Abstract
Glioma is the most common and aggressive type of primary brain tumor. MicroRNA (miR)-130b functions as a tumor-associated miR. The dysregulation of miR-130b is involved in numerous biological characteristics and properties of certain types of cancer. The present study revealed the function and possible molecular mechanism of miR-130b in glioma cells, reporting that the level of miR-130b was markedly higher, increasing progressively as the histologic grade of the glioma increased, compared with the level in normal tissues. Additionally, the present study demonstrated that patients with high miR-130b expression exhibited a poor 3-year survival rate and miR-130b was an independent factor for predicting the prognosis of patients with glioma. The downregulation of miR-130b reduced invasion and migration in U373 and U87 cells. Furthermore, the downregulation of miR-130b increased peroxisome proliferator-activated receptor-γ (PPARγ) expression and inhibited epithelial-mesenchymal transition (EMT) in glioma cells. The present study identified PPARγ as a direct target of miR-130b in glioma in vitro. Furthermore, PPARγ knockdown was revealed to reduce the effect on EMT caused by the downregulation of miR-130b in U87 cells. The present study demonstrated that miR-130b promotes glioma proliferation, migration and invasion by suppressing PPARγ and subsequently inducing EMT.
Keywords: microRNA-130b, glioma, peroxisome proliferator- activated receptor-γ, migration, invasion
Introduction
Glioma is the most common and aggressive type of malignant primary cerebral tumor, exhibiting high invasive and proliferative abilities, which typically results in the failure of conventional treatment and a poor outcome, despite the combination of multidisciplinary therapies including maximal surgical resection, radiation and chemotherapy (1–3). The median survival length of glioma in patients is <15 months under conventional treatments (4). The World Health Organization (WHO) classification grades glioma between well differentiated low grade glioma, WHO grades I–II, and high grade anaplastic glioma and glioblastoma, WHO grades III and IV, respectively (5). A higher grade glioma is associated with poor prognosis and increased mortality (6). Therefore, revealing the detailed molecular mechanism and the diagnostic and prognostic markers of glioma is crucial for the prediction of clinical outcomes.
MicroRNAs (miR/miRNA) are endogenous small non-coding single strand RNAs consisting of ~22 nucleotides, which regulate gene expression at the post-transcriptional level by binding to 3′-untranslated (3′-UTR) regions of target mRNAs through complementary binding, causing translational suppression or mRNA degradation (7–10). Due to the involvement of miRs in the control of a number of diverse biological processes including cell growth (11), apoptosis (12) and stem cell renewal (13), numerous studies have suggested that the deregulation of miRNAs may contribute to cancer progression and development by modulating the relevant tumor suppressor and oncogenic activities (14,15). Furthermore, the high stability of miRNA in serum highlights the potential for diagnostic and prognostic biomarkers using expression profiles of specific miRNAs for patients with malignancy (16).
miR-130b, a type of miR identified in previous years, has been revealed to perform an oncogenic role in liver, gastric and renal cell carcinoma, whilst being downregulated in endometrial cancer (17,18). miR-130b has been linked to mesenchymal differentiation and hypoxic response modulation (19). Colangelo et al (20) reported that miR-130b performed a pivotal role in colon tumorigenesis, and elevated miR-130b in plasma was associated with a poor response to chemotherapy in colorectal cancer. However, miR-130b inhibited the proliferation and invasion of cells in pancreatic cancer by regulating the signal transducer and activator of transcription 3 (21). It also has been demonstrated that the downregulated expression of miR-130b by p53 mutants contributes to zinc-finger E-box binding homeobox 1-dependent epithelial-mesenchymal transition (EMT) in endometrial cancer (22). Therefore, the functional significance of miR-130b in tumorigenesis and progression is hypothetically cancer-type specific. Recently, miR-130b was revealed to regulate embryonic neural progenitor cell proliferation and differentiation by targeting the X-linked fragile X mental retardation 1 gene (23). Notably, miR-130b was identified as a robust biomarker of glioma with high positive predictive value (24). However, the molecular pathways through which miR-130b modulates the development and progression of glioma have not been revealed.
