Abstract
We conducted epidemiologic and genetic analyses of family clusters of Mycobacterium ulcerans (Buruli ulcer) disease in southeastern Australia. We found that the incidence of M. ulcerans disease in family members was increased. However, the risk for exposure appeared short-term and not related to human-human transmission.
Keywords: Mycobacterium ulcerans, bacteria, tuberculosis and other mycobacteria, Mycobacterium ulcerans disease, Buruli ulcer, epidemiology, infection, exposure risk, genome sequencing, transmission family clusters, human-to-human transmission, Australia
Mycobacterium ulcerans is a slow-growing organism that causes necrotizing infections of skin and soft tissue, often requiring reconstructive surgery and resulting in long-term disability (1,2). Prevailing opinion is that humans are infected from the environment; insects, such as mosquitoes (3,4), and water-residing biting arthropods (5,6), have been proposed as vectors for transmission. In Victoria, Australia, there is evidence that native opossums might be involved in transmission (7). However, despite extensive research, the environmental reservoir of the organism and mode of transmission remain unknown.
We postulated that examination of M. ulcerans disease (Buruli ulcer) family clusters might provide useful new information about disease epidemiology. Theoretically, genetically related first-degree relatives have similar susceptibility to disease, and families share the same environment and therefore a similar exposure risk. Thus, we examined the epidemiology of M. ulcerans disease in family clusters managed in a large prospective observational cohort from the Bellarine Peninsula in southeastern Australia. We used data collected from all confirmed M. ulcerans cases managed during January 1, 1998–April 12, 2016, at Barwon Health, a tertiary referral hospital in Geelong, Australia (8).
The Study
For this study, only initial M. ulcerans lesions were analyzed. A family cluster was defined as multiple family members independently given a diagnosis of M. ulcerans disease who were living at the same residence at the time of diagnosis. Data was collected by using Epi Info 6 (Centers for Disease Control and Prevention, Atlanta, GA, USA) and analyzed by using Stata 12 (StataCorp LLC, College Station, TX, USA).
To determine the genetic relatedness of isolates derived from family clusters, we performed whole-genome sequencing and single-nucleotide polymorphism (SNP) analysis for 6 isolates derived from 3 family cluster pairs (Tables 1, 2). We sequenced DNA as 300-bp paired-end reads by using an MiSeq Sequencer (Illumina, Inc., San Diego, CA, USA). Resulting reads were mapped against the M. ulcerans Agy99 genome (9), including plasmid pMUM001 (10), by using Bowtie2 (11). Raw sequence reads for the 6 isolates have been deposited in the National Center for Biotechnology Information (Bethesda, MD, USA) Sequence Read Archive under BioProject accession no. PRJNA321660. We also performed whole-genome SNP analysis for 6 additional unrelated previously sequenced human M. ulcerans isolates (Sequence Read Archive accession no. SRP004497) obtained from the same disease-endemic region.
Table 1. Characteristics of 21 patients associated with family clusters of Mycobacterium ulcerans disease, Bellarine Peninsula, Victoria, Australia, 1998–2016*.
| Cluster | Isolate | Date of diagnosis | Time between lesions, mo | Location | Relationship | Patient age at diagnosis, y,/sex | Site of lesion | Type of lesion | WHO stage |
|---|---|---|---|---|---|---|---|---|---|
| 1a | mu179 | 2008 Jul 21 | 0.4 | PTL | Mother | 54/F | Right thigh | Ulcer | 1 |
| 1b | mu180 | 2008 Aug 4 | PTL | Daughter | 26/F | Left calf | Ulcer | 1 | |
| 2a | mu248 | 2010 Oct 24 | 20.6 | PTL | Husband | 84/M | Right forearm | Ulcer | 1 |
| 2b | mu394 | 2012 Jul 4 | PTL | Wife | 84/F | Right forearm | Ulcer | 1 | |
| 3a | NT | 2011 Jul 25 | 0.1 | QUE | Husband | 76/M | Right ankle | Ulcer | 3 |
| 3b | NT | 2011 Jul 28 | QUE | Wife | 75/F | Right elbow | Ulcer | 1 | |
| 4a | mu294 | 2011 Aug 22 | 1.3 | PTL | Wife | 65/F | Right knee | Ulcer | 1 |
| 4b | mu308 | 2011 Sep 29 | PTL | Husband | 65/M | Left calf | Ulcer | 1 | |
| 5a | NT | 2011 Aug 25 | 1.1 | BH | Father | 56/M | Right leg | Ulcer | 1 |
| 5b | NT | 2011 Sep 26 | BH | Son | 26/M | Right leg | Ulcer | 1 | |
| 6a | NT | 2012 Jun 19 | 22.7 | PTL | Wife | 34/F | Left knee | Ulcer | 1 |
| 6b | NT | 2014 Apr 30 | PTL | Husband | 37/M | Right ankle | Ulcer | 1 | |
| 7a | NT | 2012 Aug 14 | 22.9 | QUE | Wife | 74/F | Left ankle | Ulcer | 1 |
| 7b | NT | 2014 Jul 3 | QUE | Husband | 76/M | Left leg | Ulcer | 1 | |
| 8a | NT | 2012 Oct 16 | 15.9 | BH | Sister | 20/F | Right foot | Ulcer | 1 |
| 8b | NT | 2014 Feb 14 | BH | Brother | 18/M | Left leg | Ulcer | 1 | |
| 9a | NT | 2013 Apr 27 | 12.7 | QUE | Wife | 85/F | Right ankle | Ulcer | 1 |
| 9b | NT | 2014 May 12 | QUE | Husband | 90/M | Left forearm | Ulcer | 1 | |
| 10a | NT | 2013 Dec 10 | 2.8 | PTL | Father | 34/M | Left hand | Ulcer | 1 |
| 10b | NT | 2014 Mar 4 | PTL | Daughter | 4/F | Right knee | Nodule | 1 | |
| 10c | NT | 2014 Mar 5 | 0.0 | PTL | Son | 7/M | Right ankle | Nodule | 1 |
*BH, Barwon Heads; NT, not tested; PTL, Point Lonsdale; QUE, Queenscliff; WHO, World Health Organization.
