ABSTRACT
The role of the nucleolus and autophagy in maintenance of nuclear integrity is poorly understood. In addition, the mechanisms of nuclear destruction in cancer cells senesced after conventional chemotherapy are unclear. In an attempt to elucidate these issues, we studied teratocarcinoma PA1 cells treated with Etoposide (ETO), focusing on the nucleolus. Following treatment, most cells enter G2 arrest, display persistent DNA damage and activate p53, senescence, and macroautophagy markers. 2–5 µm sized nucleolar aggresomes (NoA) containing fibrillarin (FIB) and damaged rDNA, colocalized with ubiquitin, pAMPK, and LC3-II emerge, accompanied by heterochromatin fragments, when translocated perinuclearly. Microscopic counts following application of specific inhibitors revealed that formation of FIB-NoA is dependent on deficiency of the ubiquitin proteasome system coupled to functional autophagy. In contrast, the accompanying NoAs release of pericentric heterochromatin, which exceeds their frequency, is favored by debilitation of autophagic flux. Potential survivors release NoA in the cytoplasm during rare mitoses, while exit of pericentric fragments often depleted of H3K9Me3, with or without encompassing by NoA, occurs through the nucleolar protrusions and defects of the nuclear envelope. Foci of LC3-II are accumulated in the nucleoli undergoing cessation of rDNA transcription. As an origin of heterochromatin fragmentation, the unscheduled DNA synthesis and circular DNAs were found in the perinucleolar heterochromatin shell, along with activation and retrotransposition of ALU elements, colocalized with 45S rDNA in NoAs. The data indicate coordination of the basic nucleolar function with autophagy regulation in maintenance of the integrity of the nucleolus associated domains secured by inactivity of retrotransposons.
KEYWORDS: aggresome, ALU retrotransposition, autophagy, cellular senescence, LADs, NADs, nucleolus, pericentric fragments, rRNA transcription, ubiquitin-proteasome
Introduction
In addition to its specific function in ribosome synthesis, the nucleolus also has additional roles in the cell which are less explored.1 These include proteome regulation,2 sensing cellular stress,3 maintenance of genome structure and integrity,4,5 and cell aging.6 The aggresome is a large body of a few microns in diameter, enwrapped in vimentin, located near the centrosome at an indentation of the nucleus, often found in neurodegenerative diseases, progeria, and cancer.7 Aggresome formation arises from insufficient degradation of proteins by the ubiquitination-proteasomal system (UPS) and is targeted as a polyubiquitinated aggregate for selective autophagic clearance.8-11
It has been shown that the nucleolar aggresome (NoA) can be induced experimentally by inhibition of the UPS and represents a counterpart of the cytoplasmic aggresome, acting as a platform for misfolded nucleolus-associated proteins in response to proteotoxic stress.12-14 In our study of cellular senescence and autophagy induced by the genotoxic agent etoposide in the human ovarian germ cell line PA1 (PA1-ETO) we noted considerable amounts of condensed chromatin grains in the perinuclear cytoplasm accompanying the fibrillarin-positive aggresome of the nucleolar origin. Previously, the role of nucleolus releasing rDNA from cell nuclei was found in the so called piecemeal microautophagy of senescing yeasts.15,16 Release of chromatin from cell nuclei with the involvement of macroautophagy in replicative senescence or accelerated cell senescence under genotoxic and oncogenic stress has been reported by several investigators. Survey of the nuclear integrity by macroautophagy with the involvement of the nuclear envelope limited chromatin sheets,17,18 nuclear lamin B, lamin B receptors, and lamin-associated domains (LADs)19-24 have been revealed. Furthermore, the role of retrotransposon activation, particularly SINE/ALU, in the DNA damage response (DDR) has been demonstrated.25,26 In turn, DDR is a hallmark of cellular senescence.27 Therefore, it is not surprising that activated ALU elements of pericentric heterochromatin transposed with aid of LINE 1 into preferred AT-rich satellite DNA, colocalize with gamma-H2AX foci in ex vivo senescing human stem cells28 and thus can favor centromere sticking. Interestingly, forced suppression of ALU transcription was sufficient to overcome persistent DDR and re-install self-renewal of human stem cells.28 De-Cecco and colleagues further demonstrated activation of retrotransposons in senescence of mammalian tissues,29,30 while Sedivy et al.31 suggested cell death by retrotransposition, may be with involving the release of DNA from the nuclei of senescent cells.32 In accord, Sturm and colleagues33 have arrived to the conclusion that mobilization of transposable elements comprising about 50% of the human genome, plays a primary role in genome disintegration during terminal aging.
Based on this data, we used PA1-ETO cells as a model system of genotoxically treated cancer stem cells to investigate the causal relationship between the NoA formation and accompanying chromatin release, leading ultimately to nuclear disintegration. Focusing on the nucleolus and its basic function, we have studied this complex process in relation to 2 degradation pathways, the ubiquitin-proteasomic system (UPS) and macroautophagy (hereafter termed “autophagy”), and the activity of the largest class of mammalian SINE retrotransposons ALU.
Results
Etoposide treatment of PA1 cells induces persistent DNA damage response and prolonged cell arrest in the DNA damage checkpoint
Etoposide (ETO) is an inhibitor of Topoisomerase II. ETO treatment (8 µM for 20 h) induced in PA1 cells massive apoptosis and necrosis culminating by day 5,34 ∼99% of such cells detached from support. The cells still attached to support were examined and found accumulating in a prolonged late S-G2-phase (Fig. 1A, B) (detailed by us earlier,34,35 some underwent polyploidy, mitoses (mostly arrested aberrant metaphases) were also encountered. Figure 1C presents proportions of cells containing CHK2-positive foci of DNA damage in their nuclei in one of the typical experiments. Thus, on days 2–4 nearly all cells, which remained attached to support, signaled DNA damage, a hallmark of cell senescence27,36 Cell senescence and accompanying enhanced autophagy were shown in our model earlier by upregulation of LC3, p53, p21CIP21, and p16INKA4a proteins by immuno-fluorescence and Western blots.34,35 Nevertheless, after day 5 post-ETO treatment, the tiny cell portion free of DNA damage begins to increase (Fig. 1C) and in 1-2 weeks PA1-ETO cells recover the clonogenic growth.34
Figure 1.
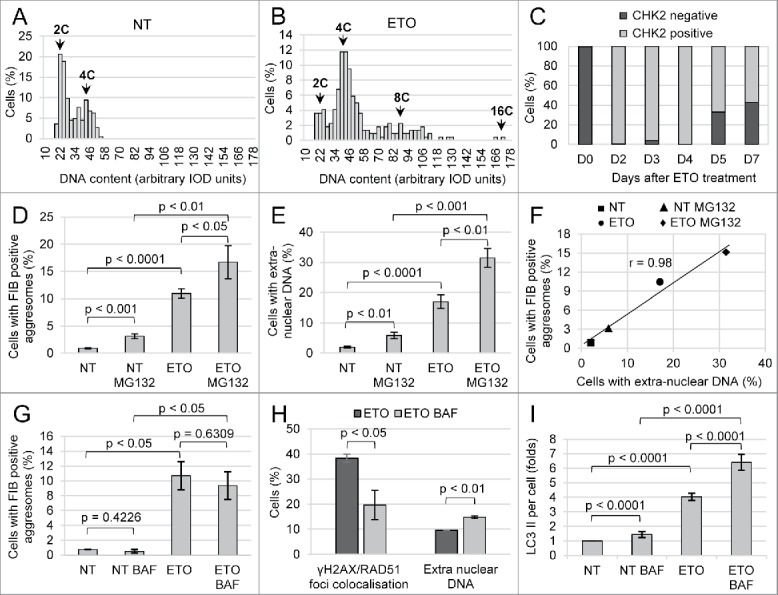
The parameters of PA1 cells response to ETO treatment. The cells were sampled on day 4 from the start of ETO (20 h) treatment (repeated at least 3 times). (A, B) Representative DNA image cytometry histograms in non-treated (NT) cells and ETO-cells; (C) The typical dynamics of DNA damage signaling by CHK2 in PA1-ETO treated cells demonstrating persistent DNA damage on days 2–4 in most cells; (D) Influence of ETO and UPS inhibitor MG132 on the proportion of cells displaying FIB-positive aggresomes (NoA) shows the significance of UPS insufficiency for NoA origin; (E) Influence of ETO and MG132 on the cell proportion releasing chromatin shows similar responses; (F) High correlation between the proportions of cells displaying FIB-NoA and showing release of extranuclear DNA in the same samples; (G) Influence of ETO and inhibitor of autophagic flux Bafilomycin A1 (BAF) on the proportion of cells displaying FIB-positive aggresomes (NoA) shows the insignificant tendency of BAF to reduce the number of FIB-NoA in ETO-cells; (H) BAF increases ∼1.5-fold the proportion of ETO-cells releasing extra-nuclear DNA and halves the proportion of cells undergoing DNA repair; (I) Accumulation of LC3II per cell as determined by immunostaining measuring integral fluorescence intensity in 200 random cells confirming inhibition of autophagic flux in ETO-treated cells induced by BAF.