The present study aimed to investigate the expression and function of miR-130b in human glioma. Furthermore, the interaction of miR-130b and peroxisome proliferator-activated receptor-γ (PPARγ) was studied in order to reveal the underlying mechanisms. The data of the present study indicated that the expression level of miR-130b is significantly higher in glioma compared with normal brain tissue, and that the expression level closely correlates with WHO grading and poor survival outcome in patients. In addition, the results of the present study demonstrate that miR-130b increases the migratory and invasive behavior of glioma cells, and may contribute to tumor metastasis by inhibiting PPARγ and promoting EMT. Therefore, the present study proposes that miR-130b may be a novel therapeutic target for glioma.
Materials and methods
Ethical review
All protocols were approved by the Ethics Committee of The Fifth Affiliated Hospital of Zhengzhou University (Zhengzhou, China) according to the Declaration of Helsinki. Informed consent was obtained from all patients.
Tissue samples and cell lines
All tissue samples, including 47 males and 38 females (age range, 31–75 years; median, 54 years) were collected from patients who were undergoing surgical resection at the Department of Neurosurgery, The Fifth Affiliated Hospital of Zhengzhou University from January 2010 to December 2010 for the prospective study None of the patients were treated with radiation or chemotherapy prior to surgery. The specimens were histologically confirmed and 10, 19, 25 and 31 patients with glioma were classified as WHO I, II, III and IV (5), respectively. Furthermore, 10 types of normal brain tissue, including 7 males and 3 females (age range, 28–69 years; median, 48 years) were resected at the Department of Neurosurgery, The Fifth Affiliated Hospital of Zhengzhou University during the treatment of non-glioma diseases from January 2010 to December 2010 for the prospective study. All samples were snap-frozen in liquid nitrogen immediately following resection and stored at −80°C.
The human glioma U87, U373, U251, T98G and SHG44 cell lines (The Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China) were cultured in high-glucose Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) with 1% penicillin and streptomycin (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) in a humidified incubator at 37°C containing 5% CO2.
The miRNA vectors, including the miR-130b inhibitor, the negative control for the miR-130b inhibitor and PPARγ small interfering (si)RNA (5′-AAUAUGACCUGAAGCUCCAAGAAUAAG-3′) were purchased from GeneCopoeia, Inc. (Guangzhou, China). The cells were transfected with the vectors and the aforementioned siRNA using Lipofectamine® 2000 according to the protocol of the manufacturer (Invitrogen; Thermo Fisher Scientific, Inc.).
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and tissues using TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the protocol of the manufacturer, and cDNA was synthesized from 2 µg RNA with the TaqMan miRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and a TaqMan Human miRNA Assay kit (Applied Biosystems). The standard PCR conditions were as follows: 95°C for 30 sec followed by 40 cycles at 95°C for 5 sec and 60°C for 34 sec with a final dissociation stage; the samples were run with an ABI 7300 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative expression of miR-130b was determined as the fold difference relative to U6. RT-qPCR primers against mature miRNA hsa-miR-130b (HmiRQP0158) and Homo sapiens small nuclear RNA U6 qPCR Primer (HmiRQP9001) were purchased from GeneCopoeia, Inc. The relative expression level of miR-130b was calculated by the 2−ΔΔCq method and normalized to U6 small nuclear RNA (25). Six independent experimental replicates were performed.
Western blot analysis
A modified radioimmunoprecipitation assay buffer with protease inhibitor was used for protein isolation (Sigma-Aldrich; Merck Millipore). The protein concentrations were determined using the bicinchoninic acid protein assay kit (Pierce, Thermo Fisher Scientific, Inc.). Cell lysates containing 40 µg total protein were separated by 10% SDS-PAGE, transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA) and blocked with 5% non-fat milk in 0.1% Tween-20 in PBS (PBST). The membranes were then incubated with the primary antibodies against PPARγ (cat no. 2435; dilution, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), E-cadherin (cat no. 14472; dilution, 1:1,000; Cell Signaling Technology, Inc.), vimentin (cat no. 5741; dilution, 1:1,000; Cell Signaling Technology, Inc.) and GAPDH (cat no. sc-293335; dilution, 1:5,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. The membrane was then washed 3 times for 10 min with PBST, followed by incubation with horseradish peroxidase-conjugated goat anti-mouse (cat no. 1662408) or goat anti-rabbit (cat no. 1706516) secondary antibodies (dilution, 1:5,000; Bio-Rad Laboratories, Inc., Hercules, CA, USA) for 2 h at room temperature. The proteins were detected using Enhanced Chemiluminescence Reagent (EMD Millipore).
