ABSTRACT
Mitosis in metazoans is characterized by abundant phosphorylation of histone H3 and involves the recruitment of condensin complexes to chromatin. The relationship between the 2 phenomena and their respective contributions to chromosome condensation in vivo remain poorly understood. Recent studies have shown that H3T3 phosphorylation decreases binding of histone readers to methylated H3K4 in vitro and is essential to displace the corresponding proteins from mitotic chromatin in vivo. Together with previous observations, these data provide further evidence for a role of mitotic histone H3 phosphorylation in blocking transcriptional programs or preserving the ‘memory’ PTMs. Mitotic protein exclusion can also have a role in depopulating the chromatin template for subsequent condensin loading. H3 phosphorylation thus serves as an integral step in the condensation of chromosome arms.
KEYWORDS: Aurora B, chromatin condensation, epigenetics, Haspin, histone reader, histone, mitosis, phoshpo/methyl switch
Introduction
A rapid increase in chromatin compaction during mitosis, known as chromosome condensation, is essential for the faithful distribution of identical genetic material between daughter cells. Chromosome condensation involves the recruitment of condensin complexes to chromatin1 and is characterized by a pattern of posttranslational modifications (PTMs) in histones.2-5 In prophase, the kinases Haspin and Aurora B phosphorylate T3, S10, and S28 of histone H3 through evolutionarily conserved mechanisms.6-8 Even though chromosome condensation becomes evident at the onset of mitosis, H3 phosphorylation continues increasing from prophase to metaphase due to transactivation of Aurora B and a positive feedback loop involving Haspin.9 In addition, other kinases can be recruited to ensure robust H3 phosphorylation.10 Histone phosphorylation is so abundant that phosphorylation-dependent conformational changes were occasionally thought to drive chromatin condensation.11 The discovery of condensins that promote condensation by physically wrapping the chromatin however has provided an alternative explanation,12 which is now widely accepted. Although experiments on chromatin condensation in vitro reveal that phosphorylation of condensin I is the sole mitosis-specific modification required for the compaction of reconstituted chromatids,13,14 accumulating evidence suggests that additional components contribute to this process in vivo.
One of the significant outcomes of chromatin condensation is the modulation of general gene transcription.15 Although production of some non-coding RNAs continues at the centromere,16 bulk transcription of spliced messengers is largely suppressed in mitosis and resumes at the end of cell division. Transcription programs require the association of histone readers in chromatin-associating proteins and complexes with regulatory PTMs such as methylated lysine.17-19 Binding of methyllysine readers can in turn be modulated by removing the corresponding PTM or through a mechanism termed phospho/methyl switch.20,21 (Fig. 1). The phospho/methyl switch can displace reader-containing proteins from chromatin or prevent priming of demethylases and therefore preserve or “memorize” the methyllysine PTM. In contrast to demethylation that permanently erases an epigenetic modification, phosphorylation of serine and threonine residues adjacent to methyllysine provides a tool to temporarily prevent binding of reader proteins without affecting the PTM itself 20,22-24 A recent study that compared the behavior of methyllysine-interacting domains in vitro and in vivo revealed novel aspects of histone H3 phosphorylation, by linking the expulsion of reader proteins and chromatin condensation.23
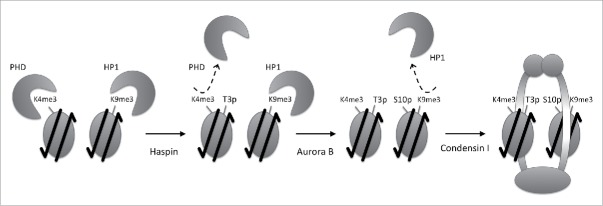
Figure 1.
Haspin- and Aurora B-targeted threonine and serine sites in histone H3 tail. A wide array of cellular events requires posttranslational modifications on H3K4, H3K9 and H3K27 in interphase, however protein complexes that bind these PTMs can interfere with mitosis. Mitotic histone H3 phosphorylation at H3T3, H3S10 and H3S28 can pause the transcriptional programs, preserve the ‘memory’ PTMs, or play a role in preparing a chromatin template for condensins.