The fibrillarin-positive bodies include damaged rDNA, autophagy markers and are released into cytoplasm accompanied by heterochromatin fragments
Fibrillarin (FIB) serves as a marker of the nucleolus. It is functioning in processing of rRNA and is localized in the dense fibrillar component of the nucleolus.37,38 FIB antibody stains the nucleoli in the punctuate-fluffy pattern in control PA1 cells (Fig. 2A). In ETO treated PA1 cells (PA1-ETO) we revealed 1-2 nucleolar or perinuclear dense bodies enriched with FIB – nucleolar aggresomes (NoA) in about 10% cells (Fig. 2B, C) on days 2 - 5 post-treatment. These round or ring-shaped FIB-positive bodies colocalized in ETO cells with the autophagy inducer LC3-II (Fig. 2D) and contained the DAPI-positive material in its core (Fig. 2C and E, F). The LC3-II-foci in cytoplasm (scarcer in non-treated samples) indicate to activate autophagy (Fig. 2D, E) in these cells. Using the FISH probe for nucleolar organizers (NORs) of the short arm of all 5 pairs of NOR-containing acrocentric chromosomes the similarly-sized dense nuclear bodies and buds are revealed beside normally looking nucleoli (Fig. 2G, H). The NoAs are also positively stained for ubiquitin (Fig. 2I) and for phosphorylated AMPK (Fig. 2J), a general autophagy inducer.23,39 Notably that the DNA core of NoAs is labeled, besides LC3-II, by CHK2 antibody indicating to rDNA damage (Fig. 2K) and the ATM-dependent targeting of this damaged DNA to autophagy.23 Another participant of the DNA damage response (DDR) response p53 is also activated34 and found accumulated in the nucleolus of PA1-ETO cells (not shown). The DNA exit from cell nuclei can be restricted only to the NoA DNA core (as on Fig. 2K). However, much more often NoA is surrounded by various amounts of the granular DAPI-positive material (Fig. 2C, J). Sometimes FIB-NoAs are found in cytoplasm of rare mitoses (Fig. 2L), which suggests that in viable, potentially surviving cell minority (comprising <1%) the NoAs can be released into cytoplasm for autophagic degradation during mitotic disintegration of the nucleolus and nuclear membrane. In summary, our first observations showed that formation of NoA in selected nucleoli and the accompanying release of the heterochromatin from cell nuclei appeared to be interdependent DDR events and have the relationship with degrading systems.
Figure 2.
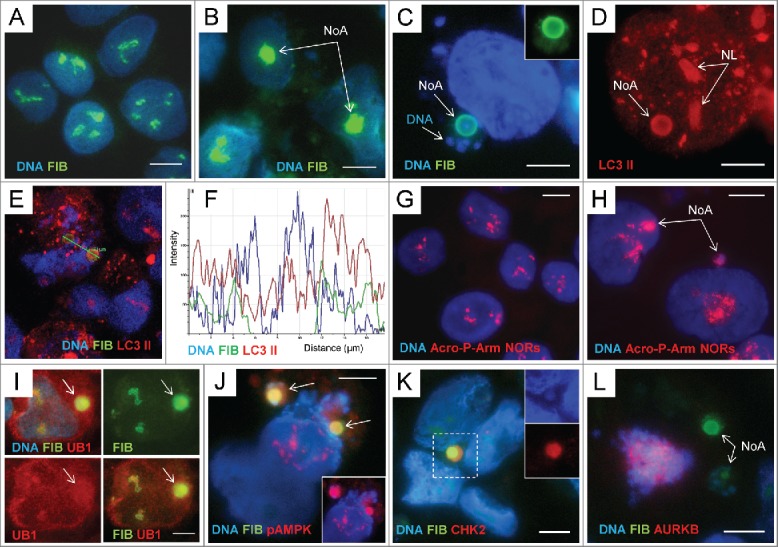
The fibrillarin-positive nucleolar aggresomes include damaged rDNA, autophagy markers and are released into cytoplasm accompanied by heterochromatin: (A) – immunostaining for FIBRILLARIN (FIB) and counterstaining with DAPI in non-treated PA1 cells; (B) the same staining of ETO-treated cells. The round nucleolar FIB-positive aggregates are designated as the nucleolar aggresomes NoA; (C, D) the perinuclear ring-shaped NoA colocalising FIB with LC3-II and DAPI (inside) and surrounded by DAPI-positive granular chromatin. The nucleoli (NL) designated on (D) are also positive for LC3-II; (E, F) confocal section through the interior of 2 NoA bodies and the plot of fluorescence intensity in the region of interest showing colocalization of all 3 components in the sample stained with 3 fluorochromes for FIB, LC3-II, and DNA (note a DAPI-peak inside); (G) FISH with the probe Acro P-arm-NORs counterstained with DAPI in NT PA1 cells; (H) the same reaction applied on ETO-treated cells where NORs are revealed both in intranuclear and perinuclear NoA (arrowed); (I) accumulation of UBIQUITIN 1(UB1) in the perinuclear FIB-positive NoA; (J) Colocalization of FIB with pAMPK in 2 NoAs (imaged in RGB optical filter); on insert, the same image in a red channel for pAMPK; (K) a cytoplasmic NoA composed of the largely colocalising FIB, CHK2, and the DNA containing DAPI-stained core (on inserts); (L) Fragment of the mitotic cell (tripolar mitosis) highlighted by DAPI and AURKB containing 2 NoAs in the cytoplasm. Bars = 10 µm.
In an attempt to elucidate the link between NoA and the chromatin release from cell nuclei, we addressed the following questions: Does NoA frequency induced by ETO correlate with DNA release from cell nuclei? Do the NoAs and DNA release events depend on ubiquitin-proteasome system (UPS) or/and autophagy (applying corresponding inhibitors MG132 and Bafilomycin A1)? In all experiments, repeated at least 3 times (if not stated otherwise), the samples of cells detached from support were evaluated on day 4 post ETO treatment.
Formation of NoA induced by ETO is increased by inhibition of UPS and highly correlates with release of extranuclear DNA, which exceeds it in frequency
The Fig. 1 D shows that the inhibitor of UPS MG132 induces NoAs (∼4-fold) in non-treated cells (NT), otherwise very rare, that ETO ∼20-fold increases the NoA frequency, and that adding of MG132 to ETO-treated cells nearly 2-fold potentiates this effect. On Fig. 1E the same proportions are seen in the frequency of DNA release from cell nuclei microscopically visualized near nuclei in the same samples (on different cytospins, not treated with detergents and specifically stained for DNA with Toluidine blue, see in METHODS). Fig. 1F shows that the correlation of both phenomena counted in 5 independent experiments is extremely high. However, in absolute numbers, the proportion of cells displaying NoAs is 1.5–2 times smaller than the proportion of cells releasing microscopically visualized clumps of extranuclear DNA (Fig. 1F). This may mean that the 2 phenomena are dependent on 2 very closely related mechanisms.
Inhibition of autophagic flux insignificantly impairs the frequency of NoAs in ETO samples but much enhances the frequency of extranuclear DNA
Then we compared the effect of suppression of autophagy on both phenomena applying the inhibitor of late autophagy (preventing fusion of autophagosome with lysosomes and stopping degradation of cargo and hence autophagic flux) Bafilomycin A1 (BAF).7,23 We found that, contrary to inhibition of UPS by MG132 substantially increasing the frequency of NoA-bearing cells (as seen on Fig. 1D), BAF insignificantly decreased the frequency of FIB-NoA-containing cells, both alone and with ETO (Fig. 1G). At the same time, BAF increased 1.5-fold the proportion of ETO-cells containing extranuclear DNA (Fig. 1H), thus confirming the role of lysosomes in autophagic degradation of the damaged nuclear DNA.40 In addition, BAF halved the proportion of cells repairing the DNA damage as detected by colocalized ϫH2AX/ RAD51 foci (Fig. 1H). The proportions of the accumulation of LC3-II in the cells after BAF measured on whole IF-stained cells in these experimental series are presented on Fig. 1I. It shows that ETO 4-fold increases the content of LC3-II per average cell, while BAF increases it additionally 2.5-fold. BAF halted autophagic flux in ETO-treated cells (shown earlier as an accumulation of LC3-II in relation to LC3-I).35 Figure 1I indicates to both functional and insufficient autophagy (apparently in different cells of the same samples) after ETO treatment.
In summary, in this series of experiments, it was elucidated that the process of NoA formation induced by ETO and exhibited by the FIB-aggregated bodies targeted for autophagy is mostly provoked by proteotoxic stress and insufficiency of UPS degradation pathway. Natural resolution of NoA apparently needs both functional autophagy and proliferative capacity thus characterizing this state as the reversible senescence. On the contrary, the deficiency of late autophagy in a proportion of the ETO-treated cells is preferentially responsible for accompanying, surpassing and lethal release of heterochromatin from their cell nuclei. In accord, our previous and current experiments also showed that PA1-ETO cells did not recover after treatment with BAF and that prolonging BAF treatment just brought to full nuclear deterioration in ETO-treated cells.35
But why the 2 phenomena, the potentially protective formation of aggresome in the nucleolus counteracting protein misfolding and potentially lethal release of the heterochromatin destructing cell nucleus correlate so tightly? Does the nucleolus topologically unite both phenomena? If so, where in particular the chromatin fragmentation is occurring? To this end, we considered the knowledge on the role of activated SINE/ALU elements in cell aging caused by genotoxic stress, particularly by ETO25 and the data on the DNA damage by retrotransposition of activated ALU elements (by LINE 1) in pericentric heterochromatin, found in senescent cells.28-32 We further attempted to explore the relation of these processes to the nucleolus.