Luciferase reporter assay
The predicted 3′-UTR sequence of PPARγ, containing the putative binding sequences for miR-130b with a corresponding mutated target site sequence, was synthesized and inserted into the pRL-TK control vector (Promega Corporation, Madison, WI, USA). The U87 cells were seeded into a 24-well plate at a density of 8,000 cells/well and were transfected with 120 ng miR-130b inhibitor or the negative control. The cells were then cotransfected with 30 ng wild type or mutant 3′-UTR PPARγ mRNA using Lipofectamine® 2000 according to the protocol of the manufacturer. The cell lysates were collected 48 h subsequent to transfection. The firefly and Renilla luciferase activities were measured according to the protocol of the manufacturer (Dual-Luciferase Assay system; Promega Corporation). pRL-TK expressing Renilla luciferase was cotransfected as an internal control.
Cell proliferation assay
Cellular proliferation was measured using the Cell Counting Kit-8 assay (CCK-8; Beckman Coulter, Brea, CA, USA) according to the protocol of the manufacturer. The cells were seeded in a 96-well plate at a density of 5×103 cells/well, and were treated with or without miR-130b transfection. At 24, 48 and 72 h, 10 µl CCK-8 solution was added to each well and incubated for 3 h at 37°C. The absorbance rate at 450 nm was recorded using a microplate reader (Bio-Rad Laboratories, Inc.). All experiments were repeated ≥3 times.
Cell migration and invasion
Cellular migration and invasion abilities were tested using 8 µm pore-size Transwell chambers (Corning Incorporated, NY, USA). The harvested cells (1×105 cells/chamber in 100 µl DMEM) were resuspended in serum-free medium and seeded in the upper chamber, 750 µl DMEM supplemented with 10% FBS was added to the lower chambers, or, for the invasion assay, the upper chambers were precoated with Matrigel solution (BD Biosciences, Franklin Lakes, NJ, USA) and incubated at 37°C for 4 h prior to the start of the invasion assay. Subsequently, the Transwell plates for the migration and invasion assays were each incubated at 37°C for 24 h. The cells on the inner layer were removed with a cotton swab and the adherent cells on the lower surface were fixed with methanol and stained with 0.1% crystal violet for 5 min. A total of five randomly selected fields were counted randomly in each well under a light microscope (Axioskop 2 plus; Carl Zeiss Co., Ltd., Jena, Germany). Six independent experimental replicates were performed.
Statistical analysis
All data are expressed as the mean ± standard error of the mean. The SPSS statistical package for Windows version 13 (SPSS, Inc., Chicago, IL, USA) was used for the Pearson χ2 tests and the multivariate Cox regression analysis. A two-tailed Student's t-test, Kaplan-Meier plot, log-rank test, Pearson's correlation coefficient analysis or analysis of variance were performed in order to evaluate the statistical significance using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Elevated miR-130b expression positively correlates with WHO grade of human glioma
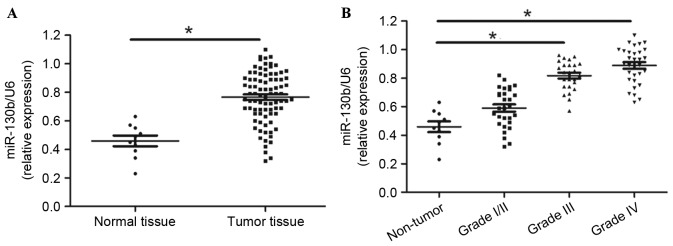
The present study evaluated miR-130b using RT-qPCR, normalized against an endogenous control (U6 RNA) in normal brain (n=10) and glioma tissues (n=85). The data of the present study suggested that miR-130b expression was significantly elevated in the glioma tissue compared with the normal tissue (Fig. 1A; P<0.01). Furthermore, the increase in miR-130b expression was associated with the increasing glioma pathological grade of the gliomas (Fig. 1B; P<0.05). Thus, these results demonstrate that the high grade glioma overexpressed miR-130b.
Figure 1.