Table 2. Description of 8 single-nucleotide polymorphisms specific to >1 of 6 family cluster isolates of Mycobacterium ulcerans disease, Bellarine Peninsula, Victoria, Australia, 1998–2016*.
| Position | Loci | Protein | Substitution | Amino acid change | Isolate | Coverage statistics |
|---|---|---|---|---|---|---|
| 398430 | Intergenic | – | G/A | – | mu179 | T: 0, A: 35, G: 0, C: 1 |
| 398430 | Intergenic | – | G/A | – | mu180 | T: 0, A: 67, G: 0, C: 0 |
| 398430 | Intergenic | – | G/A | – | mu248 | T: 0, A: 100, G: 0, C: 1 |
| 398430 | Intergenic | – | G/A | – | mu294 | T: 0, A: 75, G: 0, C: 0 |
| 398430 | Intergenic | – | G/A | – | mu308 | T: 0, A: 58, G: 0, C: 0 |
| 1758272 | MUL_1618 | Membrane protein | C/T | Synonymous | mu248 | T: 91, A: 1, G: 0, C: 0 |
| 2153447 | MUL_1947 | Thiamine pyrophosphate | A/G | Lys→Arg | mu294 | T: 0, A: 1, G: 58, C: 0 |
| 2153447 | MUL_1947 | Thiamine pyrophosphate | A/G | Lys→Arg | mu308 | T: 0, A: 0, G: 40, C: 0 |
| 2462577 | MUL_2205 | Hypothetical protein | T/C | Asp→Gly | mu179 | T: 1, A: 1, G: 0, C: 47 |
| 4359638 | MUL_3902 | Membrane protein | C/A | Ala→Ser | mu180 | T: 0, A: 60, G: 1, C: 0 |
| 4359638 | MUL_3902 | Membrane protein | C/A | Ala→Ser | mu248 | T: 0, A: 108, G: 0, C: 1 |
| 5189291 | Intergenic | – | G/T | – | mu248 | T: 76, A: 0, G: 4, C: 0 |
| 5354966 | MUL_4830 | Putative GTPase | T/C | Synonymous | mu180 | T: 2, A: 0, G: 1, C: 18 |
| 5354966 | MUL_4830 | Putative GTPase | T/C | Synonymous | mu248 | T: 0, A: 0, G: 0, C: 20 |
| 5577431 | MUL_5032 | Immunogenic protein mbt64 | A/G | Synonymous | mu394 | T: 0, A: 0, G: 28, C: 0 |
*A total of 4,918 core single-nucleotide polymorphisms were identified for all 6 isolates compared with the African Agy99 reference genome. –, not applicable (mutations were not within a coding region).
A total of 324 patients with M. ulcerans disease from the Bellarine Peninsula, Victoria, Australia, were managed in the Barwon Health observational cohort during January 1, 1998–April 12, 2016. Median age was 57 years (IQR 34–74 years), and 164 patients (50.6%) were men. For the whole cohort, a combined time of 1,968.5 years had elapsed from diagnosis of the initial M. ulcerans lesions until the time of study analysis (April 12, 2016). The median duration elapsed from initial diagnosis until study analysis was 4.7 years (IQR 2.8–9.7 years).