ALU activation, clustering, relocation, and release together with 45S rDNA after ETO treatment
qRT-PCR revealed that ALU transcription was enhanced by ETO culminating by day 5 in a ∼3-fold increase (Fig. 3A). By FISH-ALU studies we found the change of the distribution pattern from homogenous to tandemly clustered pattern in the nuclei of about 10–20% of cells, the ALU clusters tended to delineate the perinucleolar chromatin (Fig. 3C, E and compare with B). Combination of the FISH labels for 45S rDNA and ALU revealed ALU concentration in selected nucleoli (NoA) of some cells and its release from cell nuclei together with 45S rDNA-positive bodies (Fig. 3D-E), the same as the behavior seen for FIB-NoA. Finally, a proportion of cells lose any order in the ALU label distribution, which fused in large drops randomly releasing from cell nuclei (Fig. 3F). So, we have found that activation of ALU elements by ETO involved the regions of the nucleolar organizers and perinucleolar heterochromatin, participating in the formation and release of the NoA, as well as in ultimate deterioration of cell nuclei. Our data suggest the role of retrotransposon activation in the continuity of these events, from NoA to destruction of cell nuclei.
Figure 3.
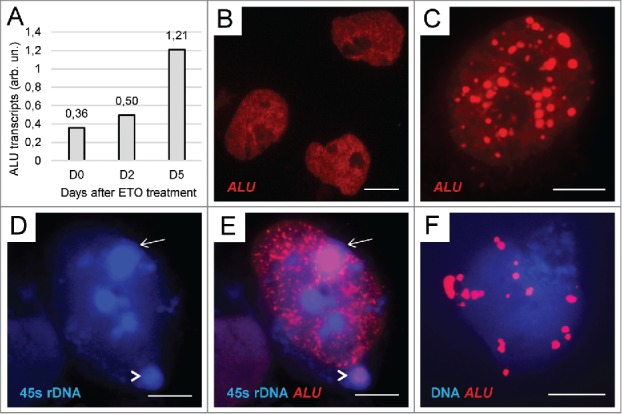
Activation, redistribution, and release of ALU elements in ETO-treated cells: (A) The result of qRT-PCR for ALU repeats in the PA1-ETO experiment showing ∼3-fold increase of ALU transcription by day 5 post-treatment; (B) NT control – the relatively even distribution of ALU in cell nuclei; (C) clustering of fused ALU elements around nucleoli after ETO treatment; (D, E) colocalization of ALU with 45S rDNA in the nuclear (arrow) and perinuclear (arrowhead) NoA after ETO-treatment as revealed by double FISH; (F) chaotic release of large fused ALU drops from the cell nucleus after ETO treatment. Bars = 10 µm.
ETO treatment induces formation of extrachromosomal circular DNAs around nucleoli, and the release of pericentric heterochromatin fragments from cell nuclei encompassed by NoA
We next sought to find which particular portion of heterochromatin was involved in the release. We found microscopically the ringlets of CHK2/DAPI-positive material forming “a torulus” concentrating this damaged DNA around large centered nucleoli (Fig. 4A) in about 10% of ETO-treated cells. The site corresponds the anchoring regions of the distal pericentric heterochromatin to ribosomal genes repeats.41 Extrachromosomal circular DNAs (eccDNA), known as characteristic for the amplification of rDNA,42 major satellite DNA,43 and also used in transposition, e.g. LINE44 result from the “rolling circle” replication mechanism. eccDNAs have been already described in senescence of mammalian cells.45,46 This mechanism is displayed as the unscheduled synthesis of the nicked DNA. Indeed, using a 90-min pre-incubation with BrdU, we could reveal in a proportion of cells the absence of the overall label (no S-phase) coupled with the sharp label of the perinucleolar rim, less intensive label inside the nucleoli, and also the same grainy material already degrading in the perinuclear vacuole (Fig. 4B) indicating to its fast release from cell nucleus. Thus, we have revealed that at least part of the released fragmented heterochromatin formed by “rolling circles” is originating in ETO cells in the nucleolus associated heterochromatin. Correspondingly, by using CREST antibody we identified this heterochromatin as predominantly pericentric. Centromeres were found accumulating near/inside the nucleoli (Fig. 4C), the clusters of pericentric fragments were leaving cell nuclei encompassed by FIB-positive NoA (Fig. 4D) or spilling from nuclei without visible NoA. In should be also noted that in S-G2 –phase, where PA1-ETO cells were arrested, centromeres tend to associate with the nucleoli,47 the same was found by us for irradiated lymphoma and HeLa cells.48 We next tried to evaluate the change of epigenetic state of the chromatin in PA1 cells after ETO treatment. In non-treated cells H3K9Me3, the label of silent pericentric heterochromatin49 marked the nuclei heterogeneously highlighting clusters of pericentric heterochromatin (Fig. 4E). In ETO-treated cells the average binding H3K9Me3 per nuclear DNA content measured by fluorescence intensity against DAPI was enhanced (1.78-fold, p < 0.001). At the same time, most centromere clusters leaving cell nuclei were depleted of the silencing H3K9Me3 label (Fig. 4F). Similarly, we also found that while in non-treated cells the DNA 5’–Methyl cytosine label revealed preferentially the clumps of heterochromatin, after ETO the DNA methylation pattern became smoothened, the concentration of the label increased 2-fold (p < 0.001), while its heterogeneity decreased as exemplified (Fig. 4G,H). The opposite showed the staining for active chromatin: when applying the epigenetic mark H3K4Me3 it revealed more bright nuclei of control non-treated cells and sombre staining of the ETO-treated cell nuclei (not shown), the concentration of H3K4Me3 decreased 1.48-fold (p < 0.001). These findings indicate to inactivation of euchromatin and a more open conformation assumed by constitutive heterochromatin in senescing cells after ETO treatment. Our observations are fully in accord with corresponding literature data on the epigenetic landscape of the chromatin in replicatively senescing cells.29,30 In turn, the de-methylation and decompaction of pericentric heterochromatin explains activation of retrotransposable elements nested in it and found above (Fig. 3).
Figure 4.
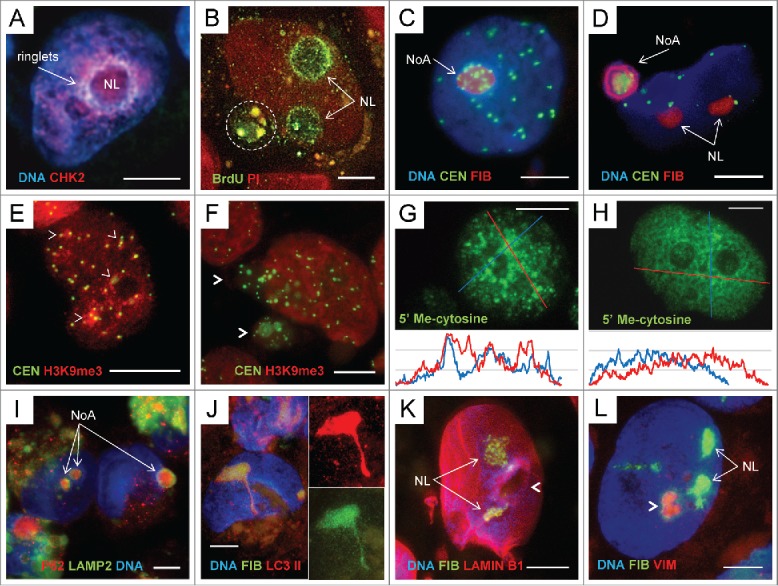
The topological relationship between NoAs and fragmented pericentric heterochromatin, change of its epigenetic profile and the ‘channelled’ release from cell nuclei in ETO cells, day 4: (A) Emergence of the ringlets of extrachromosomal circular DNA enriched with CHK2 signaling DNA damage in the ‘torulus’ around nucleolus (NL), RGB image; (B) 90 min incubation with BrdU of ETO cells revealed selective BrdU inclusion in the perinucleolar chromatin, less in the nucleoli (NL, arrows) and the same labeled material degrading in the perinucleolar vacuole (encircled) at the background of unlabelled nuclei (no S-phase). Counterstaining of DNA with Propidium Iodide (PI); (C) The tendency of pericentric heterochromatin labeled by CREST antibody to gather around and inside the FIB-enriched nucleolus (NoA) RGB image; (D) A large perinuclear NoA with a FIB-ring colocalized with DAPI-positive DNA material encompassing a cluster of multiple centromeres (CREST-antibody-positive), RGB image; (E,F) Change of the pattern in staining of pericentric heterochromatin: (E) in NT cells clusters of CEN accumulate H3K9Me3 label (arrowheads); (F) after ETO the released from cell nucleus CEN clusters are nearly depleted of H3K9Me3 (arrowheads), while the very nucleus homogenously enhance its content; (G,H) – Immunocytochemical reaction for 5’ methyl-cytosine on typical untreated (G) and ETO-treated (H) samples; the densitograms of each nucleus presented below show preferential DNA methylation of heterochromatin clumps in NT and smoothening of the DNA methylation profile after ETO treatment; (I) 2 intranuclear and one perinuclear NoAs are positively stained in the core for p62/SQSTM1, marking the aggregates for autophagic clearance, which are encircled by LAMP2 indicating to attraction of lysosomes from cytoplasm (RGB image); (J) the ETO treated cell with a nucleolar FIB/ LC3II- positive channel toward cytoplasm. Note abundance of brown lipofuscin in cytoplasm - a sign of cell overload with undigested waste, the abnormally high concentration of LC3II in the nucleolus and a channel, while LC3-II foci in cytoplasm are nearly absent; (K) A large nucleus stained for LAMIN B1, FIB and DNA (DAPI) possessing a large hole in the nuclear lamin surrounded by a “torulus” (arrowhead) with 2 nucleoli nearby (NL) conventionally stained for FIB (RGB image); (L) The hollow nucleolus surrounded by a “torulus” (arrowhead) is poor with DNA and FIB, however enriched with VIMENTIN (VIM); the 2 other nucleoli (NL) display the opposite (conventional) features: contain FIB and do not contain VIM (RGB image); The 2 other nucleoli have a conventional appearance (RGB image). Bars = 10 µm.
