ABSTRACT
The eukaryotic genome is partitioned into topologically associating domains (TADs). Despite recent advances characterizing TADs and TAD boundaries, the organization of these structures is an important dimension of genome architecture and function that is not well understood. Recently, we demonstrated that knockdown of BRG1, an ATPase driving the chromatin remodeling activity of mammalian SWI/SNF enzymes, globally alters long-range genomic interactions and results in a reduction of TAD boundary strength. We provided evidence suggesting that this effect may be due to BRG1 affecting nucleosome occupancy around CTCF sites present at TAD boundaries. In this review, we elaborate on our findings and speculate that BRG1 may contribute to the regulation of the structural and functional properties of chromatin at TAD boundaries by affecting the function or the recruitment of CTCF and DNA topoisomerase complexes.
KEYWORDS: BRG1, CTCF, Hi-C, SWI/SNF;TADs, topologically associated domains, topoisomerase
Introduction
Recent advances in molecular biology and microscopy technologies have enhanced our understanding about the higher-order folding of the genome in an unprecedented manner.1-5 It is now widely accepted that the dynamic folding of the chromatin is fundamental in regulating gene expression and DNA replication. The genome is folded in a hierarchical manner into chromosome territories, genomic compartments, and topologically associating domains (TADs), in which specific long-range looping interactions occur.6 Each of these structures can be dynamically regulated during development, and perturbations in these folding units are associated with multiple diseases and cancer.7-9
One interesting feature that was revealed upon fine mapping of the folding of the genome is the TAD structures, which range from 100kb up to 1Mb in size.7,10-13 TADs are units of chromosomes that exhibit higher frequency of physical contacts between genes and their cognate regulatory regions. The TADs have been shown to be stable across different species, cell types, and cellular conditions.12 The replication timing, the presence of different histone modifications and the expression of the genes inside a TAD are highly correlated.12,14,15 However, the underlying features that generate invariant TAD boundaries remain unknown. In human cells, TAD boundaries have been associated with the enrichment of CTCF, cohesin, DNase I hypersensitivity, certain histone marks and the timing of replication domains.7,13,14 Furthermore, in Drosophila, the combinatorial binding of different types of insulators (also referred to as architectural proteins), such as BEAF32 and TFIIIC, is associated with the strength of the TAD boundary.16,17 The boundaries serve as barriers to long- and short-range interactions between DNA sequences, thus the most likely interaction partner for a genomic region is another region within the same TAD. A strong TAD boundary limits interactions between the two adjacent TAD domains, whereas a weak TAD boundary allows a higher frequency of inter-TAD interactions.17-19 Although a relationship between TAD boundaries and the binding of insulators has been demonstrated, the effects of enzymes that modify or remodel chromatin are largely unknown.
The major ATPase subunit of the SWI/SNF chromatin remodeling complex, BRG1 (also known as SMARCA4), is required for proper nucleosome occupancy and positioning at target regions20 as well as for establishing numerous long-range chromatin interactions.21-26 BRG1 plays dual roles both as a transcriptional activator and repressor.10 In this review, we highlight our recent finding that BRG1 contributes to TAD boundary strength and discuss the implications of BRG1 loss on nucleosome occupancy and higher order chromatin organization.27
BRG1 is associated with TAD boundaries
To delineate the role of BRG1 in genome architecture, we performed RNA-seq and Hi-C in control and BRG1 knockdown mammary epithelial MCF-10A cells.27 Moreover, to probe BRG1 localization, we performed ChIP-seq in wild type parental MCF-10A cells. We reported that BRG1 regulates the expression of genes related to extracellular matrix and lipid synthesis. In addition, BRG1 knockdown resulted in the differential expression of many long non-coding RNAs, which are thought to be involved in genome organization.28 ChIP-seq analysis of BRG1 demonstrated binding primarily (∼60%) at genic regions.27 Intergenic BRG1 peaks included super-enhancers, which are defined as clusters of regulatory regions with unusual enrichment of factor binding sites and/or histone modifications that are in close proximity,29 though this classification has recently been challenged.30 Taken together, our findings showed that BRG1 binding was widespread throughout the genome.
