Abstract
The detection of circulating tumor cells (CTCs) is limited by the rarity of these cells in the peripheral blood of patients with cancer. Understanding tumor biology may be useful in the development of novel therapeutic strategies for patients with lung cancer. The present study evaluated a novel size-based filtration platform for enriching CTCs from patients with lung cancer. Blood samples were obtained from 82 patients with lung cancer for CTC analysis. CTC enrichment by size-based filtration was performed on 5-ml blood samples. The collected cells were detected by immunofluorescence using monoclonal anti-human antibodies against protein tyrosine phosphatase, receptor type C (CD45) and epithelial cell adhesion molecule (EpCAM; an epithelial cell marker), as well as a DAPI nucleic acid stain. CTCs were detected in 57 patients (69.5%) using the size-based filtration platform. The mean CTC counts, defined as the number of cells with DAPI-positive, CD45-negative and EpCAM-positive staining, were 1.48±1.71 per 5 ml blood for the 66 stage I–III patients and 8.00±9.95 per 5 ml blood for the 16 stage IV patients. The presence of ≥1 CTCs per 5-ml blood sample was significantly associated with pathological stage (stage IV vs. stage I–III, P=0.009), but not with patient age or gender, tumor histology, tumor size or lymphovascular invasion. The mean CTC count of healthy donors was 0.25±0.55 per 5 ml blood. In summary, CTCs from the blood of patients with lung cancer were enriched using a size-based filtration platform and immunofluorescent staining with DAPI, CD45 and EpCAM. The CTC counts of patients with stage IV cancer were higher than those of patients with stages I–III cancer. These results suggest that this novel platform may be a useful tool for determining the prognosis of patients with lung cancer.
Keywords: circulating tumor cells, metastasis, non-small cell lung cancer
Introduction
Lung cancer is the most common cause of cancer mortality worldwide (1). Platinum-based therapy is a first-line treatment for patients with inoperable lung cancer. The identifications the epidermal growth factor receptor (EGFR) gene mutations and anaplastic lymphoma kinase (ALK) gene rearrangement have improved the clinical outcome of patients with lung cancer by treatment with EGFR and ALK inhibitors (2). However, overall survival remains poor (3). Thus, studies on the mechanisms of carcinogenesis, clinical characteristics and targeted therapeutics are required.
The detection and characterization of circulating tumor cells (CTCs) in the blood of patients with cancer may be useful for understanding the biology of tumor metastasis and establishing novel therapeutic strategies. Among the technologies developed to isolate and analyze CTCs, only the CellSearch platform from Janssen Diagnostics, LLC (Raritan, NJ, USA) has been approved by the US Food and Drug Administration for prognostic use in patients with metastatic breast, colorectal or prostate cancer. This platform uses antibodies bound to magnetic beads to positively select for epithelial cell adhesion molecules (EpCAM) on CTCs. Although a number of studies have reported on CTC quantification in patients with lung cancer using the CellSearch system, CTC detection using this system is poor in patients with advanced non-small cell lung cancer (NSCLC) (4,5).
In addition to the CellSearch platform, other platforms have been developed, such as the Isolation by Size of Epithelial Tumor Cells (ISET) system, which is dependent on tumor cell size (6,7) or inertial focusing using a microfluidics platform (8,9). Numerous studies have compared the performances of the ISET and CellSearch systems (5,10,11). The aforementioned studies were able to isolate a greater number CTCs by ISET than by the CellSearch method. A size-based platform may be more efficient for capturing all CTCs, including CTCs undergoing epithelial-mesenchymal transition (EMT), which typically lose expression of epithelial markers and thus cannot be detected by CellSearch (12) However, size-based platforms may not detect small CTCs. The present study evaluated a novel CTC detection platform based on cell size (13). CTCs in the blood of patients with lung cancer were detected using the size-based platform and immunofluorescent staining with DAPI, and anti-protein tyrosine phosphatase, receptor type C (CD45) and anti-EpCAM antibodies.