Chromosome condensation in early mitosis
Prophase chromatin compaction coincides with histone phosphorylation6-8 and dissociation of a wide array of nuclear proteins from chromatin.20,23-26 Even organisms that divide by closed mitosis – without breakdown of the nuclear envelope – redirect a portion of nuclear proteins to the cytoplasm during chromosome segregation.27 In contrast to protein expulsion in early mitosis, recruitment to prophase chromatin is limited to a specific subset of proteins, many of which have a role in directing mitosis progression. Of particular importance are the condensin complexes that form and stabilize chromatin loops28 and the kinases that phosphorylate histone H3.6,7
Haspin is one of the main kinases to act on histones in early mitosis.6 It phosphorylates T3 of histone H3 producing the epigenetic mark H3T3ph, which is recognized by Survivin, a component of the chromosomal passenger complex.5,29,30 Survivin is required for the recruitment of Aurora B kinase and subsequent phosphorylation of H3S10 and H3S28.7,8,29 Small molecule inhibition of Haspin has a marked effect on early mitosis and chromosome condensation,23 but inhibition of Aurora B produces its effect only when decatenation and spindle attachment become important.31 These data agree with the idea that Haspin acts upstream of Aurora B; inhibition of the former affects phosphorylation of both H3T3 and H3S10 in vivo, but inhibition of the latter still permits H3T3ph accumulation.
Mitotic H3 phosphorylation first occurs close to the pericentromeric heterochromatin and subsequently spreads out over the chromosome arms.2-4 H3K4me3, a PTM enriched at transcription start sites, has been shown to decrease Haspin activity in vitro,32-34 which may account for the delayed euchromatin condensation in vivo. The H3K9ac modification, linked to gene activation, suppresses H3 phosphorylation,35 whereas the heterochromatin-associated H3K9me3 mark does not affect in vitro catalytic activities of Haspin and Aurora B.33,34 Differential mitotic condensation of hetero- and euchromatin might have important functional consequences; whereas hardly any heterochromatin along chromosome arms is actively transcribed, delayed euchromatin condensation shortens the time without general gene transcription. Even though the spatiotemporal patterns are different, many outcomes of H3T3 and H3S10 phosphorylation at the molecular level are similar – addition of a bulky negatively charged phosphate group can impede the function of the adjacent methyllysine PTM (Fig. 1). Eventually, H3T3ph and H3S10ph entirely cover the chromosomes from late prophase to metaphase. Maximum H3 phosphorylation and chromosome compaction coincide in metaphase and early anaphase,36,37 suggesting that the 2 are functionally linked in vivo.
While the importance of histone phosphorylation in vivo has been documented, experiments on chromatin condensation in vitro suggest that histone phosphorylation is not essential for the actual process of condensation, which primarily depends on the loop-forming proteins condensins.13,14 A minimal in vitro system functions without the H3 kinases and requires only core histones, topoisomerase, chaperones, and condensin.14 Depletion of condensin by RNA interference in cells leads to a delay though not the loss of prophase chromatin condensation.1 Likewise, conditional knockout cells without the SMC2 condensin subunit undergo residual albeit delayed chromatin compaction.28 The resulting metaphase chromosomes are easily disrupted, suggesting structural differences between compacted chromosomes in the presence and absence of condensin. Interestingly, studies using immunofluorescence show that efficient in vivo deposition of condensin, particularly condensin I in prometaphase, requires a prior H3 phosphorylation by Aurora B,7,38,39 (Fig. 2). Since conditional knockout cells without SMC2 undergo chromosome condensation only after nuclear envelope breakdown28 condensin II – which enters the nucleus before mitosis in contrast to condensin I – probably contributes to prophase chromatin compaction, either by acting before or by collaborating with histone kinases. In agreement with this idea, depletion of the early condensin II, but not the late condensin I, partially reduces H3 phosphorylation.40
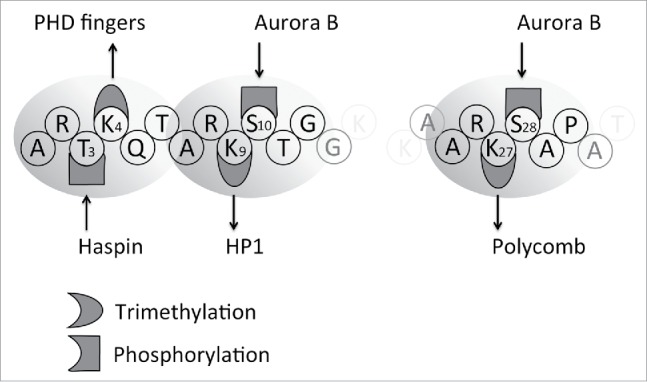
Figure 2.