Comparison of the control and BRG1 knockdown Hi-C data sets revealed that the majority of the TAD boundaries were largely overlapping between control and BRG1 knockdown cells,27 consistent with prior results.31-33 However, more than 10% of the TAD boundaries were unique to either the control or the BRG1 knockdown cells, providing evidence that BRG1 may contribute to the integrity of TAD boundaries. In support of this concept, we observed an enrichment of BRG1 binding at TAD boundaries. This result may be expected since TAD boundaries have been shown to be enriched for both housekeeping and developmentally regulated genes.13 Surprisingly, the strength of the TAD boundaries, as assessed using the insulation method,34 was significantly reduced in BRG1 knockdown human mammary epithelial MCF-10A cells. In other words, upon BRG1 knockdown, there was a higher frequency of inter-TAD genomic interactions. We further validated this result by intersecting TAD boundaries with BRG1 ChIP-seq data and categorizing the boundaries as either BRG1 bound or not bound. The strength of TAD boundaries that were bound by BRG1 was stronger than the boundaries that lacked BRG1 localization.27
An explanation for the effect of BRG1 knockdown is that BRG1 loss may disrupt chromatin accessibility and preclude the binding of transcription factors and/or chromatin modifiers at TAD boundaries. To examine this possibility, we analyzed a publicly available MNase-seq data set from wildtype and BRG1 knockdown mouse embryonic fibroblast (MEF) cells.20 By intersecting the nucleosome occupancy data with the ENCODE MEF CTCF ChIP-seq data set, we showed that there was decreased nucleosome occupancy at CTCF-bound regions in BRG1 knockdown cells when compared to control cells.27 Thus, our results suggested that BRG1 plays a role at TAD boundaries by regulating nucleosome occupancy and possibly CTCF localization.
The CTCF connection
Dixon et al. recently hypothesized that the orderly positioning and occupancy of nucleosomes at TAD boundaries renders the local chromatin at the boundary “less flexible,” and thus prevents long-range interactions surpassing the boundary.35 The orderly positioning and occupancy of nucleosomes at TAD boundaries may be achieved by enriched CTCF binding, which positions ∼20 nucleosomes around its binding sites.36 Our observation of the relationship between BRG1 knockdown and the reduction of nucleosome occupancy around the CTCF sites supports such a mechanism.27 It was recently shown that the directionality of CTCF binding is very strongly associated the formation of TADs,37-41 though the relationship between BRG1 binding and CTCF binding site orientation remains to be determined. We propose that loss or knockdown of BRG1 results in the reduction of nucleosome occupancy at CTCF sites either through loss of its ATP-dependent chromatin remodeling activity or by negatively impacting CTCF binding (Fig. 1). Such an effect of BRG1 may also occur at regions where other chromatin organizers (e.g. cohesin) are bound.
Figure 1.