Materials and methods
Patients
The present study was approved by the Institutional Review Board of the Samsung Medical Center (Seoul, Korea; no. 2011-12-024). A total of 82 patients with NSCLC undergoing surgery from December 2012 to September 2013 at the Samsung Medical Center (Seoul, Korea), from whom 5 ml blood per patient was drawn, were enrolled in the study. All patients provided informed consent to participate in the present study prior to surgery. The patients did not receive anticancer treatment prior to blood sample collection. Only patients with histologically confirmed lung cancer were included. Demographic and clinical data, including medical history, physical examination and radiographic findings were collected. The current 7th American Joint Committee on Cancer/Union for International Cancer Control Tumor-Node-Metastasis (TNM) Cancer Staging classification was used (14). All patients underwent chest computed tomography, positron emission tomography and brain magnetic resonance imaging. Distal metastases were verified in symptomatic patients by appropriate imaging studies and pathological results from biopsies. Blood samples were also collected from healthy volunteers as normal controls.
CTC collection
Whole blood samples were collected by venipuncture into BD Vacutainer tubes containing anticoagulant citrate dextrose solution A (catalog no. 364606; BD Biosciences, Franklin Lakes, NJ, USA), layered on 1.077 g/ml density Ficoll-Hypaque gradients (GE Healthcare Life Sciences, Chalfont, UK) and centrifuged for 20 min at 400 × g at room temperature (RT). The peripheral blood mononuclear cells and Ficoll layers were collected and transferred to new tubes that were then filled with PBS (pH 7.4) and centrifuged for 10 min at 400 × g at RT. The cells were resuspended in 10 ml PBS and passed through a microfabricated porous filter (Cytogen, Inc., Seoul, Korea) to obtain non-leukocyte, nucleated cells (13). The cells were recovered from the filter and harvested onto microscope slides using Shandon Cytospin 4 (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Identification of CTCs
The cells on the microscope slides were fixed with 2% paraformaldehyde in PBS for 5 min and permeabilized with 0.2% Triton X-100 (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for 10 min. The fixed cells were blocked with bovine serum albumin in PBS for 30 min at RT and incubated with an anti-human EpCAM monoclonal antibody (mAb) (clone VU1D9; catalog no. 2929; dilution, 1:200; Cell Signaling Technology, Inc., Danvers, MA, USA) for 1 h at RT. To enhance the EpCAM signal, a Tyramide Signal Amplification Kit (catalog no. MP-20911; Invitrogen; Thermo Fisher Scientific, Inc., Rockford, IL, USA) was used. The cells were washed three times with PBS and incubated with an anti-human CD45 mAb (clone H230; catalog no. sc-25590; dilution, 1:10; Santa Cruz Biotechnology, Inc., Heidelberg, Germany) for 90 min at 37°C. Following three washes in PBS, the cells were incubated with Alexa Fluor 594-conjugated goat anti-rabbit secondary antibody (cat. no. A-11012; dilution, 1:300; Invitrogen; Thermo Fisher Scientific, Inc., Rockford, IL, USA) for 90 min at 37°C and counterstained with DAPI (MaxVision Biosciences, Inc., Bothell, WA, USA) to visualize the nuclei.
Statistical analysis
The descriptive statistics for the clinical, imaging and pathological variables were determined using median and interquartile range or number with percent, as appropriate. The counts were compared using the χ2 test. Continuous data were compared using the Mann-Whitney U test. P<0.05 was considered to indicate a statistically significant difference. All statistical manipulations were performed using JMP 9.2 for Windows (SAS Institute, Inc., Cary, NC, USA).
Results
Patient characteristics
The characteristics of the enrolled patients are summarized in Table I. Age, gender, histology, tumor location, pathological stage, lymphovascular invasion and tumor size were documented. The study included 66 patients with stage I–III NSCLC and 16 patients with stage IV NSCLC, with 65 patients (79.3%) receiving major pulmonary resection and 17 (20.7%) receiving chemotherapy without surgery.
Table I.
