [17]. Nevertheless, our knowledge of CDPK and CRK gene families for many other economically important horticultural crops, such as the melon (Cucumis melo L.), still remains scanty.
CDPK genes are ubiquitously expressed in plant different organs, such as roots, stems, flowers, fruits and seeds [18,20,23,24], and their localizations have been validated in the plasma membrane, cytoplasm, nucleus, chromatin, cytoskeleton, chloroplast and mitochondrion [25–29]. Numerous studies have confirmed that CDPKs are involved not only in plant growth and development, but also in abiotic and biotic stress responses. In Arabidopsis, AtCPK24 plays an important role in pollen development by connecting pathways between the vegetative nucleus and generative cells [12]. OsCPK8, OsCPK17 and OsCPK28 are almost undetectable in the panicles of rice, but are highly abundant in vegetative tissues [30]. In sandalwood, CDPKs are transiently expressed in the endosperm, but are highly activated and accumulated during periods of sprouting and fruit ripening [31], indicating that CDPKs participate in various processes of plant growth and development. In recent years, another pivotal role of CDPKs has been confirmed in the response of plants to biotic stresses. Some grapevine CDPK genes, VpCDPK6, VpCDPK9, VpCDPK14, VpCDPK16, and VpCDPK19 play positive roles to powdery mildew pathogen caused by Erysiphe necator [32]. Expression of gene TaCPK2 can be induced by powdery mildew in wheat [20], and CaCDPK10 plays a negative role to Ralstonia solanacearum in Capsicum annuum [17]. CDPKs are also involved in the response of plants to abiotic stresses. OsCPK4, OsCPK7, OsCPK12, and OsCPK21 transcripts were reported to be up-regulated in rice in response to salt stress [33–36]. During salt stress, AtCPK23 is known to play a role in the response to salt stress by controlling K+ channels [37]. Furthermore, OsCPK12 promotes salt tolerance via a reduction in ROS accumulation [35]. During cold stress, low temperature induced expression of OsCPK13 [38], but ZmCPK1 was reported to function as a negative regulator in the signaling pathway that responds to cold stimuli [39]. During drought stress, ZmCPK4 and OsCPK9 can enhance drought tolerance via stomatal closure [40,41]. AtCPK4 and AtCPK11 interact with the ABA transcription factors ABF1 and ABF4 and phosphorylate them to regulate drought tolerance [42]. AtCPK10 is involved in plant responses to drought stress via the modulation of ABA- and Ca2+- regulated stomatal movements [43]. Moreover, CDPKs are also reported to participate in the signal transformation of hormone, such as abscisic acid (ABA) [44–46], salicylic acid (SA) [47], jasmonic acid (JA) [48], and gibberellic acid (GA) [38,49].
Despite extensive studies of CDPK and CRK genes on many other plant species, little is known about these two gene families in melon, an agriculturally and economically important crop around the world. Heavy losses in melon production are frequently caused by various biotic stresses such as powdery mildew, and abiotic stresses including drought, salt and extreme temperatures [50]. The availability of the complete melon genome sequence provides an opportunity to perform a genome-wide analysis of multigene families [51]. In this study, we identified CDPK and CRK gene families in the melon, and analyzed their genomic structures and chromosomal distributions, as well as their syntenic and phylogenetic relationships. To further elucidate their possible involvement in melon stress responses, transcriptional expression analyses were also performed under biotic (powdery mildew) and abiotic (salinity and low temperature) stresses as well as ABA treatment. Our study provides new insights into the evolutionary history of the CmCDPK and CmCRK gene families and reveals a set of potential candidate genes for future genetic modification to increase pathogen resistance and stress tolerance in the melon.
Materials and methods
Identification of melon CDPK and CRK genes
The protein sequences for 34 CDPKs and 8 CRKs of Arabidopsis were obtained from the Arabidopsis Information Resource (http://www.Arabidopsis.org/). To identify the melon CDPK and CRK gene family, Arabidopsis CDPK and CRK protein sequences were used as queries to search against the melon genome (https://melonomics.net/) using BLASTp with the E-value setting to 1e-5. In addition, HMM (Hidden Markov Model) profiles of protein kinase domain (PF00069) and EF-hand_7 domain (PF13499) [32] were downloaded from Pfam database [52] and also exploited for the identification of CDPK and CRK genes from melon genome using HMMER 3.0 (http://hmmer.janelia.org/) with default parameters. To further verify the reliability of these candidate genes, we also performed BLASTp search at NCBI using full-length amino acid of putative melon CDPK and CRK genes. Among those genes with alternative splice variants, the longest was chosen for further analysis. All non-redundant putative candidates were verified with the InterProScan program (http://www.ebi.ac.uk/interpro/) to confirm their completeness and presence of the core domain. Subsequently, all candidate gene sequences were future examined via the following online tools: SMART (http://smart.embl-heidelberg.de/) [53], Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/cdd/), and ScanProsite (http://prosite.expasy.org/scanprosite/) [54]
Genomic distribution and synteny analysis of CDPK and CRK genes in melon
Genes were mapped on chromosomes via identification of their chromosomal position, obtained from the melon database (https://melonomics.net/). MapInspect software was used to draw chromosomal distribution of CmCDPKs and CmCRKs (http://mapinspect.software.informer.com/1.0/). MCScanX software was utilized to detect the synteny relationship and duplication pattern of CDPK and CRK genes in the melon genome, as well as the synteny relationship between melon and Arabidopsis [55]. All melon protein sequences were subjected to search against themselves and proteins of Arabidopsis, respectively, using BLASTp with parameters E-value < 1e-10, and setting the output format as tabular (-m 8) [56]. The BLASTp tabular file combined with the melon gene and Arabidopsis gene location files served as input for MCScanX to analyze syntenic relationship and duplication types with default settings, visualizing them via Circos (http://circos.ca/).
Genomic structure and phylogenetic analysis
Genomic DNA sequences of CmCDPKs and CmCRKs were obtained from the melon database (https://melonomics.net/), and corresponding cDNA sequences were downloaded from PLAZA (http://bioinformatics.psb.ugent.be/plaza/). The exon- intron organization was carried out with the online tool GSDS2.0 (http://gsds.cbi.pku.edu.cn/) [57]
Full-length protein sequences of CDPK and CRK genes in Arabidopsis, tomato, and rice were obtained from the Arabidopsis Information Resource (TAIR, https://www.Arabidopsis.org/), tomato genome database (http://solgenomics.net/), and rice genome database (TIGR, http://rice.tigr.org), respectively.
Full-length CDPK protein sequences of Arabidopsis (34) [13], tomato (29) [16], rice (29) [19], and melon (18), as well as CRK protein sequences of Arabidopsis (8) [14], tomato (6) [16], rice (5) [19], and melon (7) were aligned via the ClustalX 2.0 program with default settings. Finally, a phylogenetic tree was constructed with the aligned sequences, via MEGA6.0, using the neighbor-joining method and 1000 replicates for bootstrap [58].
Plant material and stress treatments
The experiment was conducted in a greenhouse at the Northwest A&F University in China. The melon variety “Nantais Oblong”, an inbred line provided by the vegetable research center of Beijing, was used in our experiment. The seeds of melon variety “Nantais Oblong” were cultured in a greenhouse at a constant daily temperature of 25–28°C, a night temperature of 16–20°C, and a relative humidity of 60% to 80%. The roots, stems, leaves, male flowers and tendrils were sampled separately and placed into liquid nitrogen for the tissue specific analysis. When the seedlings reached the three- or four-leaf stage, uniform melon seedlings were selected and used for the future experiments. For biotic stress experiment, powdery mildew (maintained on leaves of Podosphaera xanthii race 2F) was sub-cultured onto fresh leaves every fourteen days. Then foliar portions of plants were inoculated using leaf-brushing inoculation at night and incubated in the greenhouse at a constant daily temperature of 25–28°C, a night temperature of 16–20°C, and a relative humidity of 80% to 95%. Plants cultured without inoculation were used as control. The third leaf was harvested at 0 h, 12 h, 24 h, 72 h, and 120 h post inoculation (hpi), immediately frozen in liquid nitrogen, and stored at -80°C.
For the salt stress treatment, melon seedlings at the three-leaf stage were transferred to Hoagland’s nutrient solution for hydroponics pre-culture [59], and upon development into the four-leaf stage, seedlings were treated with 250 mM NaCl, while normal plants, cultured in nutrient solution without NaCl were used as control. Low temperature stress treatment was performed by placing the plants in a growth chamber at 4°C and at 80% humidity, while plants cultured at 25°C and 80% humidity were used as control. For ABA treatment, melon seedlings at the four-leaf stage were sprayed with 100 μM ABA, while plants sprayed with water served as control. Following the respective treatments, the third leaves were harvested at 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h post treatment (hpt), which were then immediately frozen in liquid nitrogen, and stored at −80°C for further analysis.