Sequential events control chromosome condensation in vivo. Early mitosis is characterized by phosphorylation of key residues in histone H3 by Haspin (left) and Aurora B (center), which coincide with the exclusion of a set of H3K4me3-specific and H3K9me3-specific reader proteins. The resulting template, which is less populated than interphase chromatin, can be readily compacted by mitotic condensins (right). The reader-containing proteins excluded from chromatin are depicted above the nucleosomes.
Maintaining and terminating chromatin condensation
In mid-mitosis, Haspin and Aurora B translocate to centromeres and their concentration along chromosome arms decreases.9 Accordingly, histone H3 phosphorylation levels show a peak at metaphase and are gradually reduced after the metaphase-anaphase transition.41 Despite the reduction in H3 phosphorylation, chromatin condensation persists until telophase, suggesting a relaxed requirement for H3 phosphorylation once chromatin condensation has reached a threshold. Even at the time when Haspin and Aurora B localize mainly at centromeres, an experimentally induced transient loss of H3S10ph along chromosome arms is quickly restored.4 These data are consistent with the idea that metaphase re-phosphorylation of H3S10 on chromosome arms involves the continuous exchange of kinases,42 and that residual low-level phosphorylation is important for sustained condensation. The continuous evolution of chromosomes during mitosis is reflected in the dynamic behavior of proteins associated with mitotic chromosomes. A general survey identified hundreds of candidates,43 the majority of which comprise several multicomponent complexes.36 Interestingly, a large proportion of the associated proteins form part of the so-called chromosome periphery, which assembles around the chromosomes after nuclear envelope breakdown.36
The metaphase-anaphase transition marks a critical turning point in mitosis. It is controlled by the anaphase promoting complex and triggered by the proteasomal degradation of Cyclin B.44 The same mechanism eliminates a fraction of Aurora B,45 with the remaining part redistributing to the spindle midzone and midbody for the control of later events. Even though decatenation completes in early anaphase, chromatin compaction persists until late mitosis and could reduce resistance during poleward chromosome movement. During the second half of mitosis, PP1/Repo-man promotes gradual dephosphorylation of histone H3.41 Residual H3 phosphorylation is nonetheless important in the second part of mitosis, as cells subjected to sub-optimal Haspin inhibition in vivo undergo chromosome condensation until anaphase but show extensive incorporation of PHD finger-containing proteins into telophase chromosomes.23 Since Haspin inhibition reduces H3T3ph levels, its effect at the end of mitosis resembles the action of PP1/Repo-man – the removal of H3 phosphorylation. Controlling the balance between phosphorylation and dephosphorylation might thus be important until the end of mitosis and could protect chromatin from the premature recruitment of the methyllysine reader-containing proteins. In normal cells, the nuclear envelope reassembles around chromosomes when they are still condensed,46 indicating that nuclear protein import resumes before chromatin decondensation. In addition, several studies have implicated ATPases in removing residual Aurora B, and thus the final traces of H3S10ph, from chromatin.47,48 Complete chromatin decondensation thus comprises an active process instead of simple inactivation of condensation factors.