Characteristics of the NSCLC patients (n=82).
| Characteristic | Value |
|---|---|
| Age, yearsa | 64.5 (56.0–70.3) |
| Gender, n (%) | |
| Male | 51 (62.2) |
| Female | 31 (37.8) |
| Histology, n (%) | |
| Adenocarcinoma | 60 (73.2) |
| Squamous cell carcinoma | 20 (24.4) |
| Other NSCLC | 2 (2.4) |
| Location of the tumor, n (%) | |
| RUL | 20 (24.4) |
| RML | 5 (6.1) |
| RLL | 24 (29.3) |
| LUL | 25 (30.5) |
| LLL | 8 (9.8) |
| Pathological stage, n (%) | |
| I | 37 (45.1) |
| II | 16 (19.5) |
| III | 13 (15.9) |
| IV | 16 (19.5) |
| Lymphovascular invasion, n (%) | |
| Present | 16 (19.5) |
| Absent | 66 (80.5) |
| Tumor size, cma | 3.7 (2.0–5.5) |
| Treatment, n (%) | |
| Surgery | 65 (79.3) |
| Wedge resection | 6 (7.3) |
| Segmentectomy | 4 (4.9) |
| Lobectomy | 52 (63.4) |
| Pneumonectomy | 3 (3.7) |
| Chemotherapy | 17 (20.7) |
Continuous variables are presented as the median (interquartile range). NSCLC, non-small cell lung cancer; RUL, right upper lobe; RML, right middle lobe; RLL, right lower lobe; LUL, left upper lobe; LLL, left lower lobe.
CTC counts for individual patients were unchanged between two time points
To test whether CTC counts, determined by the size-based filtration platform, were consistent over a short duration, tests were performed at two time points. CTCs were counted in blood samples from 3 patients with advanced lung cancer at days 1 or 3 subsequent to the first blood draw. The CTC counts at the two time points were similar (Fig. 1). Thus, CTC enrichment with the size-based filtration platform produced consistent results.
Figure 1.
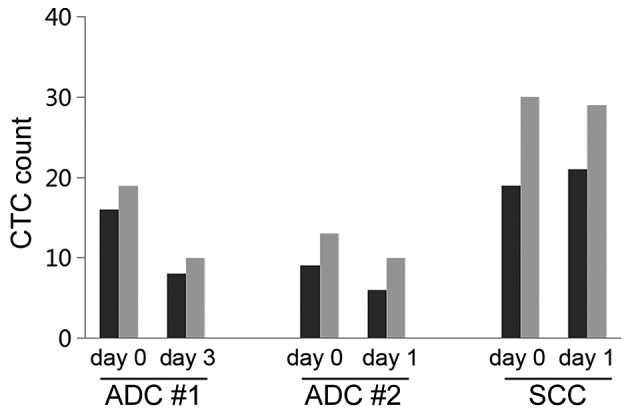
CTC counts for individual patients did not change when measured at two time points. Blood from 3 patients with advanced cancer was assayed and analyzed. ADC samples were tested twice, on the day of first blood draw and either on day 1 or day 3 subsequent to the first blood draw. SCC samples were tested twice, once on the day of first blood draw and again on day 1 subsequent to first blood draw. All blood samples were divided into two subsamples and processed individually. The light grey and dark grey bars are presented as the CTC number from 2 independent 5-ml blood samples. CTCs were counted by immunofluorescence following staining with DAPI, an anti-human protein tyrosine phosphatase, receptor type C mAb and an anti-human epithelial cell adhesion molecule mAb. CTC, circulating tumor cell; ADC, adenocarcinoma; SCC, squamous cell carcinoma; mAb, monoclonal antibody.