RNA isolation and qRT‑PCR
Total RNA from leaves was isolated using the RNASimple Total RNA Kit (TIANGEN, China) following the manufacturer’s instructions. The integrity of RNA samples was measured via 2% agar gel electrophoresis. First-strand cDNA was synthesized via reverse transcription of 1μg total RNA using the FastQuant RT Kit (TIANGEN, China) following the manufacturer’s instructions. Gene-specific primers for each CmCDPK and CmCRK were designed by Primer Premier 6.0 (S1 Table). The cDNA was diluted to 100 ng/μl for qRT-PCR, which was conducted on a Bio-Rad Real-time PCR system (Foster City, CA, USA) using the SYBR Premix Ex Taq II kit (Vazyme). The reaction mixture was prepared in a total volume of 20 μl containing: 10.0 μl SYBR Green Master mix, 0.4 μl of each forward and reverse primers (10 μM), 0.4 μl Rox Reference Day1, 2.0 μl cDNA and dilute with ddH2O to 20 μl. The PCR conditions consisted of pre-denaturing at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. To verify the specificity of the amplicons of each primer pair, melting-curve analyses of the products were conducted at the end of each PCR cycle with 95°C for 15 s, 60°C for 60 s, and then a constant increase from 60°C to 95°C with temperature increasing steps of 0.3°C/s. The melon β-actin gene (GenBank accession number AY859055) was used as internal control [50]. Each relative expression level was calculated following the 2-ΔΔCt method [60]. SPSS 21.0 software was used for statistical analysis, and the data were depicted as mean value ± SD of three biological replicate with three technical replicates. The significance of the differential expression between treatments and controls was verified via Student’s t-test. The relative expressions were log2 transformed and visualized for heat map using Mev 4.8.1 and the column diagrams were prepared using Origin 9.0 software package.
Results
Identification and distribution of CDPK and CRK genes in melon
After bioinformatic analysis, we obtained a total of 25 non-redundant sequences. After domain prediction of these sequences via InterProScan, SMART, CDD and ScanProsite, a total of 18 CmCDPKs (typically containing both STKs_CAMK protein kinase and EF-hand domain), and 7 CmCRKs (containing solely a STKs_CAMK kinase domain) were identified in the melon genome, designated as CmCDPK1 to CmCDPK18 and CmCRK1 to CmCRK7 based on their chromosomal positions, respectively (Table 1).
Table 1. Characteristics of CDPK and CRK gene family in melon.
| Gene | IDa | Chr | Gene length | No.ofaab | MW (kDa)b | pIb | EF handc | N-myristoylationd | N-Palmitoylatione | N-terminal aaf |
|---|---|---|---|---|---|---|---|---|---|---|
| CmCDPK1 | MELO3C024122 | Chr1:9620346..9633306 | 12961 | 501 | 56.25 | 5.31 | 4 | No | No | MEKPIKAS |
| CmCDPK2 | MELO3C015707 | Chr1:27325271..27331475 | 6205 | 566 | 63.36 | 5.59 | 4 | No | Yes | MGNTCRGS |
| CmCDPK3 | MELO3C015189 | Chr2:5589335..5593855 | 4521 | 531 | 59.8 | 6.46 | 4 | No | Yes | MGNCCATP |
| CmCDPK4 | MELO3C003730 | Chr4:3635832..3639934 | 4103 | 516 | 58.64 | 5.87 | 4 | Yes | Yes | MGLCFTRT |
| CmCDPK5 | MELO3C009574 | Chr4:29891445..29896126 | 4682 | 585 | 64.97 | 5.34 | 4 | No | Yes | MGNTCVGP |
| CmCDPK6 | MELO3C009565 | Chr4:29965064..29968628 | 3565 | 661 | 74.22 | 5.06 | 4 | No | Yes | MGNNCLRR |
| CmCDPK7 | MELO3C009161 | Chr4:32669721..32673288 | 3568 | 527 | 59.42 | 5.86 | 4 | No | Yes | MGNCCRSP |
| CmCDPK8 | MELO3C014588 | Chr5:1122720..1126606 | 3887 | 529 | 59.56 | 6.62 | 4 | No | Yes | MGNCCATP |
| CmCDPK9 | MELO3C014556 | Chr5:1421889..1424594 | 2706 | 535 | 59.81 | 5.65 | 4 | Yes | Yes | MGNCCSRE |
| CmCDPK10 | MELO3C025374 | Chr6:29289988..29294658 | 4671 | 519 | 58.65 | 6.34 | 4 | Yes | Yes | MGICTSKG |
| CmCDPK11 | MELO3C016330 | Chr7:22108243..22112765 | 4523 | 507 | 56.75 | 5.31 | 3 | No | Yes | MGNCSGLP |
| CmCDPK12 | MELO3C017756 | Chr7:24692093..24697309 | 5217 | 527 | 60.03 | 5.3 | 4 | Yes | Yes | MGSCVSVQ |
| CmCDPK13 | MELO3C007388 | Chr8:2507696..2511639 | 3944 | 552 | 61.65 | 6.03 | 4 | Yes | Yes | MGCCSSTQ |
| CmCDPK14 | MELO3C007904 | Chr8:6156505..6161719 | 5215 | 543 | 61.59 | 8.75 | 4 | Yes | Yes | MGVCFSAS |
| CmCDPK15 | MELO3C026727 | Chr8:16955408..16959420 | 4013 | 535 | 60.53 | 6.38 | 4 | No | Yes | MGNCCVAP |
| CmCDPK16 | MELO3C012326 | Chr10:1218591..1222869 | 4279 | 575 | 63.95 | 5.42 | 4 | No | Yes | MGNTCVGP |
| CmCDPK17 | MELO3C023209 | Chr11:233802..237889 | 4088 | 503 | 56.27 | 5.07 | 4 | No | No | MSKSSSAA |
| CmCDPK18 | MELO3C002215 | Chr12:24386696..24391834 | 5139 | 546 | 62.15 | 6.15 | 4 | Yes | Yes | MGNCNACV |
| CmCRK1 | MELO3C018436 | Chr1:261445..267908 | 6464 | 603 | 67.22 | 9.06 | 0 | Yes | Yes | MGICTSKP |
| CmCRK2 | MELO3C026598 | Chr4:25956480..25961897 | 5418 | 572 | 64 | 8.51 | 0 | Yes | Yes | MGLCHGKP |
| CmCRK3 | MELO3C007546 | Chr8:3526437..3531775 | 5339 | 604 | 67.33 | 8.72 | 0 | Yes | Yes | MGLCVSKP |
| CmCRK4 | MELO3C019099 | Chr8:11875126..11881306 | 6181 | 570 | 64.05 | 6.91 | 0 | No | Yes | MGICQAKV |
| CmCRK5 | MELO3C005914 | Chr9:24018542..24021758 | 3217 | 561 | 63.12 | 8.9 | 0 | Yes | Yes | MGHYCSKG |
| CmCRK6 | MELO3C020930 | Chr11:2732657..2737240 | 4584 | 622 | 69.44 | 9.15 | 0 | Yes | Yes | MGLCNSKP |
| CmCRK7 | MELO3C022260 | Chr11:29251128..29259710 | 8583 | 609 | 67.76 | 8.91 | 0 | No | Yes | MGQCYGKT |
a.The gene name in melon database(https://melonomics.net/).
b.The number of aa, MW, pI was predicted using ProtParam (http://web.expasy.org/protparam/).
c.The number of EF hands was predicted by ScanProsite tool(http://prosite.expasy.org/scanprosite/).
d.The myristoylation site was predicted by Myristoylator program in ExPASy (http://web.expasy.org/myristoylator/).
e.The palmitoylation site was predicted by CSS-plam 4.0 (http://csspalm.biocuckoo.org/).
f.The first eight amino acids at the N-terminal. The amino acids underlined indicate putative palmitoylation sites.
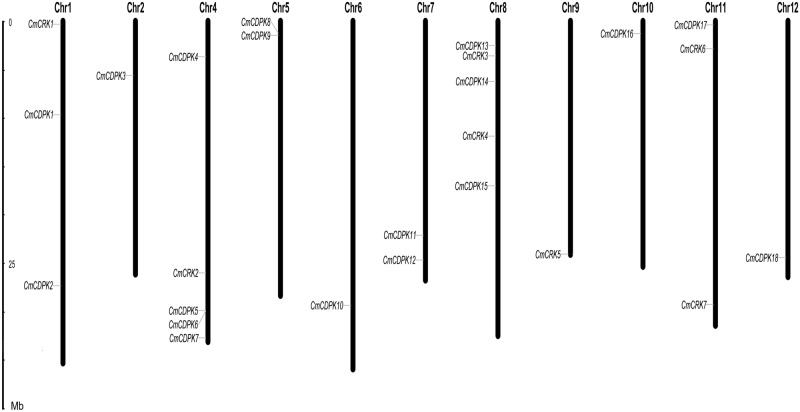
To understand the genomic distributions of CmCDPKs and CmCRKs, 25 identified genes (18 CmCDPKs and 7 CmCRKs) were mapped onto melon chromosomes. As shown in Fig 1, all of these genes were unevenly distributed across 11 melon chromosomes, with only exception for chromosome 3, which contained none of them. The largest number of CmCDPK genes (four) was located on chromosome 4, followed by chromosome 8 (three genes) and chromosome 1, 5, and 7 (two genes each). However, among 7 CmCRK genes, chromosome 1, 4, 8, 9, and 11 contained 1, 1, 2, 1, and 2 CmCRKs, respectively (Fig 1; Table 1).