Twenty-one (6.5%) patients were part of a family cluster (Table 1), 9 genetically related and 12 related by marriage. All family clusters were diagnosed after the beginning of 2008. We found that significantly fewer family clusters were diagnosed during the first half of the study period (0 of 92 cases during 1998–2007) than in the second half (21 of 232 cases during 2008–2016) (p<0.01). The median time between diagnoses of M. ulcerans lesions in an additional family member, after the initial family member was given a diagnosis, was 2.8 months (IQR 1.1–20.6 months). The rate of new diagnosis of an M. ulcerans lesion in another family member was 5.69/1,000 person-years (95% CI 3.15–10.29/1,000 person-years). We determined the cumulative proportion of patients given a diagnosis who had an affected family (Figure 1).
Figure 1.
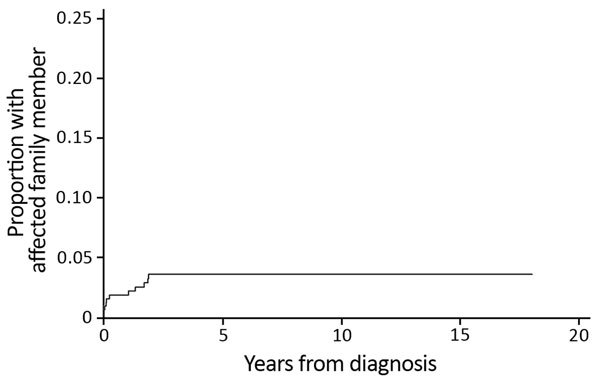
Cumulative proportion of patients with a family member affected by Mycobacterium ulcerans disease, Barwon Health cohort, Bellarine Peninsula, Victoria, Australia, 1998–2016.
Core SNPs based on common variable nucleotide positions were identified for the 6 examined family isolates by whole-genome sequencing. A total of 4,918 core SNPs ascribed to the African Agy99 reference genome were identified according to strict filtering criteria. Only 8 SNPs were specific to >1 of the 6 isolates (Table 2). Of the 8 SNPs that differed among the isolates, only 3 were nonsynonymous substitutions. The remaining 5 SNPs were either intergenic or synonymous mutations.
Pairwise comparisons of family cluster isolates showed that isolates from the 4a/4b pair were genetically identical. In contrast, isolates from the 2a/2b and 1a/1b pairs contained several isolate-specific SNPs (Table 2; Figure 2). SNP analysis of unrelated M. ulcerans isolates from the same disease-endemic area showed that 3 of the 6 isolates were also genetically identical (Figure 2), which demonstrated that unrelated isolates can share a common genotype. The remaining 3 isolates contained 1–3 unique SNPs. Thus, family cluster isolates were not any more closely genetically related than 6 random isolates from the same geographic region.
Figure 2.

Median joining network of 12 SNPs of 12 Mycobacterium ulcerans isolates from patients with Mycobacterium ulcerans disease, Barwon Health cohort, Bellarine Peninsula, Victoria, Australia, 1998–2016. Node colors indicate clusters. Blue, cluster 4a/4b; red, cluster 1a/1b; green, cluster 2a/2b. Black nodes represent 6 unrelated isolates. The size of each node is proportional to the number of genetically identical isolates with identical genotypes. Values indicate number of SNPs between each node. SNP, single-nucleotide polymorphism.
Conclusions
Our examination of family clusters of M. ulcerans disease provides useful insights into the environmental reservoir and mode of transmission of this organism. First, the median time to diagnosis between family members was short (2.8 months), and no family members were given a diagnosis of an M. ulcerans lesion >23 months apart in a cohort spanning 18 years and nearly 2,000 combined years of elapsed time since diagnosis. This finding suggests that family members have been exposed to a source in the family’s environment that persists only for a short period.
Second, with an incubation period for M. ulcerans disease estimated to be a median of 4.5 months (12), the observation that the median time between diagnoses in family clusters was <3 months suggest that infections were not being transmitted between family members. Further evidence against human-to-human transmission is apparent from whole-genome SNP analysis, which showed that pairs of isolates from 2 (2a/2b and 1a/1b) of 3 family clusters were not genetically identical. These findings support previous suggestions that M. ulcerans is unlikely to be transmitted from person to person (13).
Unknown is the type of short-term exposure that leads to the close temporal relation of family clustered infections. Opossums have been proposed as a source, either through contamination of the environment by infected feces or by an intermediate vector, such as mosquitoes, which transfer the infection from infected opossums to humans by a bite (7). Infected opossum(s) in the family environment might cause cases of human infection, then subsequently die of the disease (14), removing the source of infection. Alternatively, transmission could be related to a short-term change in the environment involving soil or foliage as a result of such events as home construction and renovation, or planting and removing trees or grasses (13). Mosquitoes in the area might be transiently infected/contaminated with M. ulcerans and infect humans through bites during this time (15).