The question remained how NoA and accompanying heterochromatin get into cytoplasm of interphase cells arrested in G2 phase? Occasionally, co-staining of the nucleolar and perinucleolar NoA for p62/SQSTM1core revealed the surrounded rim of the autophagy lysosomal marker LAMP2 (Fig. 4I), which could only result from direct contact between the nucleolus and cytoplasmic lysosomes fusing with the autophagosome-enclosed cargo. The protrusion/budding of the chromatin targeted for autophagy through the nuclear membrane fusing with lysosomes, as well as the protrusion of cytoplasmic channels into cell nucleus has been in principle described for the chromatin autophagy.50 In our material, not infrequently the narrow protrusions from the nucleoli enriched in FIB and LC3-II were observed on the nuclear side-views (Fig. 4J). They likely can also really perforate nuclear membrane, opening a wide gate for accompanying eccDNA, in particular if autophagic flux and fusion with lysosomes fails. We found not only narrow nucleolar channels but also large holes in the nuclear lamin (Fig. 4K). Interestingly, that likewise the NoA forming most often only in one selective nucleolus (as seen on Fig. 2C-D, H, I; Fig. 3E), the nuclear “hole” is also often seen beside normally shaped nucleoli (Fig. 4K); “the hole” is containing vimentin (Fig. 4L), the known carrier of eccDNA51 (as well known for caging cytoplasmic aggresomes).52,53
The 2 questions arise, if the NoA-mediated release of heterochromatin is associated with impairment of the nucleolus basic function in rRNA synthesis and how the latter is related to irreversible cell senescence?
The impairment of rDNA transcription and processing in nucleoli of ETO-treated cells causes accumulation of LC3II in them, which is prevented by BAF
LC3-II active form was reported shuttling between nucleus and cytoplasm, be present in cell nuclei,23,54 and accumulating in the nucleoli.54,55 In particular, LC3-II protein interaction network analyzed by gene ontology includes RNA processing, splicing and RNA metabolism pathways.57 However, crosstalk between autophagy and RNA processes remained unexplored.50
Here we applied the immunostaining for RPA194, the subunit of Polymerase I and combined it with the antibody for LC3-II. In non-treated cells RPA194 foci localize as due in the multiple sites of pre-rRNA transcription (located in the fibrillar center/dense fibrillar component domains),38,58 while LC3-II is stained in them weakly (Fig. 5A). In ETO-treated samples, the pattern is again heterogeneous. In some cells this normal topology between RPA194 foci and LC3-II background is largely retained (Fig. 5B, (FC) and on enlarged insert), while in other RPA194 is away from the sites of transcription coalescing at the nucleolar center or periphery (Fig. 5B, arrowheads), which is a well-known sign of rRNA synthesis suppression.38 In such cells the LC3-II is seen more intensely accumulated in the nucleoli. Notably that in some of such cells, where the RPA 194 has abandoned the transcription sites and coalesced nearby, we found instead clear and bright intranucleolar foci of LC3-II, (Fig. 5C), the presumed autophagosomes, while those were reduced in cytoplasm. By applying BAF to ETO-treated cells, we found the opposite effect (Supplemental Fig. 1): BAF reduced the presence of LC3-II in nucleoli and NoA and increased it in cytoplasm, the same shift was observed for p62/SQSTM1.
Figure 5.
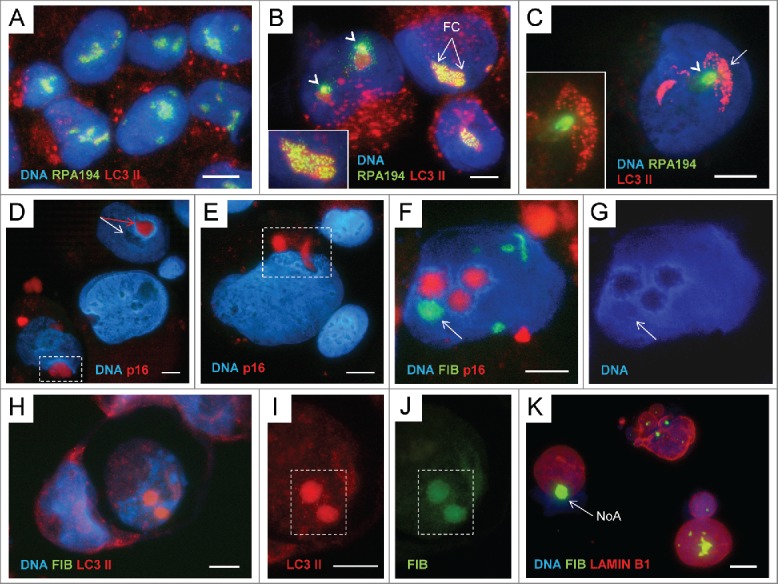
The relationship between rRNA synthesis and the autophagy inducer LC3-II in the nucleoli of ETO-treated cells and a possible role of p16INK4A in channelling of selected nucleoli: (A) NT control – RPA194 ‘punctuates’ the sites of transcription on the background of weak LC3-II presence in the nucleoli, while scarce LC3II foci in cytoplasm indicate the basic autophagy; (B) ETO - the number of cytoplasmic autophagosomes is much increased, while the nucleolar pattern is heterogeneous. In some cells RPA194 remains at the sites of transcription (FC and enlarged on insertion), while in other, it has left them and coalesced nearby (arrowheads) indicating cessation of rDNA transcription, while the presence of LC3-II in such nucleoli is increased; (C) – in some cells with coalesced RPA194 it appears to be substituted in the nucleolar sites with bright LC3-II foci; (D) p16 is seen in the selected nucleolus with ‘torulus” (red- arrowed, while a normal nucleolus is white-arrowed) and perinuclearly (enframed); (E) P16 is in the nuclear channel and perinuclearly (enframed); (F, G) p16 is found in 3 hollow DNA and FIB free nucleoli with a ‘torulus’, while the fourth arrowed nucleolus contains both FIB and DNA; (H-J) NoAs, nuclear fragmentation, and cell death: 2 ETO cells, one is engulfing the other (RGB image). The engulfed dead cell contains 2 nuclear NoAs with extremely high concentration of LC3-II, the cytoplasm is very poor with LC3-II, while the DAPI-stained nucleus is disintegrated (RGB image); (I) The enlarged fragment of the 2 NoAs in the engulfed cell, (I) in the red channel for LC3-II, (J) green channel for FIB; (K) An example of the FIB-NoA (arrow) releasing from a cell nucleus together with the DNA material in the breast cancer cell after neoadjuvant therapy (doxorubicin and paclitaxel) of the patient (from the operational material). The patchy staining for lamin B1 suggests its impairment. The tissue specimens were collected after the patients’ informed consent was obtained in accordance with the regulations of Committee of Medical Ethics of Latvia. Bars = 10 µm.
Collectively, these observations suggest that synthesis and processing of pre-rRNA require presence of a certain LC3-II nucleolar pool supported by normal autophagic flux. It can be speculated that endogenous impairment first of pre-rRNA processing and then synthesis can likely switch the formation and “soft” clearance of NoA in selective nucleoli (possibly during mitosis or by budding and scission by the nuclear membrane) into destructive process, enabling penetration of cytoplasmic autophagosomes and lysosomes into such nucleoli, opening the nuclear envelope gate for fragmented heterochromatin, and even causing NoA prolapse through it. Interestingly, the regulator of terminal senescence p16INK4a appears to be also involved in this nucleolus-associated channelling process.
The marker of terminal senescence p16INK4a is revealed in the nucleolar “holes,” channels, and perinuclear vacuoles of ETO-treated cells
The inhibitor of CDK4-kinase p16INK4A is usually considered a marker of terminal senescence. Indeed, it was activated in PA1-ETO cells in later terms post-treatment as found by Western blot.34 However, by IF we revealed that it can be also sequestered in cytoplasm in autophagic vacuoles.35 Here we paid attention that p16 can be revealed in selective nucleoli of PA1-ETO cells surrounded by a DNA “torulus” and in the nucleolar channels and perinuclear vacuoles, as well (Fig. 5D, E). Interestingly, when co-stained with fibrillarin, p16 is seen in hollow nucleoli, free of FIB and DNA (Fig. 5F, G), likewise it was found for VIM (Fig. 4L), a carrier of eccDNA. These meanwhile pilot observations provide an important key to possible contribution of p16 in-terminal senescence by the mechanism of the nucleolus-associated cell nucleus destruction. The situation with the nucleolar topology of LC3-II, nuclear integrity, and cell fate is spectacularly presented on Fig. 5H-J. Here one cell has engulfed another, which thus should be considered dead.59 In this dead cell the DAPI-stained nucleus is disintegrated, while the 2 intranuclear FIB-NoAs contain the enormously high concentration of LC3-II. On the contrary, the LC3II foci are scarce in cytoplasm indicating on the perturbed nuclear-cytoplasmic balance of this component on behalf of cell nucleus, as a possible hallmark of cell death.