Fig 1. Chromosomal distribution of melon CDPK and CRK genes.
Prediction of biochemical characteristics of CmCDPK and CmCRK genes
Although the genomic sequences of 18 CmCDPKs vary widely, from 2705 to 12960 bp, the numbers of predicted amino acid are relatively similar, ranging from 501 to 585 amino acids, with the exception of CmCDPK6 that encodes 661 amino acids (Table 1). As a result, the molecular weights of the CmCDPKs in our study ranged from 56.25 to 74.22 kDa, with a predicted pI value of less than seven for most of these (except CmCDPK14). The 18 CmCDPKs were confirmed to contain the typical CDPK structure, including an N-variable domain, a protein kinase domain, an autoinhibitory domain, and a CaM-like domain [13]. It is to be noted that the EF hand motif in the CaM-like domain recognizes and binds Ca2+ molecules [14,61]. In our study, all the CmCDPKs were predicted to contain four EF-hands, with the exception of CmCDPK11 containing three EF-hands (Table 1). Among the identified 18 CDPKs in melon, 7 CmCDPKs were predicted to contain myristoylation sites, while 16 CmCDPKs were predicted to contain palmitoylation sites (Table 1). Additionally, there were two CmCDPKs, CmCDPK1 and CmCDPK17, which had neither myristoylation nor palmitoylation sites.
In comparison, we only obtained seven CmCRK genes in the melon genome, with predicted protein molecular mass from 63.12 to 69.44 kDa (Table 1). Similar to CDPK and CRK genes in tomato [16], almost all CmCRK genes (except for CmCRK4) encode basic proteins, whereas CmCDPK genes encode either acidic or neutral proteins. According to the bioinformatic prediction, five CmCRK proteins contained myristoylation sites, while all CmCRK proteins contained palmitoylation sites.
Gene duplication and synteny analysis of CmCDPK and CmCRK
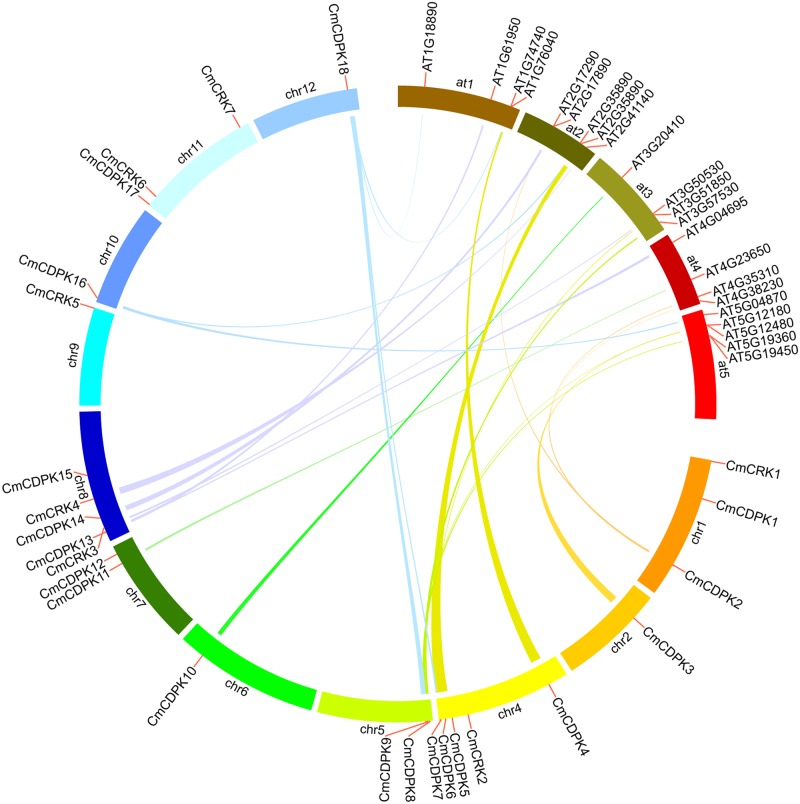
In addition to duplication of the whole genome, tandem duplication and segmental duplication also play important roles in the expansion and function of gene family [62,63]. To investigate possible evolutionary relationships between CmCDPKs and CmCRKs, we analyzed duplication events in melon using MCScanX software, visualizing the results via Circos. As shown in Fig 2, two pairs of CmCDPKs were identified as segmental duplications (CmCDPK7/ CmCDPK18 and CmCDPK9/ CmCDPK18) (S2 Table); however no tandem duplication event existed among CDPK genes in melon. Moreover, we neither found segmental duplications nor tandem duplications in CRK genes in melon.
Fig 2. Segmental duplication of melon CDPK genes, and synteny analysis of melon and Arabidopsis CDPK and CRK genes.
Chromosomes of melon and Arabidopsis were depicted in different color and circle form. The approximate distribution of each CmCDPK, CmCRK, AtCPK, and AtCRK is marked with a short red line on the circle. Coloured curves denote the details of syntenic regions between melon and Arabidopsis CDPK and CRK genes.
To further explore evolutionary connections between melon CDPK/CRK genes and Arabidopsis CDPK/CRK genes, a synteny analysis was performed via MCScanX using default parameters. As a result, we found 20 collinear gene pairs in melon CDPK and Arabidopsis CDPK gene family, including 13 CmCDPKs and 19 AtCPKs (Fig 2; S2 Table). These results imply that the syntenic relationships could be divided into three types. The first type is that a single CmCDPK corresponded to a single Arabidopsis CDPK gene, including CmCDPK3-AT5G12480 (AtCPK7), CmCDPK4-AT1G76040 (AtCPK29), CmCDPK6-AT2G35890 (AtCPK25), CmCDPK7-AT3G51850 (AtCPK13), CmCDPK10-AT3G20410 (AtCPK9), CmCDPK11-AT4G23650 (AtCPK3), and CmCDPK14-AT2G17890 (AtCPK16). For the second type, a single CmCDPK corresponded to two Arabidopsis CDPK genes, including CmCDPK8-AT5G19450 (AtCPK8)/ AT3G57530 (AtCPK32), CmCDPK9- AT5G12180 (AtCPK17)/ AT5G19360 (AtCPK34), CmCDPK13-AT1G61950 (AtCPK19) / AT4G04695 (AtCPK31), CmCDPK16-AT5G04870 (AtCPK1)/ AT2G35890, (AtCPK25), and CmCDPK18- AT1G18890 (AtCPK10)/ AT1G74740 (AtCPK30). The last type only contained one melon gene CmCDPK2, which corresponded to three Arabidopsis CDPK genes: AT4G35310 (AtCPK5), AT2G35890 (AtCPK6), and AT4G38230 (AtCPK26). Moreover, two collinear gene pairs were identified in the melon and Arabidopsis CRK gene families including CmCRK3-AT3G50530 (AtCRK5) and CmCRK4- AT2G41140 (AtCRK1).
Phylogenetic analysis and structure of the CDPK and CRK gene family
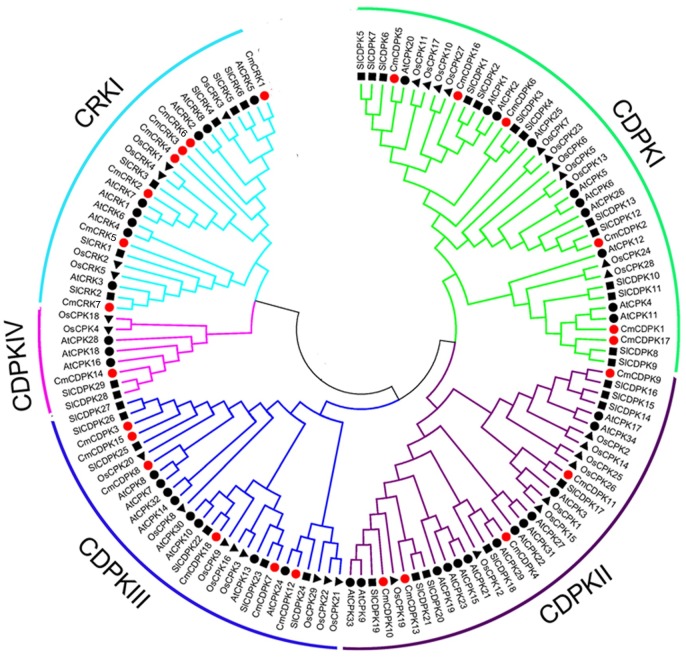
To deeply analyze the phylogenetic relationships between CDPK and CRK genes, CDPK and CRK full-length protein sequences of melon (18 CDPKs and 7 CRKs), Arabidopsis (34 CDPKs and 8 CRKs), tomato (29 CDPKs and 6 CRKs), and rice (29 CDPKs and 5 CRKs) (S1 Data Sheet) were aligned via Clustal X2.0, and then were used to construct a phylogenetic tree with MEGA6.0 (Fig 3). According to the classification of Arabidopsis CDPK gene [13,64], the 110 CDPK genes from the four species were divided into four groups (CDPK I, CDPK II, CDPK III and CDPK IV), while all CRKs fell into one group CRK I. Interestingly, group CDPK IV evidently clustered with CRK I on the phylogenetic tree. Of the four CDPK groups, CDPK I was the largest, consisting of 6 melon CDPKs combined with 10 Arabdopsis CDPKs, 13 tomato CDPKs, and 11 rice CDPKs. CDPK II contained 5 CDPKs from melon, 12 from Arabidopsis, 8 from tomato, and 8 from rice, while group CDPK III contained 6, 9, 6, and 8 CDPKs from melon, Arabidopsis, tomato, and rice, respectively. Finally, CDPK IV was the smallest group that comprised of one melon CDPK, 3 Arabidopsis CDPKs, 2 tomato CDPKs, and 2 rice CDPKs (Fig 3). However, all the 26 CRK homologues gathered into group CRK I on the phylogenetic tree. As shown in Fig 3, lots of CDPK and CRK proteins from Arabidopsis, tomato, and rice clustered into sub-branches, while there were no pairs of paralogous relationships among CmCDPKs and CmCRKs.