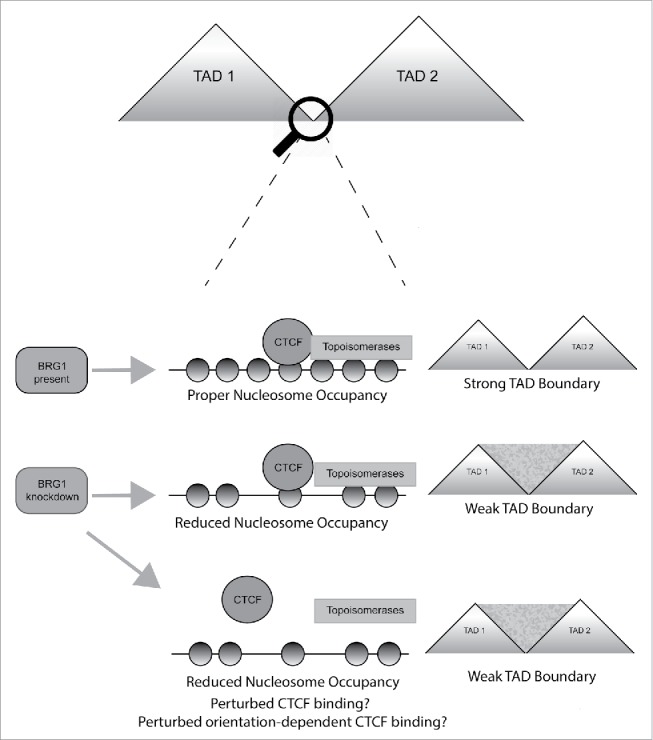
A schematic figure depicting the possible connection between BRG1, CTCF and topoisomerases. In the presence of BRG1 (top panel), CTCF and topoisomerases can efficiently bind to TAD boundaries and promote proper nucleosome occupancy and uncoiling of the DNA, resulting in a strong TAD boundary. We previously reported that nucleosome occupancy around CTCF sites was reduced upon BRG1 knockdown.27 Therefore, we propose that upon BRG1 knockdown, CTCF and topoisomerases may interact with TAD boundary sequences, but the lack of ATP-dependent remodeling activity may alter nucleosome occupancy and affect boundary strength (middle panel). Alternatively, the binding of CTCF and topoisomerases may be perturbed, resulting in altered nucleosome occupancy and reduced boundary strength (bottom panel).
The TOP connection
The model proposed by Dixon et al.,35 is reinforced by accumulating evidence that highlights the role of the topoisomerase complex as a regulator of TAD boundaries.42 Recent results demonstrate that topoisomerase II beta (TOP2B) binding overlaps with almost half of CTCF/cohesin binding sites, and that TOP2B may facilitate supercoiling at CTCF sites in a transcription-dependent manner.43 Direct links between topoisomerases and BRG1 are provided by a proteomic profiling using mass spectrometry that identified a significant interaction between BRG1 and TOP2B.44 Consistent with this finding, BRG1 also binds to topoisomerase II alpha (TOP2A).45 Furthermore, BRG1 is required for the recruitment of topoisomerase I (TOP1) to chromatin, and in the case of both TOP2A and TOP1, the ATPase activity of BRG1 has been shown to be essential for the recruitment of the topoisomerase proteins.45,46 These findings suggest an interplay and functional cooperativity between CTCF, BRG1 and topoisomerases in the organization of TADs via the active regulation of TAD boundary regions.
Additional evidence for engagement of topoisomerases in genome organization is provided by studies from bacteria and yeast. A Hi-C study in prokaryotes examined the biology of chromosomal interaction domains (CIDs), which are prokaryotic chromatin folding structures that are analogous to the eukaryotic TADs. Treatment of the prokaryotic cells with novobiocin, a drug that inhibits DNA gyrase (a homolog of topoisomerase II) and thus supercoiling, perturbed the sharpness and the positions of CID boundaries.47 In yeast, fine resolution nucleosome mapping determined that self-associating domains similar to but smaller than TADs exist and that their boundaries were bound by the RSC chromatin remodeling enzyme.48 The RSC ATPase is structurally related to BRG1, and the genetic analyses performed confirmed a role for RSC in yeast genome organization.48 Moreover, Hrp1, an ATP-dependent chromatin remodeling protein from the CHD subfamily, was shown to collaborate with Top1 to maintain open chromatin at active gene regions in yeast.49 Taken together, these findings suggest a strong link between chromatin remodeling enzymes, including the mammalian SWI/SNF complex, architectural proteins, and topoisomerases in genome organization.