Expression levels of miR-130b in glioma tissues. (A) Glioma and normal brain tissues. (B) miR-130b levels statistically correlate with the World Health Organization grades of glioma. Values are depicted as the mean ± standard error of the mean. *P<0.05, by t-test. miR-130b, microRNA-130b.
Association of miR-130b expression with clinicopathological characteristic of glioma patients
In total, 85 samples of glioma tissues underwent RT-qPCR for miR-130b expression. The present study determined the mean level of miR-130b as a cutoff value for the high or low expression level of miR-130b. As presented in Table I, the high expression of miR-130b was markedly correlated with the histological grade of the glioma (P<0.01). However, no correlations were identified between the patient gender, age or the clinical staging of the glioma (P>0.05). Thus, these results indicate that the expression of miR-130b is correlated with malignant clinicopathological characteristics in glioma.
Table I.
Association of miR-130b expression with the clinicopathological characteristics of patients with glioma.
| miR-130b expression level | ||||
|---|---|---|---|---|
| Clinical parameter | Cases, n | High (n=40) | Low (n=45) | P-value |
| Age, years | ||||
| <60 | 53 | 26 | 27 | 0.635 |
| ≥60 | 32 | 14 | 18 | |
| Gender | ||||
| Male | 47 | 22 | 25 | 0.959 |
| Female | 38 | 18 | 20 | |
| Grade | ||||
| I+II | 29 | 7 | 22 | 0.002a |
| III+IV | 56 | 33 | 23 | |
| Clinical staging | ||||
| I–II | 25 | 10 | 15 | 0.400 |
| III–IV | 60 | 30 | 30 | |
P<0.05. miR, microRNA.
miR-130b expression correlates with poor survival in glioma
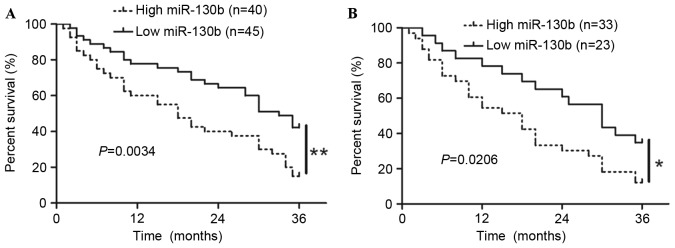
To determine the role of miR-130b in predicting the prognosis of patients with glioma, the present study constructed Kaplan-Meier survival curves using the overall 3-year survival data to analyze patients with high and low miR-130b. As presented in Fig. 2A, the overall survival length of patients with high miR-130b expression was notably shorter than those with low miR-130b expression (P<0.05). In addition, with respect to the patients with grades III or IV, the patients with high miR-130b expression exhibited a significantly reduced 3-year survival rate compared with the patients with low miR-130b (Fig. 2B; P<0.05). These data demonstrated that miR-130b expression was associated with poor survival of glioma patients.
Figure 2.
Kaplan-Meier analysis of three-year overall survival in patients with glioma according to the expression levels of miR-130b in glioma tissues. (A) Cells with high miR-130b expression exhibited a significantly worse three-year survival compared with cells with low miR-130b expression. (B) miR-130b expression exerts a marked effect on the prognosis of patients in grade III or IV glioma. *P<0.05, by log-rank test. miR-130b, microRNA-130b.
Furthermore, univariate analysis indicated no association between the patient survival rate and the patient age and gender (P>0.05). However, univariate and multivariate analysis indicated that miR-130b expression level and grade degree were independent prognostic factors for glioma (Table II).
Table II.
Univariate and multivariate analysis of prognostic factors in patients with glioma.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age | 0.565 | 0.342–0.714 | 0.334 | 1.180 | 0.815–1.709 | 0.381 |
| Gender | 1.538 | 1.227–1.869 | 0.527 | 1.024 | 0.480–2.184 | 0.951 |
| Grade | 2.318 | 1.402–3.832 | 0.001a | 2.018 | 1.287–3.165 | 0.002a |
| miR-130b | 2.034 | 1.378–2.652 | 0.001a | 2.218 | 1.576–2.817 | 0.001a |
P<0.05. HR, hazard ratio; CI, confidence interval; miR, microRNA.