In summary, the incidence rate of lesions in another family member (5.69/1,000 person-years) was higher than reported incidence rates during 2005–2009 in the general population of the Bellarine Peninsula (0.85–4.04 cases/year/1,000 population) (7). This finding suggests that genetic susceptibility or, more likely, localized exposure risk increases the likelihood of infection.
The incidence of M. ulcerans disease family clusters in an observational cohort in southeastern Australia was higher than in the general population of the disease-endemic area. However, when clusters occur, they are closely temporally related, which suggests a short-term risk for exposure and infection. Epidemiologic and genetic evidence suggests human-to-human transmission is not the source of infection.
Acknowledgment
We thank Janet Fyfe for providing the 6 isolates for whole-genome sequencing.
This study was supported by the Barwon Health Education, Training and Research Profile Fund.
Biography
Dr. O’Brien is an infectious diseases physician at University Hospital Geelong, Geelong, Victoria, Australia. His research interests are management of M. ulcerans disease, international health, HIV, tuberculosis, and travel medicine.
Footnotes
Suggested citation for this article: O’Brien DP, Wynne JW, Buultjens AH, Michalski WP, Stinear TP, Friedman ND, et al. Exposure risk for infection and lack of human-to-human transmission of Mycobacterium ulcerans disease, Australia. Emerg Infect Dis. 2017 May [date cited].
References
- 1.Asiedu K, Etuaful S. Socioeconomic implications of Buruli ulcer in Ghana: a three-year review. Am J Trop Med Hyg. 1998;59:1015–22. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien DP, Walton A, Hughes AJ, Friedman ND, McDonald A, Callan P, et al. Risk factors for recurrent Mycobacterium ulcerans disease after exclusive surgical treatment in an Australian cohort. Med J Aust. 2013;198:436–9. 10.5694/mja12.11708 [DOI] [PubMed] [Google Scholar]
- 3.Quek TY, Henry MJ, Pasco JA, O’Brien DP, Johnson PD, Hughes A, et al. Mycobacterium ulcerans infection: factors influencing diagnostic delay. Med J Aust. 2007;187:561–3. [DOI] [PubMed] [Google Scholar]
- 4.Johnson PD, Azuolas J, Lavender CJ, Wishart E, Stinear TP, Hayman JA, et al. Mycobacterium ulcerans in mosquitoes captured during outbreak of Buruli ulcer, southeastern Australia. Emerg Infect Dis. 2007;13:1653–60. 10.3201/eid1311.061369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Portaels F, Elsen P, Guimaraes-Peres A, Fonteyne PA, Meyers WM. Insects in the transmission of Mycobacterium ulcerans infection. Lancet. 1999;353:986. 10.1016/S0140-6736(98)05177-0 [DOI] [PubMed] [Google Scholar]
- 6.Marsollier L, Robert R, Aubry J, Saint André JP, Kouakou H, Legras P, et al. Aquatic insects as a vector for Mycobacterium ulcerans. Appl Environ Microbiol. 2002;68:4623–8. 10.1128/AEM.68.9.4623-4628.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fyfe JA, Lavender CJ, Handasyde KA, Legione AR, O’Brien CR, Stinear TP, et al. A major role for mammals in the ecology of Mycobacterium ulcerans. PLoS Negl Trop Dis. 2010;4:e791. 10.1371/journal.pntd.0000791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd SC, Athan E, Friedman ND, Hughes A, Walton A, Callan P, et al. Epidemiology, clinical features and diagnosis of Mycobacterium ulcerans in an Australian population. Med J Aust. 2012;196:341–4. 10.5694/mja12.10087 [DOI] [PubMed] [Google Scholar]
- 9.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17:192–200. 10.1101/gr.5942807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, Brosch R, et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A. 2004;101:1345–9. 10.1073/pnas.0305877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trubiano JA, Lavender CJ, Fyfe JA, Bittmann S, Johnson PD. The incubation period of Buruli ulcer (Mycobacterium ulcerans infection). PLoS Negl Trop Dis. 2013;7:e2463. 10.1371/journal.pntd.0002463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merritt RW, Walker ED, Small PL, Wallace JR, Johnson PD, Benbow ME, et al. Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS Negl Trop Dis. 2010;4:e911. 10.1371/journal.pntd.0000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Brien CR, Handasyde KA, Hibble J, Lavender CJ, Legione AR, McCowan C, et al. Clinical, microbiological and pathological findings of Mycobacterium ulcerans infection in three Australian Possum species. PLoS Negl Trop Dis. 2014;8:e2666. 10.1371/journal.pntd.0002666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavender CJ, Fyfe JA, Azuolas J, Brown K, Evans RN, Ray LR, et al. Risk of Buruli ulcer and detection of Mycobacterium ulcerans in mosquitoes in southeastern Australia. PLoS Negl Trop Dis. 2011;5:e1305. 10.1371/journal.pntd.0001305 [DOI] [PMC free article] [PubMed] [Google Scholar]