Fig 3. Phylogenetic analysis of CDPK and CRK genes.
Phylogenetic relationships among melon (red circle), Arabidopsis (black circle), tomato (black square) and rice (black triangle). The phylogenetic tree was constructed using the aligned protein sequences by the Neighbor-Joining method with 1,000 bootstrap replicates. Distinct colors lines are used to cluster the subfamily of CDPK and CRK genes.
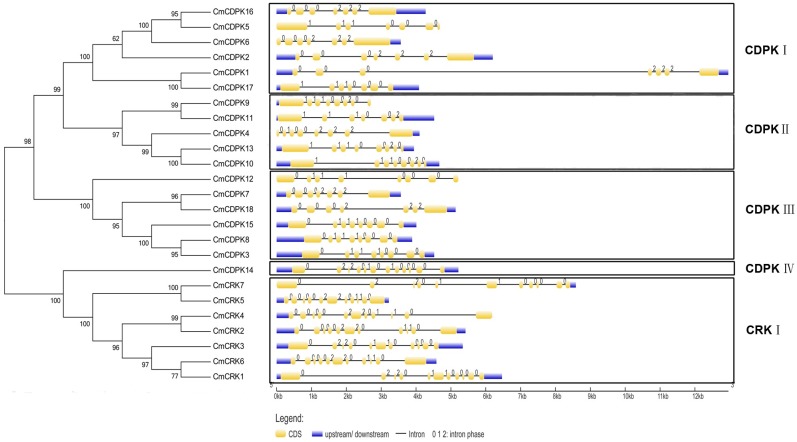
The exon-intron structures can also provide important evidence to support phylogenetic relationships within a gene family [22]. To get further insights into evolutionary relationships between the CDPK and CRK gene families, gene structures of 18 CmCDPKs and 7 CmCRKs were integratively depicted, based on the gene annotation profiles and completed melon genome sequences (Fig 4). According to the relatively high bootstrap values, five groups (CDPK I–IV and CRK I) could be observed in Fig 4, which was consistent with the topology of Fig 3. All members of group CDPK I had seven exons, with two distinct intron phase patterns (111000 and 000222), while CmCDPKs in CDPK II and III contained seven or eight exons, respectively, sharing similar intron patterns with CDPK I. For example, compared to CDPKs in group I, four genes (CmCDPK3, CmCDPK8, CmCDPK12, and CmCDPK15) in CDPK III possessed eight exons with an additional intron gain in the 5’ end, exhibiting a phase pattern 0111000, while the intron phase pattern of the remaining two genes (CmCDPK7 and CmCDPK18) in this group was 000222. Group CDPK IV contained only one gene: CmCDPK14. Strikingly, CmCDPK14 carried twelve exons with an intron phase pattern 02201010000, which was significantly above that of CmCDPKs (with seven or eight exons) in the other three groups. However, it was similar to CmCRK1, CmCRK3, and CmCRK7 of group CRK I, which had eleven exons with phase pattern 0220110000 (Fig 4), inferring that these genes likely derived from a common ancestor. The remaining four members in CRK I carried eleven exons with another intron phase pattern of 0000220110.
Fig 4. Phylogenetic relationship and exon-intron organization of melon CDPK and CRK genes.
The phylogenetic tree was constructed using the full-length protein sequences of 18 melon CDPK genes and 7 melon CRK genes by the Neighbor-Joining method with 1,000 bootstrap replicates. Gene structures were performed by online tool GSDS 2.0 (http://gsds.cbi.pku.edu.cn/). Exons and introns are represented by yellow boxes and gray lines, respectively. The intron phase numbers 0, 1 and 2 are labeled at the beginning of each intron. The diagram is drawn to scale.
Expression profiles of melon CDPK and CRK genes in different tissues
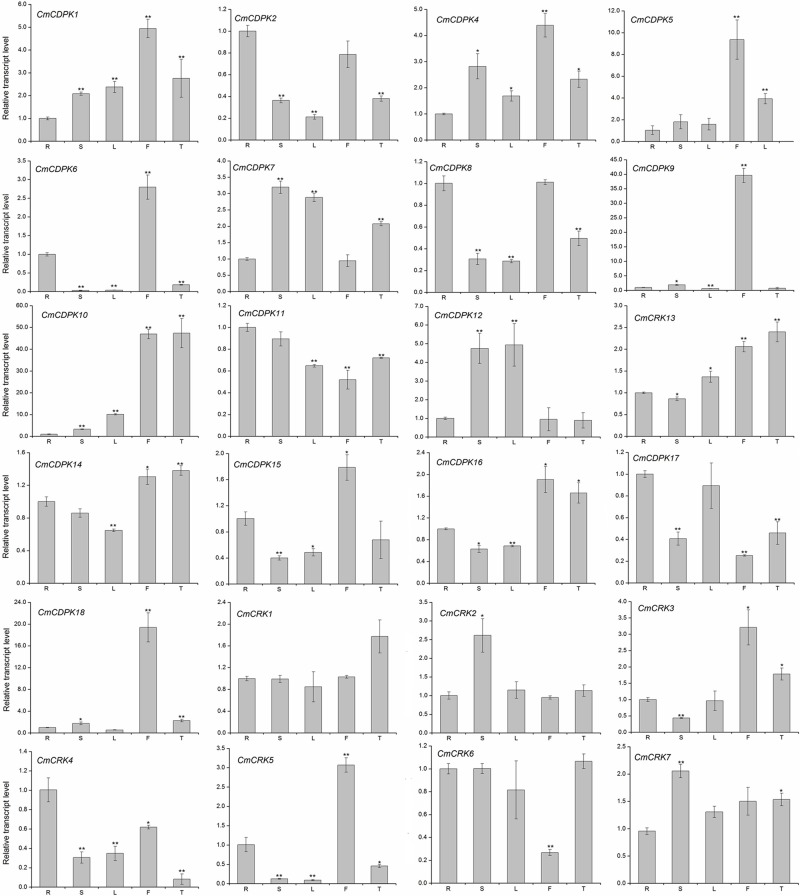
To assess the potential functions of CmCDPK and CmCRK genes during melon development, we investigated the expression patterns of all CmCDPK and CmCRK genes in five tissues (roots, stems, leave, male flowers and tendrils). As shown in Fig 5, all the identified CmCDPKs (except CmCDPK3) and CmCRKs were expressed in at least one of the five tissues, whereas the expression level of CmCDPK3 was not detected in any of the five organs. Some CmCDPKs and CmCRKs, such as CmCDPK9, CmCDPK18, and CmCRK5, were strongly expressed in male flowers, while CmCRK1 was expressed at nearly the same levels in all tissues (Fig 5). All these data implied that CmCDPK and CmCRK genes might be involved in the growth and development of different tissues of melon.
Fig 5. Expression profiles of CmCDPK and CmCRK genes in different tissues by quantitative RT-PCR.
The transcript levels of the respective genes in roots were used as reference and set to a value of 1. R, roots; S, stems; L, leave; F, male flowers; T, tendrils.