Promoting effect of miR-130b on glioma cell proliferation
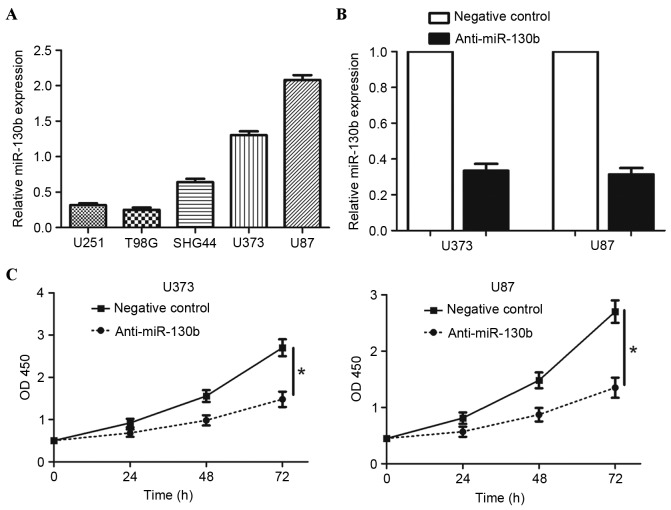
To investigate the role of miR-130b in glioma, the present study measured the levels of miR-130b in five glioma cell lines and observed that miR-130b expression levels were high in U373 and U87 cells, compared with in the U251, T98 G and SHG44 cell lines (Fig. 3A; P<0.05). Therefore, U373 and U87 cells were used for the subsequent analysis, and the expression level of miR-130b in the two cells was suppressed. As assessed by RT-qPCR, the expression of miR-130b was downregulated by ectopically expressing miR-130b inhibitors in the two cell lines (Fig. 3B; P<0.05). The present study examined cell proliferation using a CCK-8 assay, revealing that the downregulation of miR-130b led to a significant reduction of cell proliferation in the U373 and U87 cells (Fig. 3C; P<0.05). These results indicate that miR-130b promotes proliferation in glioma cells.
Figure 3.
Downregulation of miR-130b expression inhibited cell proliferation in glioma cells. (A) Expression of miR-130b in glioma cell lines. (B) U373 and U87 cells that were transfected with miR-130b inhibitors (anti-miR-130b) and negative control underwent RT-qPCR for miR-130b. *P<0.05, by t-test. (C) Cell proliferation, as measured by Cell Counting Kit-8 assays, was inhibited by the downregulation of miR-130b in U373 and U87 cells, compared with in the control cells. *P<0.05, by t-test. Values are depicted as the mean ± standard error of the mean. miR-130b, microRNA-130b; OD, optical density; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Downregulation of miR-130b inhibits the migration and invasion of glioma cells
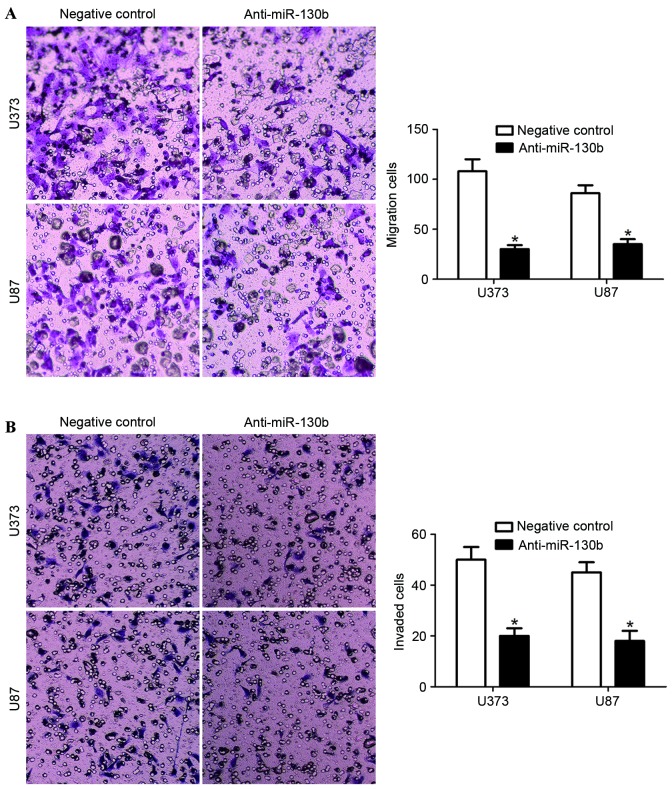
Transwell assays were carried out to determine the effect of miR-130b knockdown on cell migration and invasion, with the upper chamber coated with Matrigel to mimic the extracellular matrix. The present study identified that the downregulation of miR-130b significantly reduced the cell migration and invasion ability in U373 and U87 cells (Fig. 4A and B; P<0.05). These data suggest that miR-130b promotes glioma cell migration and invasion.