Conclusions and future perspectives
We propose that the association between BRG1 knockdown and reduction in TAD boundary strength may be due to perturbations in the recruitment of CTCF and topoisomerases, and may therefore affect integrity of the chromatin structure and the “stiffness” of the chromatin at TAD boundaries (Fig. 1). In further support of the chromatin “stiffness” hypothesis, a recent study showed that a small deletion of a TAD boundary was not sufficient to disrupt the TAD domain, as it remained stable.50 This result suggests that the boundary is not defined by the exact boundary sequence or length, but instead depends either on the supercoiling or the overall composition of the factors present at the boundary. Our recent data indicating that BRG1, and hence the mammalian SWI/SNF chromatin remodeling enzyme, binds to TAD boundaries and promotes boundary strength adds a novel biochemical activity, ATP-dependent chromatin remodeling, to the complex structure that regulates TAD formation and function. Other chromatin remodeling complexes may play similar roles, as these enzymes can function in a redundant manner.51 Continued examination of the factors found at TAD boundaries will yield important insights into the biophysical properties of TADs and their boundaries, as well as into chromatin folding and overall genome organization that supports biological control.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by P01 CA82834.
References
- [1].de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev 2012; 26:11-24; PMID:22215806; http://dx.doi.org/ 10.1101/gad.179804.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Denker A, de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev 2016; 30:1357-82; PMID:27340173; http://dx.doi.org/ 10.1101/gad.281964.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell 2016; 164:1110-21; PMID:26967279; http://dx.doi.org/ 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barutcu AR, Fritz AJ, Zaidi SK, van Wijnen AJ, Lian JB, Stein JL, Nickerson JA, Imbalzano AN, Stein GS. C-ing the genome: a compendium of chromosome conformation capture methods to study higher-order chromatin organization. J Cell Physiol 2016; 231:31-5; PMID:26059817; http://dx.doi.org/ 10.1002/jcp.25062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fritz AJ, Barutcu AR, Martin-Buley L, van Wijnen AJ, Zaidi SK, Imbalzano AN, Lian JB, Stein JL, Stein GS. Chromosomes at work: organization of chromosome territories in the interphase nucleus. J Cell Biochem 2016; 117:9-19; PMID:26192137; http://dx.doi.org/ 10.1002/jcb.25280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell 2013; 49:773-82; PMID:23473598; http://dx.doi.org/ 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Barutcu AR, Lajoie BR, McCord RP, Tye CE, Hong D, Messier TL, Browne G, van Wijnen AJ, Lian JB, Stein JL, et al. Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome Biol 2015; 16:214; PMID:26415882; http://dx.doi.org/ 10.1186/s13059-015-0768-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 2013; 23:260-9; PMID:23152449; http://dx.doi.org/ 10.1101/gr.138032.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Taberlay PC, Achinger-Kawecka J, Lun ATL, Buske FA, Sabir K, Gould CM, Zotenko E, Bert SA, Giles KA, Bauer DC, et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res 2016; 26:719-31; PMID:27053337; http://dx.doi.org/ 10.1101/gr.201517.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Attanasio C, Nord AS, Zhu Y, Blow MJ, Biddie SC, Mendenhall EM, Dixon J, Wright C, Hosseini R, Akiyama JA, et al. Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res 2014; 24:920-9; PMID:24752179; http://dx.doi.org/ 10.1101/gr.168930.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012; 485:381-5; PMID:22495304; http://dx.doi.org/ 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays 2013; 35:818-28; PMID:23832846; http://dx.doi.org/ 10.1002/bies.201300040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dixon JR, Siddarth S, Feng Y, Audrey K, Yan L, Yin S, Ming H, Liu JS, Bing R. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485:376-80; PMID:22495300; http://dx.doi.org/ 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. Topologically associating domains are stable units of replication-timing regulation. Nature 2014; 515:402-5; PMID:25409831; http://dx.doi.org/ 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Le Dily F, D Baù, Pohl A, Vicent GP, Serra F, Soronellas D, Castellano G, Wright RHG, Ballare C, Filion G, et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev 2014; 28:2151-62; PMID:25274727; http://dx.doi.org/ 10.1101/gad.241422.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vogelmann J, Le Gall A, Dejardin S, Allemand F, Gamot A, Labesse G, Cuvier O, Nègre N, Cohen-Gonsaud M, Margeat E, et al. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet 2014; 10:e1004544; PMID:25165871; http://dx.doi.org/ 10.1371/journal.pgen.1004544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Van Bortle K, Nichols MH, Li L, Ong C-T, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol 2014; 15:R82; PMID:24981874; http://dx.doi.org/ 10.1186/gb-2014-15-5-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cubeñas-Potts C, Caelin C-P, Corces VG. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett 2015; 589:2923-30; http://dx.doi.org/ 10.1016/j.febslet.2015.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cubeñas-Potts C, Caelin C-P, Corces VG. Topologically associating domains: an invariant framework or a dynamic scaffold? Nucleus 2015; 6:430-4; http://dx.doi.org/ 10.1080/19491034.2015.1096467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans JA, Wilson BG, Park PJ, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci U S A 2013; 110:10165-70; PMID:23723349; http://dx.doi.org/ 10.1073/pnas.1302209110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim S-I, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res 2009; 37:6019-27; PMID:19696073; http://dx.doi.org/ 10.1093/nar/gkp677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim S-I, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc Natl Acad Sci U S A 2009; 106:2259-64; PMID:19171905; http://dx.doi.org/ 10.1073/pnas.0806420106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Harada A, Mallappa C, Okada S, Butler JT, Baker SP, Lawrence JB, Ohkawa Y, Imbalzano AN. Spatial re-organization of myogenic regulatory sequences temporally controls gene expression. Nucleic Acids Res 2015; 43:2008-21; PMID:25653159; http://dx.doi.org/ 10.1093/nar/gkv046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Imbalzano A, Imbalzano KM, Nickerson JA. BRG1, a SWI/SNF chromatin remodeling enzyme ATPase, is required for maintenance of nuclear shape and integrity. Commun Integr Biol 2013; 6:e25153; PMID:24228137; http://dx.doi.org/ 10.4161/cib.25153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Imbalzano KM, Cohet N, Wu Q, Underwood JM, Imbalzano AN, Nickerson JA. Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One 2013; 8:e55628; PMID:23405182; http://dx.doi.org/ 10.1371/journal.pone.0055628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hill DA, Chiosea S, Jamaluddin S, Roy K, Fischer AH, Boyd DD, Nickerson JA, Imbalzano AN. Inducible changes in cell size and attachment area due to expression of a mutant SWI/SNF chromatin remodeling enzyme. J Cell Sci 2004; 117:5847-54; PMID:15537831; http://dx.doi.org/ 10.1242/jcs.01502 [DOI] [PubMed] [Google Scholar]
- [27].Barutcu AR, Lajoie BR, Fritz AJ, McCord RP, Nickerson JA, van Wijnen AJ, Lian JB, Stein JL, Dekker J, Stein GS, et al. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res 2016; 26:1188-201; PMID:27435934; http://dx.doi.org/ 10.1101/gr.201624.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].M Melé, Rinn JL. “Cat's Cradling” the 3D genome by the act of LncRNA transcription. Mol Cell 2016; 62:657-64; PMID:27259198; http://dx.doi.org/ 10.1016/j.molcel.2016.05.011 [DOI] [PubMed] [Google Scholar]