Expression profiles of melon CDPK and CRK genes under P. xanthii inoculation
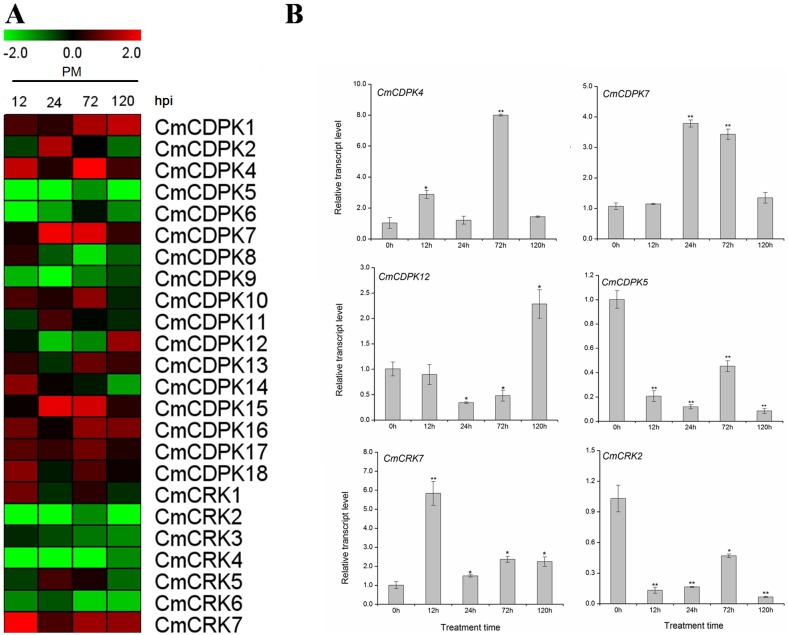
Powdery mildew is one of the most devastating biotic stresses that decreases melon yield and quality [65]. To get insight into the response of melon CDPK and CRK genes to powdery mildew, we designed specific primers for all CmCDPKs and CmCRKs, and examined their expression patterns in melon following inoculation with P. xanthii race 2F (Fig 6). However, the expression of CmCDPK3 had not been detected in leaves after using three independent pairs of specific primers. Therefore, it was discarded in further analysis. Results showed that the majority of CmCDPKs and CmCRKs responded to P. xanthii inoculation, differing in their expression patterns (Fig 6A). Using fold-changes above two fold or below 0.5-fold as thresholds, 10 CmCDPK genes and one CmCRK gene were found to be up-regulated upon P. xanthii inoculation. Among of these, the transcription level of CmCDPK7 reached a peak of nearly 3.8-fold at 24 h post inoculation (hpi), and rapidly decreased to around 1.3-fold at 120 hpi, similar with the expression pattern of CmCDPK15. Moreover, CmCDPK4 was up-regulated to 2.8-fold at 12 hpi, and then increased rapidly to 8.0-fold at 72 hpi. The only CmCRK gene induced by P. xanthii inoculation was CmCRK7, which peaked 5.8-fold at 12 hpi, but then fluctuated around 2.3-fold at 72 hpi (Fig 6B). By contrast, we found a total of ten genes with repressed expression levels following P. xanthii inoculation. Among them, six were CmCDPK genes (CmCDPK5, CmCDPK6, CmCDPK8, CmCDPK9, CmCDPK12, and CmCDPK19) and four were CmCRKs (CmCRK2, CmCRK3, CmCRK4, and CmCRK6). The transcript abundance of CmCDPK9 as well as that of CmCRK4 decreased to around 0.30-fold at 12 hpi, which remained at a significantly lower level than that of control for the rest of the treatment time Intriguingly, CmCDPK12 was down-regulated at 24 hpi but increased to 2.3-fold at 120 hpi (Fig 6B). All these results imply that CmCDPK and CmCRK genes likely played important roles in mediating the responses of melon to P. xanthii induced stress.
Fig 6. Expression of CDPK and CRK gene in melon following inoculation with P. xanthii by quantitative RT-PCR.
(A) Expression of CmCDPK and CmCRK following inoculation with P. xanthii. The relative transcript level was log2 transformed and visualized as heat map by Mev4.8.1. Gene highly or lowly expressed was colored by red or green, respectively. (B) Detailed expression of selected CmCDPKs and CmCRKs with different expression patterns following P. xanthii inoculation. Means ± standard deviation of three biological replicates each with three technical replicates were plotted as histogram by Origin 9.0. “*,**” significantly differ from the 0 h control at p<0.05 and 0.01, respectively, according to the Student’s test.
Expression profiles of melon CDPK and CRK genes under salt, low temperature, and ABA treatment
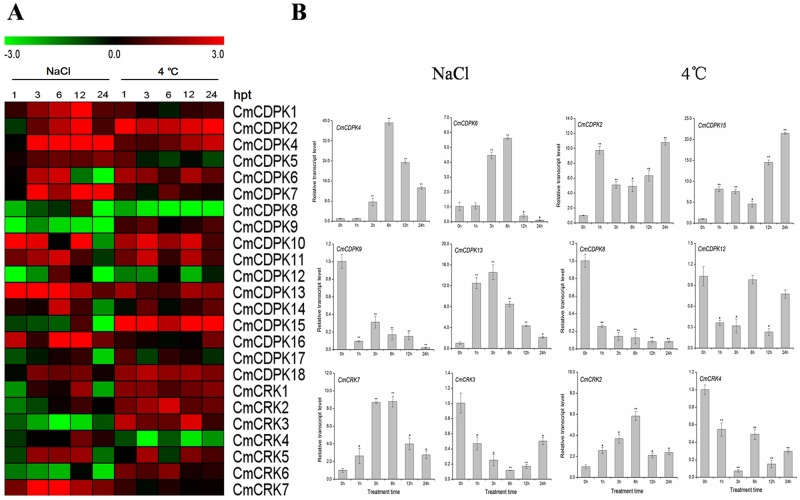
Previous research showed that CDPK genes are widely involved in the adaptation of plants to environment stimuli, and that the expressions of CDPK genes are effected by salt, cold and exogenous phytohormones [7]. However, little is known about how melon CDPK and CRK genes respond to these stimuli. To detect potential functions of melon CDPK and CRK genes under abiotic stresses and hormone treatment, we investigated the expression of CmCDPKs and CmCRKs under salt (250 mM NaCl), cold (4°C), and abscisic acid (ABA 100 μM) treatments. As shown in Figs 7 and 8, almost all CmCDPKs and CmCRKs (except CmCDPK3 as described above) responded to at least one stress; however some responded only slightly. Compared to the salt and ABA treatment, cold stress showed a stronger and more complex effect on the transcript levels of CmCDPKs and CmCRKs. Nonetheless, some genes showed similar expression patterns under different treatments. CmCDPK2, CmCDPK4, CmCDPK13, and CmCDPK18 were up-regulated under both NaCl and cold stresses, while CmCDPK8 showed lower transcription levels in all treatments. Furthermore, converse expression patterns were also observed, such as the down-regulation of the transcription level of CmCDPK11 due to ABA treatment, which was up-regulated under NaCl and cold stresses.
Fig 7. Expression of CDPK and CRK gene in melon under salt (250 mM NaCl) and cold (4°C) by quantitative RT-PCR.
(A) Expression of CmCDPK and CmCRK under salt (250 mM NaCl) and cold (4°C). The relative transcript level was log2 transformed and visualized as heat map by Mev 4.8.1. Gene highly or lowly expressed was colored by red or green, respectively. (B) Detailed expression of selected CmCDPK and CmCRK with different expression patterns under salt (250 mM NaCl) and cold (4°C). Means ± standard deviation of three biological replicates each with three technical replicates were plotted as histogram by Origin 9.0. “*,**” significantly differ from the 0h control at p<0.05 and 0.01, respectively, according to the Student’s test.
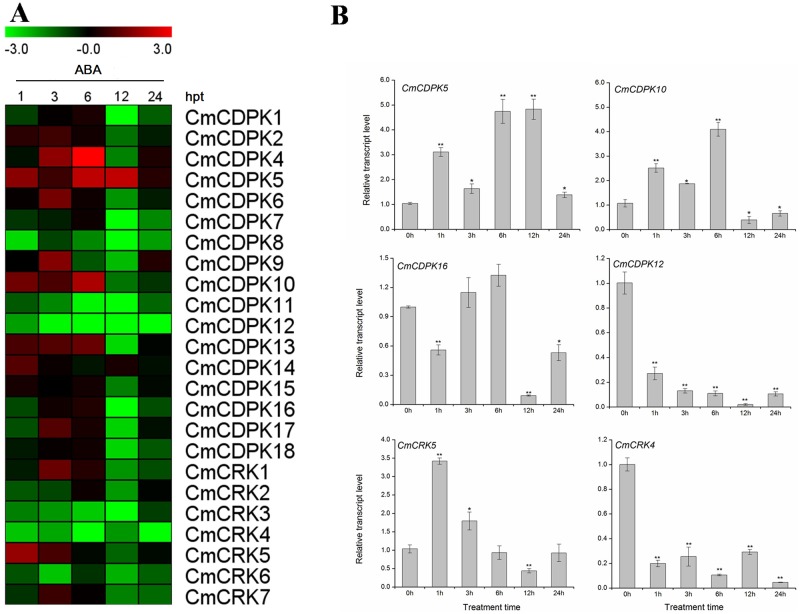
Fig 8. Expression of CDPK and CRK gene in melon under ABA (100 μM) treatment by quantitative RT-PCR.
(A) Expression of CmCDPK and CmCRK under ABA (100 μM) treatment. The relative transcript level was log2 transformed and visualized as heat map by Mev 4.8.1. Gene highly or lowly expressed was colored by red or green, respectively. (B) Detailed expression of selected CmCDPK and CmCRK with different expression patterns under ABA (100 μM) treatment. Means ± standard deviation of three biological replicates each with three technical replicates were plotted as histogram by Origin 9.0. “*,**” significantly differ from the 0h control at p<0.05 and 0.01, respectively, according to the Student’s test.