Figure 4.
miR-130b regulates the migration and invasion ability of glioma cells. (A) Cellular migration, as measured by Transwell assays, was inhibited by the downregulation of miR-130b in U373 and U87 cells, compared with in the control cells. *P<0.05, by t-test. (B) miR-130b downregulated U373 and U87 cells yielded a lower number of invaded cells, compared with the control cells. *P<0.05, by t-test. Values are depicted as the mean ± standard error of the mean. miR-130b, microRNA-130b.
miR-130b promotes EMT by inhibiting PPARγ in glioma
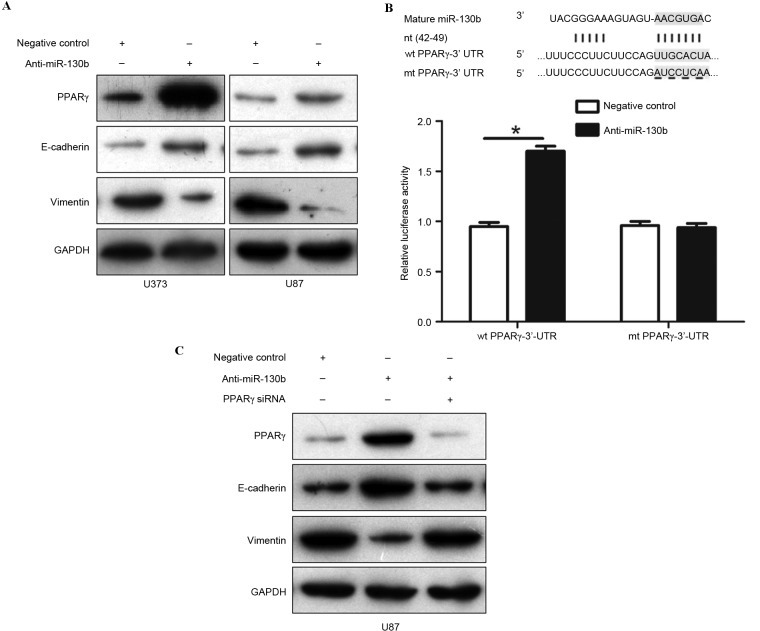
A previous study reported that miR-130b promotes EMT progression by targeting PPARγ in colorectal cancer (17). The present study performed western blotting to detect the presence of PPARγ, E-cadherin and vimentin in U373 and U87 cells that were transfected with miR-130b inhibitors or the negative control. The downregulation of miR-130b was revealed to markedly restore the expression of PPARγ and increase the level of E-cadherin, but reduced vimentin expression in the two cells (Fig. 5A; P<0.05). To confirm that PPARγ is directly targeted by miR-130b in glioma cells, the present study performed a dual-luciferase reporter assay to explore whether miR-130b directly interacted with the 3′-UTR of PPARγ mRNA. The results of the present study showed that endogenous miR-130b inhibition increased the luciferase activity of the wild-type reporter, but not of the mutant reporter (Fig. 5B; P<0.05). In addition, the knockdown of PPARγ expression by a specific siRNA negated the results induced by the downregulation of miR-130b in U87 cells (Fig. 5C; P<0.05). These results demonstrate that miR-130b promotes EMT progression in glioma by directly suppressing PPARγ expression.