In conclusion, we found that the mediated or started by NoAs release of pericentric fragments activated by retrotransposition in senescent PA1-ETO cells is topologically related to the nucleolus associated pericentric heterochromatin involving rDNA and activated ALU elements. These nucleolus rDNA associated domains should be identified as NADs, recently revealed by Hi-C technology, known as being enriched in centric/pericentric heterochromatin and retrotransposable ALU elements, found in most (or all) chromosomes.60-62
The described phenomena, the release of NoA accompanied by the DNA exit from cell nuclei were also seen by us in breast cancer samples after conventional neoadjuvant therapy with doxorubicin and paclitaxel63 (Fig. 5K).
Discussion
Our data showed that formation of the nuclear FIB-enriched dense bodies (NoA) targeted for selective autophagy in perinuclear cytoplasm (as co-labeled for ubiquitin 1, p62/SQSTM1, LC3-II, pAMPK, and LAMP2) in senescing PA1-ETO treated cells is caused by DDR and insufficiency of UPS. These NoAs apparently arise from disturbance of processing of pre-rRNA in the nucleolus as manifested by misfolding and accumulation in them of fibrillarin. The attraction of p62/SQSTM1 to polyubiquitinated aggregates recruiting LC3-II for selective autophagic degradation is a cell preventive measure to reduce proteotoxic stress.8,11,64-66 We have found that NoAs in ETO-treated cells also include the damaged rDNA. In this the phenomenon is similar to the piecemeal autophagy in senescing yeasts.15,16 rDNA is the “Achilles’ pied” of the nucleus and is the first damaged by replicative stress in response to adverse conditions. 12 However, the emergence of NoA was found here very tightly correlating with release of heterochromatic pericentric DNA fragments accompanying NoAs and exceeding their microscopically registered frequency. Contrary to NoA clearance needing functional autophagy and preservation of proliferation capacity as manifested by entrance into mitosis, the heterochromatin release from interphase cells arrested in the DNA damage checkpoint G2 and its accumulation in cytoplasm was favored by debilitation of late autophagy and often by the nucleolar channelling into cytoplasm or even just a prolapse of NoA through the nuclear envelope. The preliminary insight also revealed that the regulator of terminal senescence p16INK4A could be involved in this terminal process. Therefore, it appears that the response of PA1 cells to ETO consisted of 2 stages, whose schematic is presented on Fig. 6. The first, protective stage, with the insufficiency of UPS compensated by still efficient autophagy is executing selective autophagic clearance of FIB-NoAs in potentially surviving cells. As suggested by Latonen,13 such NoAs can serve as a platform for clearance of many misfolded proteins. During the second, destructive stage partly overlapping with the first, the release of pericentric fragments (the apparent reason of the persisting secondary DNA damage) is associated with increasing disability of late autophagy, leading to nuclear destruction and death of most ETO-treated senescent cells. Importantly, we found here that both, FIB-NoA formation and fragmentation of pericentric/centric heterochromatin with origination of extrachromosomal circular DNAs are focused in these G2 arrested cells onto the nucleolus, mostly likely on NADs, thus explaining the tight numerical correlation of the 2 phenomena. Our observations using the marks for rRNA transcription and processing, the mark for autophagy initiation p62/LC3-II, and BAF impairing the autophagic flux suggest that the basic nucleolar function is coordinated with the autophagy activities. Moreover, the mechanism, which is responsible for fragmentation of pericentric heterochromatin, involves, as found here, its de-repression heralded by loss of H3K9Me3 mark. Subsequent activation of retrotransposons and formation of eccDNA fragments by the “rolling circle” was revealed. Importantly, Guo et al.67 earlier found that functional autophagy supports the genome stability by degradation of transposon RNA in cytoplasm. The activation of transposable elements due to de-repression of pericentric heterochromatin of senescent cells is a known phenomenon.29,30 However, here we found for the first time that these changes are associated with the nucleolus and its functional state. The nucleolus detention function retaining DNA (cytosine-5′-methyltransferase 1 [DNMT1]) in the intergenic regions of 28S rDNA and thus keeping it away from its targets in heterochromatin has been described by Audas et al.68,69 By our hypothesis, de-repression of transposons locked in NADs, in case if DNMT1 is sequestered in NoA, may provide a link between the formation of NoA and fragmentation of pericentric nucleolus-associated heterochromatin induced by ETO (Fig. 6).
Figure 6.
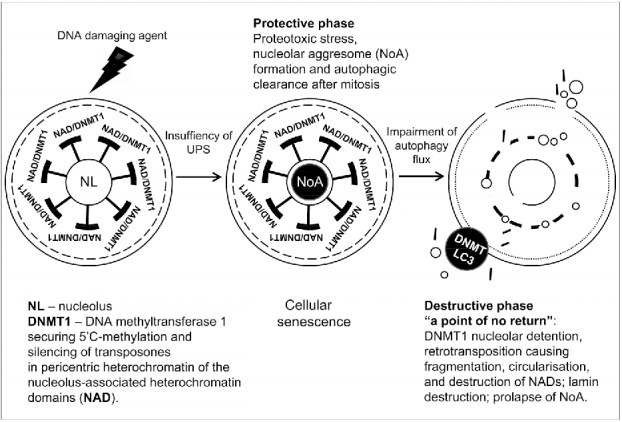
Schematic showing 2 stages of cellular senescence in the response of PA1 cells to ETO, protective and destructive, and the relationship between NoA formation in the first, and release of heterochromatin fragments, in the second phase. These fragments originate from the nucleolus associated domains (NADs). It is hypothesized that dysfunction of the nucleolus in response to stress may lead to detention of DNA (cytosine-5)-methyltransferase 1 (DNMT1) in NoA, whose removal from targets in NADs activate retrotransposons resulting in fragmentation of pericentric heterochromatin and ultimate terminal disintegration of cell nuclei.
In this article we skip the detailed discussion of the data on the nuclear lamin, lamin B receptors (LBR), lamin associated domains (LADs), and participation and traffic of nuclear membranes reported by several authors,20,22, 56,70 as involved in replicative, genotoxic and oncogenic senescence, describing the relationship with the chromatin autophagy, also recently reviewed.17,23,50,70 In our PA1-ETO model, we have also detected thinning of lamin B1 and disappearance of epichromatin (the 30-nm granular DNA layer embedded in LBR, revealed by a specific antibody71) in the cells with multiple CHK2-positive foci.35 However, the non-contradictory discussion of the relationship between the changes in cell nucleus periphery with those in the nucleolus and with the activity and topology of LC3-II in cellular senescence would be too much speculative here, it still needs more research. The consensus promising data indicate that NADs of non-acrocentric chromosomes are “communicating” the nuclear periphery, LADs relocate after mitosis to the periphery of reconstructing nucleolus,60,72 while ALU is enriched in the epichromatin of the nuclear lamin.73
The most important message of the current study is that “other” nucleolar functions in sensing cellular stress, proteome regulation, maintenance of genome integrity, and cell aging are very likely linked to the basic nucleolar function in transcription and processing of pre-rRNA. This makes a lot of sense. We conclude that in co-operation with the protein degrading, recycling and energy supplying cellular systems the nucleolus is ensuring the integrity of NAD network, secured by silenced retrotransposons.
Materials and methods
Cell culture and treatment
PA-1 (ATCC) cells were cultured in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% foetal bovine serum (FBS, Sigma). Cells were grown without antibiotics in 5% CO2 incubator at 37°C. Exponentially growing PA-1 cells were incubated with an 8 μM dose of etoposide (ETO) for 20 h. After ETO removal the cells were maintained by replenishing culture medium every 48 h and sampled on day 4. For testing the unscheduled DNA replication in senescent cells, the cells were incubated with 5 µM BrdU for 90 min before sampling. For studies of the contribution of the protein degrading systems, MG132 (Cayman) 20 μM was added to the culture medium for 12 h between day 3 to day 4 after ETO removal, before sampling. Bafilomycin A1 (Baf A, Cayman Chemicals) 50 nM was added to the culture medium for 24 h between day 3 to 4 after ETO removal, and sampled.
The microscopic counts for aggresome and extra nuclear DNA positive cells were performed in 5 independent experiments, collecting 400 cells in each. Extra nuclear DNA was considered if histochemically stained with Toluidine blue (see below) DNA-positive material was seen adjusted to cell nucleus.
Immunofluorescence
Cells were suspended in warm FBS and cytospun onto glass slides. Cytospins were fixed in methanol for 7 min at −20 °C and dipped 10 times in ice cold acetone (or fixed in 2% paraformaldehyde/PBS). Slides were then washed thrice in TBS 0.01% Tween 20 (TBST) for 5 min. Slides were subsequently blocked for 15 min in TBS, 0.05% Tween 20%, 1% BSA at room temperature. Samples were covered with TBS, 0.025% Tween 20%, 1% BSA containing primary antibody and incubated overnight at 4 °C in a humidified chamber. Samples were then washed thrice in TBST and covered with TBST containing the appropriate secondary antibodies (Goat anti-mouse IgG Alexa Fluor 488 (A31619, Invitrogen) and Goat anti-Rabbit-IgG Alexa Fluor 594 (A31631, Invitrogen)) and incubated for 40 min at room temperature in the dark. Slides were washed thrice for 5 min with TBST and once for 2 min in PBS. Samples were then counterstained with 0.25 μg/ml DAPI for 2 min, and finally embedded in Prolong Gold (Invitrogen). Primary antibodies and their source are listed in Table 1.