Following NaCl treatment, the transcription abundance of CmCDPK13 sharply increased to 12.5-fold at 1 h post treatment (hpt) and reached to a maximum of 14.6-fold at 3 hpt. However, it decreased gradually for remaining treatment duration, similar to the expression pattern of CmCRK7 (Fig 7B). Four genes (CmCDPK1, CmCDPK2, CmCDPK4, and CmCDPK7) have been found to be strongly up-regulated at 3 hpt and remained at significantly elevated levels (Fig 7A). However, the expression level of CmCDPK9 dramatically reduced to 0.09-fold at 1 hpt and remained at a stable low level, similar to the expression pattern of CmCRK3 (Fig 7B). Moreover, CmCDPK6 showed a much more complex expression pattern, with 1.0-fold at 1 hpt, which increased from 4.5-fold to 5.6-fold at 3 to 6 hpt, then rapidly decreased from 0.39-fold to 0.11-fold at 12 to 24 hpt, respectively (Fig 7B).
The expression profiles of CmCDPK and CmCRK genes in response to 4°C treatment were shown in Fig 7. Strikingly, only three genes (CmCRK8, CmCRK12, and CmCRK4) were significantly down-regulated due to cold stress. All of other genes were up-regulated, differing at expression levels and treatment times, with the exception for CmCDPK1 and CmCRK7 that displayed no obvious changes. The transcript level of CmCDPK15 rapidly increased to 8.1-fold at 1 hpt, but gradually decreased to 4.6-fold at 6 hpt, which was then up-regulated to a considerable 21.5-fold at 24 hpt, similar to the expression pattern of CmCDPK2 (Fig 7B). Moreover, cold stress significantly up-regulate CmCDPK2 at all treatment times, compared to its expression abundance in response to high salinity (Fig 7A). These expression analyses suggest that the majority of CmCDPKs and CmCRKs were involved in the low temperature response of melon.
As a phytohormone, ABA widely participates in the response of plant to biotic and abiotic stresses [66]. In the current study, we found that all CmCDPK and CmCRK genes (except for CmCDPK5 and CmCDPK14) were down-regulated at 12 hpt with ABA treatment, such as CmCDPK16 reaching 0.09-fold at 12 hpt (Fig 8B). The expression levels of CmCDPK12 and CmCRK4 were significantly decreased after ABA treatment at all treatment times, while only a few genes, such as CmCDPK5 were up-regulated by exogenous ABA (Fig 8B). CmCRK5 in particular responded rapidly to ABA, reaching 3.4-fold at 1 hpt, afterward sharply decreased to 0.44-fold at 12 hpt, similar to the expression trend of CmCDPK10 (Fig 8B). When compared to NaCl and 4°C treatments, the majority of CmCDPK and CmCRK genes likely acted as negative regulators in the signal transduction induced by exogenous ABA.
Discussion
With the completion of the sequencing of the whole genome, an increasing number of studies have focused on the analysis of a particular gene family. Compared to CRK genes, the CDPK gene family has been investigated in far more plants. However, the relationship between the CDPK and CRK gene families in melon still remain ambiguous. In the present study, we investigated the physico-chemical characteristics, gene structures, chromosome distributions, synteny relationships, duplication events, and phylogenetic relationships between CmCDPK and CmCRK genes. Furthermore, we analyzed gene expression patterns in response to biotic and abiotic stresses. Our work provides a better understanding of CDPK and CRK gene families in melon, which will serve as a molecular genetic basis for the genetic improvement of melon.
Identification of CDPK and CRK gene families in melon
To the best of our knowledge, the CDPK and CRK gene families in melon have never been studied before. In the present study, we identified 18 CDPK and 7 CRK genes in the melon sequenced genome (Fig 1; Table 1), and all CmCDPKs had both STKs_CAMK protein kinase and EF-hand domains, whereas CmCRKs had only STKs_CAMK protein kinase [16]. Compared to CDPK genes in Arabidopsis (34), tomato (29), rice (31), poplar (30) and cotton (41) [13,16,18,21,22], we found a lower amountof CDPK genes in melon. However, this is similar to cucumber (19 CsCDPKs) [23], an important vegetable crops from Cucurbitaceae family.
Gene duplication events play a major role in genomic rearrangements and expansions. Whole genome duplication (γ, β, α) events often occur in angiosperms leading to an increasing numbers of whole genome genes [63,67]. Meanwhile, tandem and segmental duplications also play important roles in CDPK gene family expansion [68,69]. Two tandem and 12 segmental duplications have been reported in the CDPK gene family of poplar [21], 13 segmental duplications in cotton [22], and seven segmental duplications in rice [19], indicating that segmental duplications play a predominant role in CDPK gene expansion in a majority of plants. Similarly, the low number of segmental and tandem duplications explains the low CDPK gene numbers in cucumber [23]. In our study, only two segmental duplication events were identified among CmCDPKs (CmCDPK7/ CmCDPK18 and CmCDPK9/ CmCDPK18) and no tandem duplication was detected (Fig 2; S2 Table), which is a plausible reason for the relatively low number of CDPKs in melon. Taken together, it can be postulated that 18 CmCDPKs and 7 CmCRKs are sufficient for mediating Ca2+ signals in melon.
Structure analysis and evolutionary relationships of CmCDPKs and CmCRKs
The CDPK genes show a high combination with Ca2+ via EF-hands, and allosteric properties of Ca2+ binding and the activation threshold may cause the differences in number and position of EF hands [70]. Furthermore, the special structure of EF-hand domain can be used to distinguish the CRK gene family from the CDPK gene family. In general, but not in all cases, the CDPK genes in plants have four EF-hands. However, exceptions have been reported for several CDPK genes in Arabidopsis and maize that contain two or three EF-hands [13,71]. In our study, all CmCDPKs (except for CmCDPK11) contain four EF-hands and CmCRKs contain no EF-hand domains (Table 1). Hence, it will be interesting to explore differences in the biological function between CmCDPK11 (three EF-hands) and other CmCDPKs (four EF-hands) in the future.
The N-terminus of a subset of CDPK proteins contained a myristoylation motif, which is supposed to promote protein-membrane and protein—protein interactions [72]. Palmitoylation is reversible and provides a regulatory mechanism of the subcellular localization [70,72,73]. Among the identified 18 CmCDPKs in melon, 7 CmCDPKs were predicted to contain myristoylation sites, while 16 CmCDPKs were predicted to contain palmitoylation sites and two CmCDPKs have neither myristoylation nor palmitoylation sites (Table 1). All the seven CmCRK proteins contained palmitoylation sites, and five of them contained myristoylation sites. This is consistent with the previous findings that most CRKs have N-terminal modifications [74].
In our study, we investigated phylogenetic relationships to reveal the evolutionary history of the CDPK and CRK gene families. Our results revealed that all CDPK and CRK genes from melon, Arabidopsis, tomato, and rice were distributed among all four groups, which is consistent with an origin of the CDPK gene before the divergence of eudicot and monocot [16,32]. Apart from degeneration of EF-hands, CRKs showed high similarities with CDPKs. CDPK genes were identified throughout the whole plant kingdom, even in some ancient plants, such as algae [75]. However CRK genes are absent in algae [16]. Previously, it was demonstrated that CRK and CDPK IV originated from a common ancestor, and that they separated after the split of green algae and the last common ancestor of the land plant lineage [16,22]. In agreement with this, a close phylogenetic relationship was found between the CDPK IV and CRK groups in our study (Fig 3). Compared to CDPK and CRK proteins from Arabidopsis, tomato, and rice clustering into sub-branches, there was no pairs of paralogous relationships among CmCDPKs and CmCRKs in phylogentic tree (Fig 3), which may be a possible reason for the number variations of CDPK/CRK genes between melon and the other three species.
The exon-intron structure reflects the evolution, expansion, and functional relationships of a gene family [22]. A change in the number of introns reflects the rate of gene divergence. Moreover, the insertion of a small DNA fragment alters the phase, finally leading to variations in gene function [76]. Three main types of mechanisms can cause the differences in the exon-intron structure (exon/ intron gain/ loss, exonization/ pseudoexonization and insertion/ deletion) [32,77]. To get further insights into evolutionary relationships between the CDPK and CRK gene families, we analyzed the exon-intron structure and intron phases of CmCDPKs and CmCRKs. As shown in Fig 4, the CDPKs were divided into four subgroups, which was consistent with the topology of the phylogenetic tree (Fig 3). The exon numbers of CmCDPKs in CDPK I–III were seven or eight. However, gene CmCDPK14 in group CDPK IV contained 12 exons, which was similar to the exon numbers of CmCRKs. Furthermore, group CDPK IV clustered with CRK I rather than with the other three CDPK groups in Figs 3 and 4, indicating that they originated from a common ancestor [16,22]. Previous studies reported that intron loss happens more easily than intron gain during evolution [78]. The distance tree and gene structure (intron number and intron phase) suggest that the ancient ancestor of CmCDPK genes may have a similar intron number with CmCRKs in the early time, but experienced intron lost events during the process of evolution. Therefore, we propose that CDPK IV, which contains 12 exons, may be a more ancient lineage of the CDPK gene family.