CTC count correlates with pathological stage
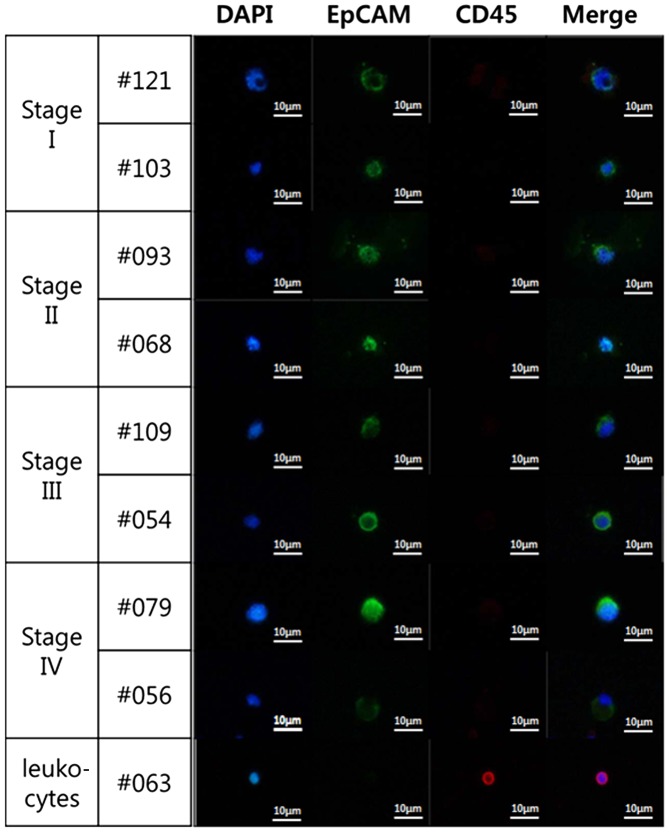
CTCs in blood from the patients with lung cancer (n=82) were counted using a triple-color staining protocol, including DAPI staining for nucleated cells, CD45 mAb staining as a leukocyte marker, and EpCAM mAb staining as an epithelial cell marker. CTCs of epithelial origin are negative for CD45 and positive for EpCAM, while blood cells are positive for CD45. All DAPI-positive, CD45-negative and EpCAM-positive cells were counted as CTCs (Fig. 2). The mean CTC count in the 16 patients with stage IV cancer (8.00±9.95 per 5 ml) was significantly higher compared with that in patients with stage I (1.22±1.51 per 5 ml; n=37; P<0.001), stage II (1.81±1.94 per 5 ml; n=16; P=0.022) or stage III cancer (1.85±1.95 per 5 ml; n=13; P=0.035) (Fig. 3). The mean CTC count of the healthy donors was 0.25±0.55 per 5 ml blood.
Figure 2.
Representative immunofluorescent images at different cancer stages. Images show different cell sizes and shapes detected in two representative samples from each cancer stage. Collected cells were stained with DAPI (blue staining) and antibodies against CD45 (red staining) and EpCAM (green staining). Leukocytes (bottom row) were stained as a control for DAPI- and CD45-positive cells. Magnification, ×400. Merge, merged images; CD45, protein tyrosine phosphatase, receptor type C; EpCAM, epithelial cell adhesion molecule.
Figure 3.
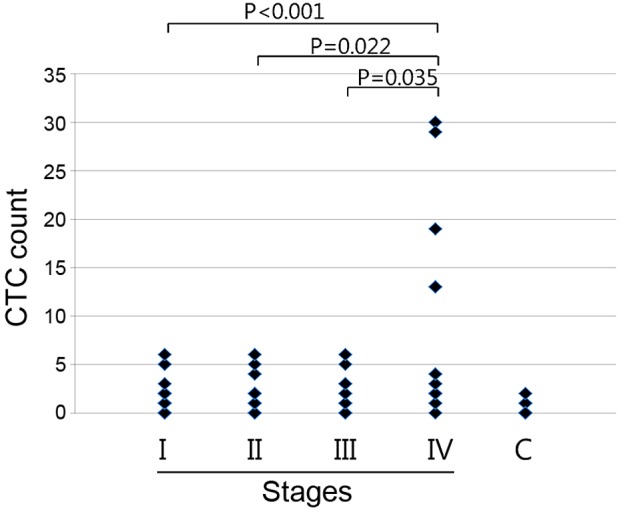
CTC count correlated with pathological stage. Pathological stage was determined by a pathologist at the Samsung Medical Center (Seoul, Korea) from tissue biopsies. Subsequent to the blood draw, the cells were processed directly using the proposed platform, and the collected cells were immunofluorescently stained with an anti-human protein tyrosine phosphatase, receptor type C, CD45 mAb and an anti-human epithelial cell adhesion molecule, EpCAM mAb, as well as DAPI. All DAPI-positive, CD45-negative and EpCAM-positive cells were counted under a fluorescence microscope. Data are presented as the CTC numbers from 5 ml blood samples. Stage I, n=37; stage II, n=16; stage III, n=13; stage IV, n=16; C, n=20. CTC, circulating tumor cell; mAb, monoclonal antibody; C, healthy donors.