Table 1.
Primary antibodies and their source.
| Antibody against | Description | Specifity/Immunogen | Used conc. | Catalog Nr., Manufacturer |
|---|---|---|---|---|
| Bromo-deoxyuridine | Mouse monoclonal | Detects BrdU incorporated into DNA as well as bromouridine incorporated into RNA | 1:200 | A21300, Invitrogen |
| Centromere Protein | Human | Derived from human CREST patient serum | 1:50 | 15–234, Antibodies Inc. |
| CHK2 (phospho T68) | Rabbit polyclonal | Epitope around the phosphorylation site of Threonine 68 (VSTpQE) of human Chk2 | 1:100 | ab38461, Abcam |
| Fibrillarin | Mouse monoclonal | Corresponding to Saccharomyces cerevisiae Fibrillarin | 1:50 | ab4566, Abcam |
| Histone H3K4me3 | Mouse monoclonal | Synthetic peptide corresponding to Human Histone H3 aa 1–100 (tri methyl K4) conjugated to KLH | 1:50 | ab1012 |
| Histone H3K9me3 | Rabbit polyclonal | Synthetic peptide within Human Histone H3 aa 1–100 (N-terminal) (tri methyl K9) conjugated to KLH | 1:400 | ab8898 |
| LAMIN B1 | Rabbit polyclonal | Synthetic peptide conjugated to KLH derived from within residues 400 - 500 of mouse Lamin B1 | 1:200 | ab16048, Abcam |
| LAMP2 | Mouse monoclonal | The details of the immunogen for this antibody are not available | 1:500 | 555803, BD PharMingen™ |
| LC3B | Rabbit polyclonal | Peptide derived from within residues 1 - 100 of Human LC3B | 1:100 | ab63817, Abcam |
| 5′methyl-cytosine | Mouse monoclonal | Recognizes 5-methylcytosine in methylated DNA | 1:100 | NA81, Calbiochem |
| p16INK4a | Rabbit polyclonal | Corresponding to C-terminus of human p16 | 1:100 | ab7962, Abcam |
| p-AMPKα1/2 (Thr183/172) | Rabbit polyclonal | Epitope corresponding to phosphorylated Thr172 of AMPKα1 of human origin | 1:50 | sc-101630, Santa Cruz |
| p53(SP5) | Rabbit monoclonal | Recombinant human full-length wild type p53 protein | 1:50 | MA5–14516, Pierce |
| RPA194 | Mouse monoclonal | Raised against amino acids 1–300 of RPA194 of human origin | 1:50 | sc-48385, Santa Cruz |
| SQSTM1 (p62) | Rabbit polyclonal | Epitope corresponding to amino acids 151–440 of SQSTM1 of human origin. | 1:200 | sc-25575, Santa Cruz |
| Ub (FL-76) | Rabbit polyclonal | Epitope corresponding to amino acids 1–76 representing full length Ub of human origin | 1:50 | sc-9133, Santa Cruz |
| VIMENTIN | Rabbit polyclonal | Epitope corresponding to amino acids 1–84 mapping at the N-terminus of Vimentin of human origin | 1:50 | sc-5565, Santa Cruz |
When staining for 5′methylcytosine, treatment with 2N HCl at 37C for 20 min was used after fixation step, followed by 5 washes in PBS for 1 min.
For microscopic observations - a fluorescence light microscope (Leitz Ergolux L03–10) equipped with a color video camera (Sony DXC 390P) and a confocal laser microscope (Leica DM600) were used. To capture fluorescent images, in addition to separate optical filters, a 3-band BRG (blue, red, green) optical filter (Leica) was used. Image cytometry was performed by semi-automatic measuring of fluorescence values for each cell in all 3 channels and analyzed with Image-Pro Plus 4.1 software (Media Cybernetics).
DNA image cytometry
For DNA cytometry measurements, stoichiometric Toluidine blue DNA staining was performed as stated previously.74 In brief, cytospins were fixed in ethanol: acetone (1:1) for > 30 min at 4 °C and air-dried. Slides were then treated with 5N HCl for 20 min at room temperature, washed in distilled water 5 times for 1 min and stained for 10 min with 0.05% toluidine blue in 50% citrate-phosphate McIlvain buffer pH 4. Slides were rinsed with distilled water, blotted dry and dehydrated in butanol twice for 3 min at 37°C. Samples were then incubated twice in xylene for 3 min at room temperature and embedded in DPX (Sigma).
Digital images of at least 200 randomly selected cell nuclei were collected using a Sony DXC 390P color video camera calibrated in the green channel. DNA content per cell nucleus was measured as the integral optical density (IOD) using Image-Pro Plus 4.1 software (Media Cybernetics). The stoichiometry of DNA staining was verified using the values obtained for metaphases compared with anaphases and telophases (ratio 2.0); the summary error of the method and device was estimated to be less than 5%. Metaphase and ana-telophase counts were performed on TB-DNA or DAPI-stained specimens per 1,000 cells in 5 independent experiments.
Amplification, cloning, sequencing and ALU and rDNA probe labeling for in situ hybridization
Degenerative oligonucleotide primers were used for PCR amplification from human genomic DNA of ALU elements fragments. Primers were designed on the base of known sequences in GeneBank (Z30996.1, Z30987.1, Z31000.1). The primers were: 5’ GTCAGRAGWTCRAGACCA, 3’ MTCYYRRCTCACTGCAA. The primers allow amplification of the majority of targets in the genome. PCR with degenerative primers amplified a mix of ALU fragments of approximately 150 bp size ranges. Amplification products were gel separated, isolated, cloned into pGEM-T Easy Vector (Promega) according to standard manufactory protocol and sequenced.
For amplification from human genomic DNA of 45S rDNA fragments the following primers were designed on the base of known sequences in GeneBank (NR_046235): 5’ CCGTGGTCTCTCGTCTTCTC, 3’ TGCTACTGGCAGGATCAACC. PCR with these primers amplified a fragment of 990 bp size.
ALU fragments were labeled with Cy3 (Amersham, red fluorescence) and rDNA fragments with ATTO 390 (Jena Bioscience, blue fluorescence) according to a standard oligo-labeling protocol.
Fluorescence in situ hybridization (FISH)
Cells were trypsinized, washed with warm PBS, treated for 10 min with 75 mM KCl, suspended in foetal bovine serum, cytocentrifuged on slides. For ALU and rDNA FISH experiments methanol/acetone (1:1) fixation for 20 min was used and for probe for nucleolar organizers (NORs) methanol/glacial acetic acid (3:1) fixation for 1 h at −20°C was performed.
FISH was performed on ThermoBrite programmable temperature controlled slide processing system at 63°C for 3 h (ALU and rDNA). Slides were embedded in antifade mount (Vector Laboratories).
Commercial Acro-P-Arm Probe specific for rRNA genes located in the short arms of the acrocentric chromosomes (13, 14, 15, 21 and 22) was applied according to the manufacturer's instructions (LPE NOR, Cytocell) carrying out denaturation step for 2 min at 75°C and hybridization at 37°C overnight. Cells were embedded under coverslips in Prolong Gold with DAPI (Invitrogen).
ALU Real-time RT-PCR
Total RNA was extracted from PA-1 cells (2 × 106 cells) by using TRIZOL (Invitrogen). First-strand cDNA was synthesized using 2.5 µg of RNA, random hexamers and RevertAid™ M-MuLV Reverse Transcriptase (Fermentas, Lithuania) according to the manufacturer's protocols and subsequently diluted with nuclease-free water (Fermentas) 10 to 100 times. The absence of contamination with chromosomal DNA was verified by standard PCR using actin primers.75 Real-time PCR was run on a ViiA7 (Applied Biosystems). Amplification mixtures (25 µl) contained 2 µl template cDNA, 2xSYBR Green Master Mix buffer (12.5 µl) (Life Technologies, Carlsbad, CA, USA) and 2 µM forward and reverse primer. The cycling conditions comprised 10 min polymerase activation at 95°C and 40 cycles at 95°C for 15 sec and 60°C for 60 sec. A melt curve was also performed after the assay to check for specificity of the reaction. This consistent of 60 sec at 60°C followed by a ramp up of 0.05°/sec. Every cDNA sample was normalized against 4 housekeeping genes: GAPDH, ACTB, B2M, and LRP10 using geNorm.76 The calculated gene expression stability coefficient M was applied to ALU assay results. Primer sequences are listed in Table 2.
Table 2.
Primer sequences used for qRT-PCR study of ALU elements.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | GGGTCTTACTCCTTGGAGGC | GTCATCCCTGAGCTAGACGG |
| ACTB | AATCTCATCTTGTTTTCTGCGC | AGTGTGACGTGGACATCCG |
| B2M | TCTCGCTCCGTGGCCTTAGC | GCCTACCTACTTTGGGTCTGTGT |
| LRP10 | ACCGCTGCAACTACCAGACT | ACGTCGACATTACTGGAAC |
| Alu77 | GTCAGGAGATCGAGACCATCCC | TCCTGCCTCAGCCTCCCAAG |
Statistical analysis
Statistical analysis was performed in GraphPad (GraphPad Software Ltd). Student's t-test for unpaired samples was used to calculate the statistical significance of difference of means where appropriate. Statistical significance was accepted when p < 0.05.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Dr. Jamie Honeychurch is acknowledged for editing parts of the manuscript and Elina Zandberga for providing the housekeeping genes primers.