Function of CDPK and CRK genes in response to stimuli
An increasing number of studies have confirmed the crucial role of CDPKs and CRKs in plant stress response and in related signaling pathways [64]. Consequently, we investigated the expression of CmCDPKs and CmCRKs under biotic stress (powdery mildew), abiotic stress (salt and cold), and hormonal (ABA) treatment. In our study, the majority of CmCDPKs and CmCRKs were transcriptionally modified by salt, cold, ABA, and powdery mildew inoculation (Figs 6–8). Some genes showed similar expression patterns under a specific stress. As shown in S1 Fig, CmCDPK5, CmCDPK6, CmCDPK8, and CmCDPK9 were strongly down-regulated due to P. xanthii inoculation, CmCDPK9, CmCDPK12, and CmCDPK17 were down-regulated in response to salt stress, CmCDPK2, CmCDPK4, CmCDPK10, CmCDPK13, and CmCDPK18 were up-regulated due to cold stress, and most CmCDPKs were down-regulated after ABA treatment. These observations indicate that some CDPK genes could cooperatively regulate a specific stimulation, consistent with the findings of many plants, such as CDPK gene in tomato [16], poplar [21], cucumber [23], canola [29] and grape [32].
Some CmCDPKs and CmCRKs were found to respond to various stresses with different expression patterns in the current study. For instance, CmCDPK15 was up-regulated by cold stress, but down-regulated by salt stress (Fig 7). Transcriptional abundance of CmCDPK9 was down-regulated by both salt and ABA treatments, while cold stress did not result in any changes (S1 Fig). CmCRK2 was up-regulated under cold stress, but strongly down-regulated after powdery mildew inoculation (S1 Fig). Notably, CmCDPK8 was down-regulated in all the responses to powdery mildew inoculation, salt, cold, and ABA treatment, indicating its multiple functions in plant response to diverse stimuli (S1 Fig). These results imply complex functionality of CmCDPKs and CmCRKs in multiple signaling pathways, which is consistent with previous studies [7].
There are twenty-two collinear blocks of CDPK and CRK genes between melon and Arabidopsis (Fig 2; S2 Table). Intriguingly, we found correlated functional connections in syntenic genes. For example, AtCPK6 was found to significantly improve salt tolerance [79], and its collinear gene CmCDPK2 was also up-regulated during salt stress. Both CmCDPK8 and its collinear gene AtCPK32 were involved in the response to ABA stress [80]. The synteny analysis of melon and Arabidopsis may provide insights into the prediction of gene function for CDPK and CRK genes.
It has been widely known that CDPKs play critical roles in different aspects of plant response to abiotic stresses. In cucumber, almost all CsCDPKs were up-regulated under cold stress, but only half of them were induced at the transcriptional level after exposure to high salinity [23]. However, the majority of CmCDPKs in melon were up-regulated under both cold and salt stresses (Fig 7). It is interesting to point out that all the identified CmCDPK genes (except for CmCDPK5 and CmCDPK14) were strongly down-regulated at 12 hpt with ABA treatment, while most cucumber CsCDPKs still remain at high expression levels at 12 hpt with exposure to ABA stimuli [23].
In the present study, we performed an in-deep analysis of two pairs of segmental duplications, CmCDPK7/ CmCDPK18 and CmCDPK9/ CmCDPK18. Although CmCDPK7 and CmCDPK18 had similar exon-intron structures in terms of exon number and intron phase (Fig 4), their expression profiles were different in response to stress. They were both up-regulated by salt stress but the extent was stronger in CmCDPK7 than in CmCDPK18, which was similar for powdery mildew treatment (Figs 6 and 7). Based on the observations mentioned above, we suggest that CmCDPK7 and CmCDPK18 may have undergone subfunctionalization, which greatly adds gene specific functionality [81]. We found distinct expression profiles between CmCDPK9 and CmCDPK18. For instance, CmCDPK9 was down-regulated but CmCDPK18 was up-regulated due to salt stress and powdery mildew inoculation (Figs 6 and 7). Furthermore, their exon numbers and intron phases were apparently different, and CmCDPK9 belonged to CDPK II while CmCDPK18 belonged to CDPK III in the phylogenetic tree (Figs 3 and 4). Above all, the two genes might undergo a divergent process of structure and function in their evolutionary history.
Conclusions
We identified 18 CDPK and 7 CRK genes in the melon genome that were unevenly mapped among 11 chromosomes. Based on gene structure and phylogenetic tree analyses, CmCDPKs were divided into four groups with conserved domain. Synteny analysis identified two segmental duplication events of CmCDPKs and 22 syntenic blocks of CDPK and CRK genes among melon and Arabidopsis. Expression profile analysis of CmCDPKs and CmCRKs due to various stresses implied their crucial roles in response to multiple signaling pathways. This study provides important insights into the evolution and function of CDPK and CRK genes in melon, which will be vitally useful to melon breeding programs.
Supporting information
(DOCX)
(TIF)
(XLS)
(XLSX)
Acknowledgments
We are grateful to Changming Liu (Shangluo University, China) for powdery mildew inoculation, Jalal (Zhejiang University, China) for polishing our article and the vegetable research center of Beijing for providing the plant material.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Scientific Startup Foundation for Doctors of Northwest A&F University (Z109021604), Basal Research Foundation of Northwest A&F University (Z109021612), the Shaanxi Provincial Science and Technology Research and Development Project Fund, China (No. 2015NY091), and the Modern Agro-industry Technology Research System of China (No. CARS-26-18). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hepler PK. Calcium: a central regulator of plant growth and development. Plant Cell. 2005;17: 2142–2155. 10.1105/tpc.105.032508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002;14: S401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1: 11–21. 10.1038/35036035 [DOI] [PubMed] [Google Scholar]
- 4.Zielinski RE. Calmodulin and calmodulin-binding proteins in plants. Annu Rev Plant Biol. 1998; 49: 697–725. [DOI] [PubMed] [Google Scholar]
- 5.Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell. 2002;14: S389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouché N, Yellin A, Snedden WA, Fromm H. Plant-specific calmodulin-binding proteins. Annu Rev Plant Biol. 2005;56: 435–466. 10.1146/annurev.arplant.56.032604.144224 [DOI] [PubMed] [Google Scholar]
- 7.Boudsocq M, Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013;18: 30–40. 10.1016/j.tplants.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig AA, Romeis T, Jones JD. CDPK-mediated signalling pathways: specificity and cross-talk. J Exp Bot. 2004;55: 181–188. 10.1093/jxb/erh008 [DOI] [PubMed] [Google Scholar]
- 9.Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, et al. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464: 418–422. 10.1038/nature08794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper JF, Harmon A. Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol. 2005;6: 555–566. 10.1038/nrm1679 [DOI] [PubMed] [Google Scholar]
- 11.Hamel LP, Sheen J, Séguin A. Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 2014;19: 79–89. 10.1016/j.tplants.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valmonte GR, Arthur K, Higgins CM, MacDiarmid RM. Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol. 2014;55: 551–569. 10.1093/pcp/pct200 [DOI] [PubMed] [Google Scholar]
- 13.Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129: 469–485. 10.1104/pp.005645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003; 132: 666–680. 10.1104/pp.102.011999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper JF, Huang JF, Lloyd SJ. Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33: 7267–7277. [DOI] [PubMed] [Google Scholar]
- 16.Wang JP, Xu YP, Munyampundu JP, Liu TY, Cai XZ. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol Genet Genomics. 2016;291: 661–676. 10.1007/s00438-015-1137-0 [DOI] [PubMed] [Google Scholar]
- 17.Cai H, Cheng J, Yan Y, Xiao Z, Li J, Mou S, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in Capsicum annuum. Front Plant Sci. 2015;6: 737 10.3389/fpls.2015.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray S, Agarwal P, Arora R, Kapoor S, Tyagi AK. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol Genet Genomics. 2007;278: 493–505. 10.1007/s00438-007-0267-4 [DOI] [PubMed] [Google Scholar]
- 19.Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 2005; 46: 356–366. 10.1093/pcp/pci035 [DOI] [PubMed] [Google Scholar]
- 20.Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol; 66: 429–443. 10.1007/s11103-007-9281-5 [DOI] [PubMed] [Google Scholar]
- 21.Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, et al. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol Biol Rep. 2013;40: 2645–2662. 10.1007/s11033-012-2351-z [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Li W, He Q, Daud MK, Chen J, Zhu S. Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS One. 2014;9: e98189 10.1371/journal.pone.0098189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Liu M, Lu L, He M, Qu W, Xu Q, et al. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol Genet Genomics. 2015;290: 1403–1414. 10.1007/s00438-015-1002-1 [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Che Z, Zeng X, Zhou X, Sitoe HM, Wang H, et al. Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct Integr Genomics. 2016;16: 481–493. 10.1007/s10142-016-0498-8 [DOI] [PubMed] [Google Scholar]