Concluding remarks
Over the past decade substantial progress has been made in our understanding of the physiologic importance of mitotic chromatin condensation, however many questions remain. For example, the precise role of histone H3 phosphorylation sites in chromatin condensation remains unclear, and we do not fully understand the antagonistic or cooperative effects and functional crosstalk involving phosphorylation and other histone PTMs. Further studies are also needed to examine whether the exclusion of reader-containing protein complexes from mitotic chromatin depends on collaboration between phosphorylation sites. Finally, a set of PHD finger-containing proteins show a notable tendency to accumulate on spindle microtubules, particularly on those adjacent to the spindle poles in metaphase and spindle midzone in late mitosis.23 It will be interesting to investigate whether protein accumulation on microtubules is a mechanism to avert premature reassociation of histone readers with chromatin or to ensure equal distribution of important factors between daughter cells during mitosis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Research in K.v.W. laboratory is supported in part by grant SAF2013–42289-R from the Spanish Ministry of Economics and Competitiveness. C.M.G. is a CSIC predoctoral fellow financed by FPI grant BES-2014–068580. Research in the T.G.K. laboratory is supported by grants from the NIH, GM101664, GM106416, and GM100907.
References
- [1].Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. Journal of cell science 2004; 117:6435-45; PMID:15572404; http://dx.doi.org/ 10.1242/jcs.01604 [DOI] [PubMed] [Google Scholar]
- [2].Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997; 106:348-60; PMID:9362543; http://dx.doi.org/ 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- [3].Polioudaki H, Markaki Y, Kourmouli N, Dialynas G, Theodoropoulos PA, Singh PB, Georgatos SD. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett 2004; 560:39-44; PMID:14987995; http://dx.doi.org/ 10.1016/S0014-5793(04)00060-2 [DOI] [PubMed] [Google Scholar]
- [4].Van Hooser A, Goodrich DW, Allis CD, Brinkley BR, Mancini MA. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. Journal of cell science 1998; 111 (Pt 23):3497-506; PMID:9811564 [DOI] [PubMed] [Google Scholar]
- [5].Wang F, Higgins JM. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol 2013; 23:175-84; PMID:23246430; http://dx.doi.org/ 10.1016/j.tcb.2012.11.005 [DOI] [PubMed] [Google Scholar]
- [6].Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev 2005; 19:472-88; PMID:15681610; http://dx.doi.org/ 10.1101/gad.1267105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol 2001; 152:669-82; PMID:11266459; http://dx.doi.org/ 10.1083/jcb.152.4.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 2002; 7:11-7; PMID:11856369; http://dx.doi.org/ 10.1046/j.1356-9597.2001.00498.x [DOI] [PubMed] [Google Scholar]
- [9].Wang F, Ulyanova NP, van der Waal MS, Patnaik D, Lens SM, Higgins JM. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol 2011; 21:1061-9; PMID:21658950; http://dx.doi.org/ 10.1016/j.cub.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kang TH, Park DY, Choi YH, Kim KJ, Yoon HS, Kim KT. Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol Cell Biol 2007; 27:8533-46; PMID:17938195; http://dx.doi.org/ 10.1128/MCB.00018-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sauve DM, Anderson HJ, Ray JM, James WM, Roberge M. Phosphorylation-induced rearrangement of the histone H3 NH2-terminal domain during mitotic chromosome condensation. J Cell Biol 1999; 145:225-35; PMID:10209020; http://dx.doi.org/ 10.1083/jcb.145.2.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 1997; 89:511-21; PMID:9160743; http://dx.doi.org/ 10.1016/S0092-8674(00)80233-0 [DOI] [PubMed] [Google Scholar]