Figure 5.
miR-130b promotes epithelial-mesenchymal transition by suppressing PPARγ expression in glioma cells. (A) Representative western blotting reveals that the downregulation of miR-130b markedly increased the protein expression of PPARγ and E-cadherin, and reduced vimentin expression in U373 and U87 cells. (B) miR-130b and the putative binding of the miRNA sequence in the 3′-UTR of PPARγ. Anti-miR-130b led to an increase in the luciferase activity of wt 3′-UTR of PPARγ in U87 cells. Data were normalized by the ratio of firefly and Renilla luciferase activities measured 48 h post-transfection. *P<0.05. (C) Representative western blotting demonstrates that PPARγ knockdown eradicated the expression changes of PPARγ, E-cadherin and vimentin induced by miR-130b suppression in U87 cells. Data are representative of multiple repeats with similar results. miR/miRNA, microRNA; PPARγ, peroxisome proliferator-activated receptor-γ; 3′-UTR, 3′-untranslated region; wt, wild-type; mt, mutant type; nt, nucleotide target; siRNA, small interfering RNA.
Discussion
Glioma is the most common and aggressive type of primary brain tumor worldwide (2). Despite developments in the diagnosis and treatment of glioma, the incidence and mortality rates remain at a high level (4). Therefore, a complete understanding of the molecular mechanisms underlying glioma tumor initiation and progression is required to develop novel prognostic and therapeutic strategies aimed at improving the outcomes of patients with cancer. Over previous years, miRNAs have emerged as a novel class of gene regulator involved in various malignancies. Aberrant expression of a growing number of miRNAs has also been linked to the development of glioma (8,14).
The present study provides initial support for the hypothesis that miR-130b performs a pivotal role in glioma tumorigenesis. The present study detected the expression of miR-130b in patients, and the data revealed that the expression of miR-130b was significantly higher in glioma tissues compared with in the normal tissues. Furthermore, miR-130b expression was significantly correlated with the WHO grade degree. The data of the present study indicates that a high expression of miR-130b is significantly correlated with a poor 3-year survival for patients. Multivariate Cox repression analysis demonstrated that miR-130b is an independent factor in predicting 3-year overall survival. These results indicate that a high expression of miR-130b is critical for prognosis determination in patients with glioma. In addition, the present study demonstrated that miR-130b was elevated in glioma cell lines, and the knockdown of miR-130b suppressed certain malignant behaviors including proliferation, migration and invasion of glioma cells. These results suggest that miR-130b performs a critical role in the progression of glioma.
It has emerged that the EMT is essential for migration and invasion, and the progress of EMT affects physiological conditions during tumorigenesis (26). In particular, EMT performs a critical role in tumor dissemination (26,27). Therefore, the present study determined the expression of E-cadherin, an epithelial marker, and vimentin, a mesenchymal marker, using loss of function experiments. Notably, the present study found that the downregulation of miR-130b increased the E-cadherin expression level and reduced the vimentin expression level. These data indicate that miR-130b may be critical for the regulation of tumor invasion in glioma, which is consistent with the observation that the downregulation of miR-130b significantly reduced the levels of migration and invasion in U373 and U87 cells.
PPARγ is a transcription factor controlling cellular proliferation and differentiation (28), and has been identified as a possible novel target for glioma treatment (29,30). Previously, it has been reported that PPARγ is a direct target of miR-130b in colorectal cancer and contributes to EMT (20). The present study demonstrated that miR-130b binds the 3′-UTR of PPARγ mRNA in order to downregulate PPARγ protein expression, as the knockdown of miR-130b increased the PPARγ expression level and suppressed EMT in glioma U373 and U87 cells. In addition, the regulatory effect of miR-130b on PPARγ, E-cadherin and vimentin was reversed by PPARγ siRNA in U87 cells, which confirmed that miR-130b may promote EMT by inhibiting PPARγ expression in glioma.
In conclusion, the present study revealed that the expression of miR-130b is elevated in glioma, particularly in high grade tumors. High miR-130b expression is significantly correlated with poor prognosis and is an independent factor in predicting 3-year overall survival. The present study demonstrated that the downregulation of miR-130b inhibited cell proliferation, migration and invasion by restoring PPARγ expression and suppressing EMT in glioma cells. Therefore, miR-130b may be a therapeutic target in glioma.
Acknowledgements
The present study was supported by Scientific Research Foundation of Henan (grant no. 092102310090).