Funding
This work was supported by the Latvian Scientific Council, grant 2012/341.
References
- [1].Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8(7):574-85; PMID:17519961; http://dx.doi.org/ 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- [2].Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature 2005; 433(7021):77-83; PMID:15635413; http://dx.doi.org/ 10.1038/nature03207 [DOI] [PubMed] [Google Scholar]
- [3].Olson MOJ. Sensing cellular stress: another new function for the nucleolus? Sci STKE 2004; 2004(224):pe10; PMID:15026578; http://dx.doi.org/ 10.1126/stke.2242004pe10 [DOI] [PubMed] [Google Scholar]
- [4].Kobayashi T. A new role of the rDNA and nucleolus in the nucleus–rDNA instability maintains genome integrity. Bioessays 2008; 30(3):267-72; PMID:18293366; http://dx.doi.org/ 10.1002/bies.20723 [DOI] [PubMed] [Google Scholar]
- [5].Németh A, Längst G. Genome organization in and around the nucleolus. Trends Genet 2011; 27(4):149-56; PMID:21295884; http://dx.doi.org/ 10.1016/j.tig.2011.01.002 [DOI] [PubMed] [Google Scholar]
- [6].Dillinger S, Straub T, Nemeth A. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. bioRxiv 2016; http://dx.doi.org/ 10.1101/054908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016; 12(1):1-222; PMID:26799652; http://dx.doi.org/ 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171(4):603-14; PMID:16286508; http://dx.doi.org/ 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guarente L. Link between aging and the nucleolus. Genes Dev 1997; 11(19):2449-55; PMID:9334311; http://dx.doi.org/ 10.1101/gad.11.19.2449 [DOI] [PubMed] [Google Scholar]
- [10].Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008; 454(7208):1088-95; PMID:18756251; http://dx.doi.org/ 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 2010; 12(9):836-41; PMID:20811356; http://dx.doi.org/ 10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- [12].Lindstrom MS, Latonen L. The nucleolus as a stress response organelle. In: O'Day DH, Catalano A (eds.), Proteins of the Nucleolus: Regulation, Translocation, & Biomedical Functions. Dordrecht, The Netherlands: Springer Science & Business Media; 2013:251-72. [Google Scholar]
- [13].Latonen L. Nucleolar aggresomes as counterparts of cytoplasmic aggresomes in proteotoxic stress. Proteasome inhibitors induce nuclear ribonucleoprotein inclusions that accumulate several key factors of neurodegenerative diseases and cancer. Bioessays 2011; 33(5):386-95; PMID:21425306; http://dx.doi.org/ 10.1002/bies.201100008 [DOI] [PubMed] [Google Scholar]
- [14].Latonen L, Moore HM, Bai B, Jäämaa S, Laiho M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 2011; 30(7):790-805; PMID:20956947; http://dx.doi.org/ 10.1038/onc.2010.469 [DOI] [PubMed] [Google Scholar]
- [15].Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen E-L, Millen J, Goldfarb DS, Thumm M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell 2008; 19(10):4492-505; PMID:18701704; http://dx.doi.org/ 10.1091/mbc.E08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 2003; 14(1):129-41; PMID:12529432; http://dx.doi.org/ 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Erenpreisa J, Huna A, Salmina K, Jackson TR, Cragg MS. Macroautophagy-aided elimination of chromatin: sorting of waste, sorting of fate? Autophagy 2012; 8(12):1877-81; PMID:22935563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Erenpreisa J, Ivanov A, Cragg M, Selivanova G, Illidge T. Nuclear envelope-limited chromatin sheets are part of mitotic death. Histochem Cell Biol 2002; 117(3):243-55; PMID:11914922; http://dx.doi.org/ 10.1007/s00418-002-0382-6 [DOI] [PubMed] [Google Scholar]
- [19].Changou CA, Chen Y-R, Xing L, Yen Y, Chuang FYS, Cheng RH, Bold RJ, Ann DK, Kung H-J. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc Natl Acad Sci U S A 2014; 111(39):14147-52; PMID:25122679; http://dx.doi.org/ 10.1073/pnas.1404171111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dou Z, Ivanov A, Adams PD, Berger SL. Mammalian autophagy degrades nuclear constituents in response to tumorigenic stress. Autophagy 2016; 12(8):1416-7; PMID:26654219; http://dx.doi.org/ 10.1080/15548627.2015.1127465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Duarte LF, Young ARJ, Wang Z, Wu HA, Panda T, Kou Y, Kapoor A, Hasson D, Mills NR, Ma'ayan A, et al.. Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat Commun 2014; 5:5210; PMID:25394905; http://dx.doi.org/ 10.1038/ncomms6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al.. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 2013; 202(1):129-43; PMID:23816621; http://dx.doi.org/ 10.1083/jcb.201212110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sica V, Galluzzi L, Bravo-San Pedro JM, Izzo V, Maiuri MC, Kroemer G. Organelle-Specific Initiation of Autophagy. Mol Cell 2015; 59(4):522-39; PMID:26295960; http://dx.doi.org/ 10.1016/j.molcel.2015.07.021 [DOI] [PubMed] [Google Scholar]
- [24].Lukasova E, Kovařík A, Bačíková A, Falk M, Kozubek S. Loss of lamin B receptor is necessary to induce cellular senescence. Biochem J 2017; 474(2):281-300. http://dx.doi.org/ 10.1042/BCJ20160459 [DOI] [PubMed] [Google Scholar]
- [25].Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat Genet 2003; 35(3):219-20; PMID:14578886; http://dx.doi.org/ 10.1038/ng1259 [DOI] [PubMed] [Google Scholar]
- [26].Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer 2001; 30(1):64-71; PMID:11107177; http://dx.doi.org/ 10.1002/1098-2264(2000)9999:9999%3c::AID-GCC1066%3e3.0.CO;2-F [DOI] [PubMed] [Google Scholar]
- [27].Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol 2013; 75:685-705; PMID:23140366; http://dx.doi.org/ 10.1146/annurev-physiol-030212-183653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, et al.. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 2011; 10(17):3016-30; PMID:21862875; http://dx.doi.org/ 10.4161/cc.10.17.17543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 2013; 12(2):247-56; PMID:23360310; http://dx.doi.org/ 10.1111/acel.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 2013; 5(12):867-83; PMID:24323947; http://dx.doi.org/ 10.18632/aging.100621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sedivy JM, Kreiling JA, Neretti N, De Cecco M, Criscione SW, Hofmann JW, Zhao X, Ito T, Peterson AL. Death by transposition - the enemy within? Bioessays 2013; 35(12):1035-43; PMID:24129940; http://dx.doi.org/ 10.1002/bies.201300097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Baker DJ, Sedivy JM. Probing the depths of cellular senescence. J Cell Biol 2013; 202(1):11-3; PMID:23816622; http://dx.doi.org/ 10.1083/jcb.201305155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sturm Á, Ivics Z, Vellai T. The mechanism of ageing: primary role of transposable elements in genome disintegration. Cell Mol Life Sci 2015; 72(10):1839-47; PMID:25837999; http://dx.doi.org/ 10.1007/s00018-015-1896-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jackson TR, Salmina K, Huna A, Inashkina I, Jankevics E, Riekstina U, Kalnina Z, Ivanov A, Townsend PA, Cragg MS, et al.. DNA damage causes TP53-dependent coupling of self-renewal and senescence pathways in embryonal carcinoma cells. Cell Cycle 2013; 12(3):430-41; PMID:23287532; http://dx.doi.org/ 10.4161/cc.23285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huna A, Salmina K, Erenpreisa J, Vazquez-Martin A, Krigerts J, Inashkina I, Gerashchenko BI, Townsend PA, Cragg MS, Jackson TR. Role of stress-activated OCT4A in the cell fate decisions of embryonal carcinoma cells treated with etoposide. Cell Cycle 2015; 14(18):2969-84; PMID:26102294; http://dx.doi.org/ 10.1080/15384101.2015.1056948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mosieniak G, Sliwinska MA, Przybylska D, Grabowska W, Sunderland P, Bielak-Ż mijewska A, Sikora E. Curcumin-treated cancer cells show mitotic disturbances leading to growth arrest and induction of senescence phenotype. Int J Biochem Cell Biol 2016; 74:33-43; PMID:26916504; http://dx.doi.org/ 10.1016/j.biocel.2016.02.014 [DOI] [PubMed] [Google Scholar]
- [37].Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell 1985; 54(2):123-33; PMID:2933102; http://dx.doi.org/ 10.1111/j.1768-322X.1985.tb00387.x [DOI] [PubMed] [Google Scholar]
- [38].Schwarzacher HG, Wachtler F. The nucleolus. Anat Embryol (Berl) 1993; 188(6):515-36; PMID:8129175; http://dx.doi.org/ 10.1007/BF00187008 [DOI] [PubMed] [Google Scholar]