- [13].de la Barre AE, Angelov D, Molla A, Dimitrov S. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J 2001; 20:6383-93; PMID:11707409; http://dx.doi.org/ 10.1093/emboj/20.22.6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shintomi K, Takahashi TS, Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol 2015; 17:1014-23; PMID:26075356; http://dx.doi.org/ 10.1038/ncb3187 [DOI] [PubMed] [Google Scholar]
- [15].Parsons GG, Spencer CA. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol Cell Biol 1997; 17:5791-802; PMID:9315637; http://dx.doi.org/ 10.1128/MCB.17.10.5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Scott KC. Transcription and ncRNAs: at the cent(rome)re of kinetochore assembly and maintenance. Chromosome Res 2013; 21:643-51; PMID:24190519; http://dx.doi.org/ 10.1007/s10577-013-9387-3 [DOI] [PubMed] [Google Scholar]
- [17].Andrews FH, Strahl BD, Kutateladze TG. Insights into newly discovered marks and readers of epigenetic information. Nat Chem Biol 2016; 12:662-8; PMID:27538025; http://dx.doi.org/ 10.1038/nchembio.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 2012; 19:1218-27; PMID:23211769; http://dx.doi.org/ 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016; 352:aad9780; PMID: 27257261. [DOI] [PubMed] [Google Scholar]
- [20].Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005; 438:1116-22; PMID:16222246; http://dx.doi.org/ 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- [21].Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature 2003; 425:475-9; PMID:14523437; http://dx.doi.org/ 10.1038/nature02017 [DOI] [PubMed] [Google Scholar]
- [22].Andrews FH, Gatchalian J, Krajewski K, Strahl BD, Kutateladze TG. Regulation of Methyllysine Readers through Phosphorylation. ACS Chem Biol 2016; 11:547-53; PMID:26726824; http://dx.doi.org/ 10.1021/acschembio.5b00802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gatchalian J, Gallardo CM, Shinsky SA, Ospina RR, Liendo AM, Krajewski K, Klein BJ, Andrews FH, Strahl BD, M van Wely KH, et al.. Chromatin condensation and recruitment of PHD finger proteins to histone H3K4me3 are mutually exclusive. Nucleic Acids Res 2016; 44:6102-12; PMID:27016734; http://dx.doi.org/ 10.1093/nar/gkw193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 2005; 438:1176-80; PMID:16222244; http://dx.doi.org/ 10.1038/nature04254 [DOI] [PubMed] [Google Scholar]
- [25].Kadauke S, Blobel GA. Mitotic bookmarking by transcription factors. Epigenetics Chromatin 2013; 6:6; PMID:23547918; http://dx.doi.org/ 10.1186/1756-8935-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Varier RA, Outchkourov NS, de Graaf P, van Schaik FM, Ensing HJ, Wang F, Higgins JM, Kops GJ, Timmers HT. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J 2010; 29:3967-78; PMID:20953165; http://dx.doi.org/ 10.1038/emboj.2010.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Asakawa H, Hiraoka Y, Haraguchi T. Physical breakdown of the nuclear envelope is not necessary for breaking its barrier function. Nucleus 2011; 2:523-6; PMID:22064471; http://dx.doi.org/ 10.4161/nucl.2.6.16117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell 2003; 5:323-36; PMID:12919682; http://dx.doi.org/ 10.1016/S1534-5807(03)00199-0 [DOI] [PubMed] [Google Scholar]