References
- 1.Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379:1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 2.Woolf EC, Scheck AC. Metabolism and glioma therapy. CNS Oncol. 2012;1:7–10. doi: 10.2217/cns.12.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mack F, Schäfer N, Kebir S, Stuplich M, Schaub C, Niessen M, Scheffler B, Herrlinger U, Glas M. Carmustine (BCNU) plus Teniposide (VM26) in recurrent malignant glioma. Oncology. 2014;86:369–372. doi: 10.1159/000360295. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng YS, Lin C, Cheng YP, Yu YL, Tang CT, Hueng DY. Epithelial cell transformation sequence 2 is a potential biomarker of unfavorable survival in human gliomas. Neurol India. 2014;62:406–409. doi: 10.4103/0028-3886.141278. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long JM, Lahiri DK. Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders. Exp Neurol. 2012;235:402–418. doi: 10.1016/j.expneurol.2011.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu B, Hsu PK, Karayiorgou M, Gogos JA. MicroRNA dysregulation in neuropsychiatric disorders and cognitive dysfunction. Neurobiol Dis. 2012;46:291–301. doi: 10.1016/j.nbd.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 13.Utikal J, Abba M, Novak D, Moniuszko M, Allgayer H. Function and significance of MicroRNAs in benign and malignant human stem cells. Semin Cancer Biol. 2015;35:200–211. doi: 10.1016/j.semcancer.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Patil PA, Magi-Galluzzi C. MicroRNA in prostate cancer: Practical aspects. Histol Histopathol. 2015;30:1379–1396. doi: 10.14670/HH-11-647. [DOI] [PubMed] [Google Scholar]
- 16.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu K, Zheng X, Dou C, Li C, Yang W, Yao Y, Liu Q. MicroRNA-130b promotes cell aggressiveness by inhibiting peroxisome proliferator-activated receptor gamma in human hepatocellular carcinoma. Int J Mol Sci. 2014;15:20486–20499. doi: 10.3390/ijms151120486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun B, Li L, Ma W, Wang S, Huang C. MiR-130b inhibits proliferation and induces apoptosis of gastric cancer cells via CYLD. Tumour Biol. 2016;37:7981–7987. doi: 10.1007/s13277-015-4632-3. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Huang Z, Lin L, Fu M, Song Y, Shen Y, Ren D, Gao Y, Su Y, Zou Y, et al. miRNA-130b is required for the ERK/FOXM1 pathway activation-mediated protective effects of isosorbide dinitrate against mesenchymal stem cell senescence induced by high glucose. Int J Mol Med. 2015;35:59–71. doi: 10.3892/ijmm.2014.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colangelo T, Fucci A, Votino C, Sabatino L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Maggi CA, Parente D, et al. MicroRNA-130b promotes tumor development and is associated with poor prognosis in colorectal cancer. Neoplasia. 2013;15:1218–1231. doi: 10.1593/neo.13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao G, Zhang JG, Shi Y, Qin Q, Liu Y, Wang B, Tian K, Deng SC, Li X, Zhu S, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS One. 2013;8:e73803. doi: 10.1371/journal.pone.0073803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H, Sudo S, Ju J, Sakuragi N. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2013;32:3286–3295. doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong X, Zhang K, Wang Y, Wang J, Cui Y, Li S, Luo Y. MicroRNA-130b targets Fmr1 and regulates embryonic neural progenitor cell proliferation and differentiation. Biochem Biophys Res Commun. 2013;439:493–500. doi: 10.1016/j.bbrc.2013.08.096. [DOI] [PubMed] [Google Scholar]
- 24.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 27.Nakaya Y, Sheng G. EMT in developmental morphogenesis. Cancer Lett. 2013;341:9–15. doi: 10.1016/j.canlet.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 28.Koeffler HP. Peroxisome proliferator-activated receptor gamma and cancers. Clin Cancer Res. 2003;9:1–9. [PubMed] [Google Scholar]
- 29.Grommes C, Landreth GE, Heneka MT. Antineoplastic effects of peroxisome proliferator-activated receptor gamma agonists. Lancet Oncol. 2004;5:419–429. doi: 10.1016/S1470-2045(04)01509-8. [DOI] [PubMed] [Google Scholar]
- 30.Grommes C, Conway DS, Alshekhlee A, Barnholtz-Sloan JS. Inverse association of PPARγ agonists use and high grade glioma development. J Neurooncol. 2010;100:233–239. doi: 10.1007/s11060-010-0185-x. [DOI] [PubMed] [Google Scholar]