- [39].Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13(2):132-41; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lan YY, Londoño D, Bouley R, Rooney MS, Hacohen N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep 2014; 9(1):180-92; PMID:25284779; http://dx.doi.org/ 10.1016/j.celrep.2014.08.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley ARD, McStay B. The shared genomic architecture of human nucleolar organizer regions. Genome Res 2013; 23(12):2003-12; PMID:23990606; http://dx.doi.org/ 10.1101/gr.157941.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rochaix J-D, Bird A, Bakken A. Ribosomal RNA gene amplification by rolling circles. J Mol Biol 1974; 87(3):473-87; PMID:4444033; http://dx.doi.org/ 10.1016/0022-2836(74)90098-9 [DOI] [PubMed] [Google Scholar]
- [43].Rossi MS, Reig OA, Zorzópulos J. Evidence for rolling-circle replication in a major satellite DNA from the South American rodents of the genus Ctenomys. Mol Biol Evol 1990; 7(4):340-50; PMID:1974692 [DOI] [PubMed] [Google Scholar]
- [44].Thomas J, Phillips CD, Baker RJ, Pritham EJ. Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol 2014; 6(10):2595-610; PMID:25223768; http://dx.doi.org/ 10.1093/gbe/evu204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gaubatz JW. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat Res 1990; 237(5-6):271-92; PMID:2079966; http://dx.doi.org/ 10.1016/0921-8734(90)90009-G [DOI] [PubMed] [Google Scholar]
- [46].Kunisada T, Yamagishi H, Ogita Z, Kirakawa T, Mitsui Y. Appearance of extrachromosomal circular DNAs during in vivo and in vitro ageing of mammalian cells. Mech Ageing Dev 1985; 29(1):89-99; PMID:3982086; http://dx.doi.org/ 10.1016/0047-6374(85)90050-8 [DOI] [PubMed] [Google Scholar]
- [47].Weimer R, Haaf T, Krüger J, Poot M, Schmid M. Characterization of centromere arrangements and test for random distribution in G0, G1, S, G2, G1, and early S' phase in human lymphocytes. Hum Genet 1992; 88(6):673-82; PMID:1551672; http://dx.doi.org/ 10.1007/BF02265296 [DOI] [PubMed] [Google Scholar]
- [48].Erenpreisa J, Kalejs M, Ianzini F, Kosmacek EA, Mackey MA, Emzinsh D, Cragg MS, Ivanov A, Illidge TM. Segregation of genomes in polyploid tumour cells following mitotic catastrophe. Cell Biol Int 2005; 29(12):1005-11; PMID:16314119; http://dx.doi.org/ 10.1016/j.cellbi.2005.10.008 [DOI] [PubMed] [Google Scholar]
- [49].Peters AHFM, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al.. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 2003; 12(6):1577-89; PMID:14690609; http://dx.doi.org/ 10.1016/S1097-2765(03)00477-5 [DOI] [PubMed] [Google Scholar]
- [50].Luo M, Zhao X, Song Y, Cheng H, Zhou R. Nuclear autophagy: An evolutionarily conserved mechanism of nuclear degradation in the cytoplasm. Autophagy 2016; 12(11):1973-83; PMID:27541589; http://dx.doi.org/ 10.1080/15548627.2016.1217381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hartig R, Shoeman RL, Janetzko A, Tolstonog G, Traub P. DNA-mediated transport of the intermediate filament protein vimentin into the nucleus of cultured cells. J Cell Sci 1998; 111(24):3573-84; PMID:9819349 [DOI] [PubMed] [Google Scholar]
- [52].Johnston JA, Ward CL, Kopito RR. Aggresomes: A Cellular Response to Misfolded Proteins. J Cell Biol 1998; 143(7):1883-98; PMID:9864362; http://dx.doi.org/ 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell 2001; 12(5):1393-407; PMID:11359930; http://dx.doi.org/ 10.1091/mbc.12.5.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kraft LJ, Dowler J, Kenworthy AK. The Autophagosome Marker LC3 Undergoes Regulated Targeting to the Nucleus and Nucleolus. Biophys J 2015; 108(2):571a-2a; http://dx.doi.org/ 10.1016/j.bpj.2014.11.3126 [DOI] [Google Scholar]
- [55].Kraft LJ, Manral P, Dowler J, Kenworthy AK. Nuclear LC3 Associates with Slowly Diffusing Complexes that Survey the Nucleolus. Traffic 2016; 17(4):369-99; PMID:26728248; http://dx.doi.org/ 10.1111/tra.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dou Z, Xu C, Donahue G, Shimi T, Pan J-A, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al.. Autophagy mediates degradation of nuclear lamina. Nature 2015; 527(7576):105-9; PMID:26524528; http://dx.doi.org/ 10.1038/nature15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466(7302):68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Smirnov E, Cmarko D, Mazel T, Hornáč ek M, Raška I. Nucleolar DNA: the host and the guests. Histochem Cell Biol 2016; 145(4):359-72; PMID:26847178; http://dx.doi.org/ 10.1007/s00418-016-1407-x [DOI] [PubMed] [Google Scholar]
- [59].Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny M V, El-Deiry WS, Golstein P, Green DR, et al.. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 2009; 16(1):3-11; PMID:18846107; http://dx.doi.org/ 10.1038/cdd.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 2010; 21(21):3735-48; PMID:20826608; http://dx.doi.org/ 10.1091/mbc.E10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G. Initial Genomics of the Human Nucleolus. Akhtar A, ed. PLoS Genet 2010; 6(3):e1000889; PMID:20361057; http://dx.doi.org/ 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Németh A, Längst G. Chromatin Organization and the Mammalian Nucleolus In: Proteins of the Nucleolus. Dordrecht: Springer Netherlands; 2013:119-48; http://dx.doi.org/ 10.1007/978-94-007-5818-6_6 [DOI] [Google Scholar]
- [63].Gerashchenko BI, Salmina K, Eglitis J, Huna A, Grjunberga V, Erenpreisa J. Disentangling the aneuploidy and senescence paradoxes: a study of triploid breast cancers non-responsive to neoadjuvant therapy. Histochem Cell Biol 2016; 145(4):497-508; PMID:26860864; http://dx.doi.org/ 10.1007/s00418-016-1415-x [DOI] [PubMed] [Google Scholar]
- [64].Benbrook DM, Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol 2012; 34(3):286-97; PMID:23070014 [PubMed] [Google Scholar]
- [65].Kawaguchi T, Miyazawa K, Moriya S, Ohtomo T, Che XF, Naito M, Itoh M, Tomoda A. Combined treatment with bortezomib plus bafilomycin A1 enhances the cytocidal effect and induces endoplasmic reticulum stress in U266 myeloma cells: crosstalk among proteasome, autophagy-lysosome and ER stress. Int J Oncol 2011; 38(3):643-54; PMID:21174067; http://dx.doi.org/ 10.3892/ijo.2010.882 [DOI] [PubMed] [Google Scholar]
- [66].Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The Deacetylase HDAC6 Regulates Aggresome Formation and Cell Viability in Response to Misfolded Protein Stress. Cell 2003; 115(6):727-38; PMID:14675537; http://dx.doi.org/ 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- [67].Guo H, Chitiprolu M, Gagnon D, Meng L, Perez-Iratxeta C, Lagace D, Gibbings D. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun 2014; 5:5276; PMID:25366815; http://dx.doi.org/ 10.1038/ncomms6276 [DOI] [PubMed] [Google Scholar]
- [68].Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 2012; 45(2):147-57; PMID:22284675; http://dx.doi.org/ 10.1016/j.molcel.2011.12.012 [DOI] [PubMed] [Google Scholar]
- [69].Audas TE, Jacob MD, Lee S. The nucleolar detention pathway: A cellular strategy for regulating molecular networks. Cell Cycle 2012; 11(11):2059-62; PMID:22580471; http://dx.doi.org/ 10.4161/cc.20140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Boban M, Foisner R. Degradation-mediated protein quality control at the inner nuclear membrane. Nucleus 2016; 7(1):41-9; PMID:26760377; http://dx.doi.org/ 10.1080/19491034.2016.1139273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Olins AL, Langhans M, Monestier M, Schlotterer A, Robinson DG, Viotti C, Zentgraf H, Zwerger M, Olins DE. An epichromatin epitope: persistence in the cell cycle and conservation in evolution. Nucleus 2(1):47-60; PMID:21647299; http://dx.doi.org/ 10.4161/nucl.13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 2015; 16(4):245-57; PMID:25757416; http://dx.doi.org/ 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- [73].Olins AL, Ishaque N, Chotewutmontri S, Langowski J, Olins DE. Retrotransposon Alu is enriched in the epichromatin of HL-60 cells. Nucleus 5(3):237-46; PMID:24824428; http://dx.doi.org/ 10.4161/nucl.29141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Erenpreisa J, Freivalds T. Anisotropic staining of apurinic acid with toluidine blue. Histochemistry 1979; 60(3):321-5; PMID:89109; http://dx.doi.org/ 10.1007/BF00500660 [DOI] [PubMed] [Google Scholar]
- [75].Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, Ivanov A, Jascenko E, Scherthan H, Cragg M, et al.. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res 2010; 316(13):2099-112; PMID:20457152; http://dx.doi.org/ 10.1016/j.yexcr.2010.04.030 [DOI] [PubMed] [Google Scholar]
- [76].Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3(7):RESEARCH0034; PMID:12184808; http://dx.doi.org/ 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nicklas JA, Buel E. Development of an Alu-based, real-time PCR method for quantitation of human DNA in forensic samples. J Forensic Sci 2003; 48(5):936-44 PMID:14535658; http://dx.doi.org/ 10.1520/JFS2002414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