- [29].Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 2010; 330:235-9; PMID:20705815; http://dx.doi.org/ 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sawicka A, Hartl D, Goiser M, Pusch O, Stocsits RR, Tamir IM, Mechtler K, Seiser C. H3S28 phosphorylation is a hallmark of the transcriptional response to cellular stress. Genome Res 2014; 24:1808-20; PMID:25135956; http://dx.doi.org/ 10.1101/gr.176255.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 2003; 161:267-80; PMID:12719470; http://dx.doi.org/ 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JM, Knapp S. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci U S A 2009; 106:20198-203; PMID:19918057; http://dx.doi.org/ 10.1073/pnas.0901989106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Han A, Lee KH, Hyun S, Lee NJ, Lee SJ, Hwang H, Yu J. Methylation-mediated control of aurora kinase B and Haspin with epigenetically modified histone H3 N-terminal peptides. Bioorg Med Chem 2011; 19:2373-7; PMID:21397507; http://dx.doi.org/ 10.1016/j.bmc.2011.02.011 [DOI] [PubMed] [Google Scholar]
- [34].Karimi-Ashtiyani R, Houben A. In vitro phosphorylation of histone H3 at threonine 3 by Arabidopsis haspin is strongly influenced by posttranslational modifications of adjacent amino acids. Mol Plant 2013; 6:574-6; PMID:23220945; http://dx.doi.org/ 10.1093/mp/sss149 [DOI] [PubMed] [Google Scholar]
- [35].Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev 2006; 20:2566-79; PMID:16980585; http://dx.doi.org/ 10.1101/gad.1455006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Booth DG, Beckett AJ, Molina O, Samejima I, Masumoto H, Kouprina N, Larionov V, Prior IA, Earnshaw WC. 3D-CLEM Reveals that a Major Portion of Mitotic Chromosomes Is Not Chromatin. Mol Cell 2016; 64:790-802; PMID:27840028; http://dx.doi.org/ 10.1016/j.molcel.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gurley LR, D'Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem 1978; 84:1-15; PMID:206429; http://dx.doi.org/ 10.1111/j.1432-1033.1978.tb12135.x [DOI] [PubMed] [Google Scholar]
- [38].Collette KS, Petty EL, Golenberg N, Bembenek JN, Csankovszki G. Different roles for Aurora B in condensin targeting during mitosis and meiosis. Journal of cell science 2011; 124:3684-94; PMID:22025633; http://dx.doi.org/ 10.1242/jcs.088336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lipp JJ, Hirota T, Poser I, Peters JM. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. Journal of cell science 2007; 120:1245-55; PMID:17356064; http://dx.doi.org/ 10.1242/jcs.03425 [DOI] [PubMed] [Google Scholar]
- [40].Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 2004; 15:3296-308; PMID:15146063; http://dx.doi.org/ 10.1091/mbc.E04-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr Biol 2011; 21:766-73; PMID:21514157; http://dx.doi.org/ 10.1016/j.cub.2011.03.047 [DOI] [PubMed] [Google Scholar]
- [42].Wang E, Ballister ER, Lampson MA. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J Cell Biol 2011; 194:539-49; PMID:21844210; http://dx.doi.org/ 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ohta S, Bukowski-Wills JC, Sanchez-Pulido L, Alves Fde L, Wood L, Chen ZA, Platani M, Fischer L, Hudson DF, Ponting CP, et al.. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 2010; 142:810-21; PMID:20813266; http://dx.doi.org/ 10.1016/j.cell.2010.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol 2007; 8:894-903; PMID:17912263; http://dx.doi.org/ 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- [45].Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res 2005; 65:8730-5; PMID:16204042; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-1500 [DOI] [PubMed] [Google Scholar]
- [46].Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell 2003; 14:1043-57; PMID:12631722; http://dx.doi.org/ 10.1091/mbc.E02-10-0669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Magalska A, Schnellhaus AK, Moreno-Andrés D, Zanini F, Schooley A, Sachdev R, Schwarz H, Madlung J, Antonin W. RuvB-like ATPases Function in Chromatin Decondensation at the End of Mitosis. Dev Cell 2014; 301:305-18; http://dx.doi.org/ 10.1016/j.devcel.2014.09.001 [DOI] [PubMed] [Google Scholar]
- [48].Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 2007; 450:1258-62; PMID:18097415; http://dx.doi.org/ 10.1038/nature06388 [DOI] [PubMed] [Google Scholar]