- [29].Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153:307-19; PMID:23582322; http://dx.doi.org/ 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hay D, Hughes JR, Babbs C, Davies JOJ, Graham BJ, Hanssen LLP, Kassouf MT, Oudelaar AM, Sharpe JA, Suciu MC, et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat Genet 2016; 48:895-903; PMID:27376235; http://dx.doi.org/ 10.1038/ng.3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, Brouwer RWW, van de Corput MPC, van de Werken HJG, Knoch TA, van IJcken WFJ, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 2014; 111:996-1001; PMID:24335803; http://dx.doi.org/ 10.1073/pnas.1317788111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ing-Simmons E, Elizabeth I-S, Seitan VC, Faure AJ, Paul F, Thomas C, Job D, Fisher AG, Boris L, Matthias M. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res 2015; 25:504-13; PMID:25677180; http://dx.doi.org/ 10.1101/gr.184986.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Geeven G, Zhu Y, Kim BJ, Bartholdy BA, Yang S-M, Macfarlan TS, Gifford WD, Pfaff SL, Verstegen MJAM, Pinto H, et al. Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol 2015; 16:289; PMID:26700097; http://dx.doi.org/ 10.1186/s13059-015-0857-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 2015; 523:240-4; PMID:26030525; http://dx.doi.org/ 10.1038/nature14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dixon JR, Gorkin DU, Ren B. Chromatin Domains: The Unit of Chromosome Organization. Mol Cell 2016; 62:668-80; PMID:27259200; http://dx.doi.org/ 10.1016/j.molcel.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard JK. Controls of nucleosome positioning in the human genome. PLoS Genet 2012; 8:e1003036; PMID:23166509; http://dx.doi.org/ 10.1371/journal.pgen.1003036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Vietri Rudan M, Rudan MV, Christopher B, Stephen H, Christina E, Odom DT, Amos T, Suzana H. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 2015; 10:1297-309; PMID:25732821; http://dx.doi.org/ 10.1016/j.celrep.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vietri Rudan M, Rudan MV, Suzana H. Genetic tailors: CTCF and cohesin shape the genome during evolution. Trends Genet 2015; 31:651-60; PMID:26439501; http://dx.doi.org/ 10.1016/j.tig.2015.09.004 [DOI] [PubMed] [Google Scholar]
- [39].Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 2015; 162:900-10; PMID:26276636; http://dx.doi.org/ 10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 2015; 163:1611-27; PMID:26686651; http://dx.doi.org/ 10.1016/j.cell.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014; 159:1665-80; PMID:25497547; http://dx.doi.org/ 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gilbert N, Allan J. Supercoiling in DNA and chromatin. Curr Opin Genet Dev 2014; 25:15-21; PMID:24584092; http://dx.doi.org/ 10.1016/j.gde.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Uusküla-Reimand L, Hou H, Samavarchi-Tehrani P, Rudan MV, Liang M, Medina-Rivera A, Mohammed H, Schmidt D, Schwalie P, Young EJ, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol 2016; 17:182; PMID:27582050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell 2012; 150:1068-81; PMID:22939629; http://dx.doi.org/ 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dykhuizen EC, Hargreaves DC, Miller EL, Kairong C, Andrey K, Marcel K, Stefan P, Yoon-Jae C, Keji Z, Crabtree GR. BAF complexes facilitate decatenation of DNA by topoisomerase IIα;. Nature 2013; 497:624-7; PMID:23698369; http://dx.doi.org/ 10.1038/nature12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Husain A, Begum NA, Taniguchi T, Taniguchi H, Kobayashi M, Honjo T. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat Commun 2016; 7:10549; PMID:26842758; http://dx.doi.org/ 10.1038/ncomms10549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Le TBK, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 2013; 342:731-4; PMID:24158908; http://dx.doi.org/ 10.1126/science.1242059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hsieh T-HS, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell 2015; 162:108-19; PMID:26119342; http://dx.doi.org/ 10.1016/j.cell.2015.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Durand-Dubief M, Mickaël D-D, Jenna P, Ulrika N, Edgar H, Karl E. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J 2010; 29:2126-34; PMID:20526281; http://dx.doi.org/ 10.1038/emboj.2010.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Franke M, Martin F, Ibrahim DM, Guillaume A, Wibke S, Verena H, Robert S, Katerina K, Rieke K, Ivana J, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature [Internet] 2016; 538:265-9; Available from: http://dx.doi.org/ 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- [51].Morris SA, Baek S, Sung M-H, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol 2014; 21:73-81; PMID:24317492; http://dx.doi.org/ 10.1038/nsmb.2718 [DOI] [PMC free article] [PubMed] [Google Scholar]