- 25.Lu SX, Hrabak EM. An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol. 2002;128: 1008–1021. 10.1104/pp.010770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, et al. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 2003;132: 1840–1848. 10.1104/pp.103.020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chehab EW, Patharkar OR, Hegeman AD, Taybi T, Cushman JC. Autophosphorylation and subcellular localization dynamics of a salt- and water deficit-induced calcium-dependent protein kinase from ice plant. Plant Physiol. 2004; 135: 1430–1446. 10.1104/pp.103.035238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers C, Romanowsky SM, Barron YD, Garg S, Azuse CL, Curran A, et al. Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009;59: 528–539. 10.1111/j.1365-313X.2009.03894.x [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Liu WZ, Zhang Y, Deng M, Niu F, Yang B, et al. Identification, expression and interaction analyses of calcium-dependent protein kinase (CPK) genes in canola (Brassica napus L.). BMC Genomics. 2014;15: 211 10.1186/1471-2164-15-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye S, Wang L, Xie W, Wan B, Li X, Lin Y. Expression profile of calcium-dependent protein kinase (CDPKs) genes during the whole lifespan and under phytohormone treatment conditions in rice (Oryza sativa L. ssp. indica). Plant Mol Biol. 2009;70: 311–325. 10.1007/s11103-009-9475-0 [DOI] [PubMed] [Google Scholar]
- 31.Anil VS, Harmon AC, Rao KS. Spatio-temporal accumulation and activity of calcium-dependent protein kinases during embryogenesis, seed development, and germination in sandalwood. Plant Physiol. 2000;122: 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, Han YT, Zhao FL, Hu Y, Gao YR, Ma YF, et al. Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 2015;15: 164 10.1186/s12870-015-0552-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000; 23: 319–327. [DOI] [PubMed] [Google Scholar]
- 34.Asano T, Hakata M, Nakamura H, Aoki N, Komatsu S, Ichikawa H, et al. Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol Biol. 2011;75: 179–191. 10.1007/s11103-010-9717-1 [DOI] [PubMed] [Google Scholar]
- 35.Asano T, Hayashi N, Kobayashi M, Aoki N, Miyao A, Mitsuhara I, et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012;69: 26–36. 10.1111/j.1365-313X.2011.04766.x [DOI] [PubMed] [Google Scholar]
- 36.Campo S, Baldrich P, Messeguer J, Lalanne E, Coca M, San Segundo B. Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 2014; 165: 688–704. 10.1104/pp.113.230268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma SY, Wu WH. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol Biol. 2007;65: 511–518. 10.1007/s11103-007-9187-2 [DOI] [PubMed] [Google Scholar]
- 38.Abbasi F, Onodera H, Toki S, Tanaka H, Komatsu S. OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol Biol. 2004;55: 541–552. 10.1007/s11103-004-1178-y [DOI] [PubMed] [Google Scholar]
- 39.Weckwerth P, Ehlert B, Romeis T. ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2015;38: 544–558. 10.1111/pce.12414 [DOI] [PubMed] [Google Scholar]
- 40.Jiang S, Zhang D, Wang L, Pan J, Liu Y, Kong X, et al. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2013;71: 112–120. 10.1016/j.plaphy.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 41.Wei S, Hu W, Deng X, Zhang Y, Liu X, Zhao X, et al. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 2014;14: 133 10.1186/1471-2229-14-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007; 19: 3019–3036. 10.1105/tpc.107.050666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou JJ, Wei FJ, Wang C, Wu JJ, Ratnasekera D, Liu WX, et al. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010; 154: 1232–1243. 10.1104/pp.110.157545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Y, Cao J, Ni L, Zhu Y, Zhang A, Tan M, et al. ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. J Exp Bot. 2013;64: 871–884. 10.1093/jxb/ers366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asano T, Hakata M, Nakamura H, Aoki N, Komatsu S, Ichikawa H, et al. Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol Biol. 2011; 75: 179–191. 10.1007/s11103-010-9717-1 [DOI] [PubMed] [Google Scholar]
- 46.Zhao R, Sun HL, Mei C, Wang XJ, Yan L, Liu R, et al. The Arabidopsis Ca (2+) -dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 2011; 192: 61–73. 10.1111/j.1469-8137.2011.03793.x [DOI] [PubMed] [Google Scholar]
- 47.Coca M, San Segundo B. AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 2010; 63: 526–540. 10.1111/j.1365-313X.2010.04255.x [DOI] [PubMed] [Google Scholar]
- 48.Yang DH, Hettenhausen C, Baldwin IT, Wu J. Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiol. 2012; 159: 1591–1607. 10.1104/pp.112.199018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang M, Liang S, Lu YT. Cloning and functional characterization of NtCPK4, a new tobacco calcium-dependent protein kinase. Biochim Biophys Acta. 2005; 1729: 174–185. 10.1016/j.bbaexp.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 50.Cheng H, Kun W, Liu D, Su Y, He Q. Molecular cloning and expression analysis of CmMlo1 in melon. Mol Biol Rep. 2012; 39: 1903–1907. 10.1007/s11033-011-0936-6 [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, et al. The genome of melon (Cucumis melo L.). Proc Natl Acad Sci U S A. 2012; 109: 11872–11877. 10.1073/pnas.1205415109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alex B, Lachlan C, Richard D, Robert D. Finn, Volker H, Sam GJ, et al. The Pfam protein families database. Nucleic Acids Res. 2004; 38: 211–222. [Google Scholar]
- 53.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015; 43: 257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006; 34: 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40: e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geetika S, Poonam S, Preeti A, Parul G, Gulshan K, Vishal A, et al. Genome-wide characterization and expression profiling of TIFY gene family in pigeonpea (Cajanus cajan (L.) Millsp.) under copper stress. J. Plant Biochem. Biotechnol. 2016; 25: 301–310. [Google Scholar]
- 57.Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007; 29:1023–1026. [PubMed] [Google Scholar]
- 58.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Epstein Emanuel. Plant Physiology: Mineral Nutrition of Plants. 5th ed New York: John Wiley and Sons; 1972. [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001; 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 61.Liese A, Romeis T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim Biophys Acta. 2013; 1833: 1582–1589. 10.1016/j.bbamcr.2012.10.024 [DOI] [PubMed] [Google Scholar]
- 62.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000; 290: 2114–2117. [DOI] [PubMed] [Google Scholar]
- 63.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004; 4: 10 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamel LP, Sheen J, Séguin A. Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 2014; 19: 79–89. 10.1016/j.tplants.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng H, Kong W, Hou D, Lv J, Tao X. Isolation, characterization, and expression analysis of CmMLO2 in muskmelon. Mol Biol Rep. 2013; 40: 2609–2615. 10.1007/s11033-012-2347-8 [DOI] [PubMed] [Google Scholar]
- 66.Ben-Ari G. The ABA signal transduction mechanism in commercial crops: learning from Arabidopsis. Plant Cell Rep. 2012; 31: 1357–1369. 10.1007/s00299-012-1292-2 [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005;5:1 10.1186/1471-2148-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo C, Guo R, Xu X, Gao M, Li X, Song J, et al. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J Exp Bot. 2014; 65: 1513–1528. 10.1093/jxb/eru007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu C, Wang X, Xu Y, Deng X, Xu Q. Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Mol Biol Rep. 2014; 41: 6769–6785. 10.1007/s11033-014-3563-1 [DOI] [PubMed] [Google Scholar]
- 70.Klimecka M, Muszyńska G. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim Pol. 2007; 54: 219–233. [PubMed] [Google Scholar]
- 71.Kong X, Lv W, Jiang S, Zhang D, Cai G, Pan J, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics. 2013; 14: 433 10.1186/1471-2164-14-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu SX, Hrabak EM. The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol Biol. 2013; 82: 267–278. 10.1007/s11103-013-0061-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martín ML, Busconi L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 2000; 24: 429–435. [DOI] [PubMed] [Google Scholar]
- 74.Harmon AC. Calcium-regulated protein kinases of plants. Gravit Space Biol Bull. 2003; 16:83–90. [PubMed] [Google Scholar]
- 75.McCurdy DW, Harmon AC. Calcium-dependent protein kinase in the green alga Chara. Planta. 1992; 188:54–61. 10.1007/BF00198939 [DOI] [PubMed] [Google Scholar]
- 76.Sverdlov AV, Rogozin IB, Babenko VN, Koonin EV. Conservation versus parallel gains in intron evolution. Nucleic Acids Res. 2005; 33: 1741–1748. 10.1093/nar/gki316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci U S A. 2012; 109: 1187–1192. 10.1073/pnas.1109047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roy SW and Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006; 7: 211–221. 10.1038/nrg1807 [DOI] [PubMed] [Google Scholar]
- 79.Xu J, Tian YS, Peng RH, Xiong AS, Zhu B, Jin XF, et al. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta. 2010;231:1251–1260. 10.1007/s00425-010-1122-0 [DOI] [PubMed] [Google Scholar]
- 80.Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, et al. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 2005;139:1750–1761. 10.1104/pp.105.069757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005; 5:28 10.1186/1471-2148-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
(XLS)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.