CTCs were detected in 57 patients (69.5%; Table II). When patients were grouped according to disease stage, the percentages of samples with ≥1 CTC per 5 ml blood were 59.5% for stage I, 68.7% for stage II, 69.2% for stage III and 93.7% for stage IV. The proportion of CTC-positive patients grouped according to gender, age, histology, pathological stage, tumor size and lymphovascular invasion are presented in Table III. Among patients positive for ≥1 CTC per sample, no significant associations with patient characteristics were identified, with the exception of tumor stage (P=0.009). Additionally, the mean CTC count in patients with stage IV cancer was significantly higher than in patients with stage I–III cancer (8.00±9.95 vs. 1.48±1.71 per 5 ml blood; P=0.009).
Table II.
Number of CTCs per 5-ml blood sample stratified by cancer stage.
| Number of CTCs | Stages I–III (n=66) | Stage I (n=37) | Stage II (n=16) | Stage III (n=13) | Stage IV (n=16) | Control (n=20) |
|---|---|---|---|---|---|---|
| 0 | 24 (36.4) | 15 (40.5) | 5 (31.3) | 4 (30.8) | 1 (6.3) | 16 (80.0) |
| 1 | 17 (25.8) | 10 (27.0) | 4 (25.0) | 3 (23.1) | 2 (12.5) | 3 (15.0) |
| 2 | 13 (19.7) | 8 (21.3) | 3 (18.8) | 2 (15.4) | 4 (25.0) | 1 (5.0) |
| 3 | 3 (4.6) | 1 (2.7) | 0 (0) | 2 (15.4) | 2 (12.5) | 0 (0) |
| 4 | 2 (3.0) | 0 (0) | 2 (12.5) | 0 (0) | 2 (12.5) | 0 (0) |
| 5 | 4 (6.1) | 2 (5.4) | 1 (6.3) | 1 (7.7) | 0 (0) | 0 (0) |
| >5 | 3 (4.6) | 1 (2.7) | 1 (6.3) | 1 (7.7) | 5 (31.3) | 0 (0) |
Data are presented as the number of patients (%). CTCs, circulating tumor cells; Control, healthy donors.
Table III.
Number of circulating tumor cells according to patient characteristics.
| Circulating tumor cell count ≥1 | |||
|---|---|---|---|
| Characteristic | Total number of patients | n (%) | P-value |
| Gender | 0.470 | ||
| Male | 51 | 34 (66.7) | |
| Female | 31 | 23 (74.2) | |
| Age, years | 0.117 | ||
| <70 | 59 | 44 (74.6) | |
| ≥70 | 23 | 13 (56.5) | |
| Histology | 0.069 | ||
| Adenocarcinoma | 60 | 44 (73.3) | |
| Squamous cell carcinoma | 20 | 13 (65.0) | |
| Other NSCLC | 2 | 0 (0.0) | |
| Pathological stage | 0.009a | ||
| I–III | 66 | 42 (63.6) | |
| IV | 16 | 15 (93.8) | |
| Tumor size, cm | 0.252 | ||
| ≤3 | 35 | 21 (60.0) | |
| >3 | 40 | 29 (72.5) | |
| Unknown | 7 | – | |
| Lymphovascular invasion | 0.158 | ||
| Positive | 16 | 13 (81.3) | |
| Negative | 57 | 36 (63.2) | |
| Unknown | 9 | – | |
Statistically significant (P<0.05); NSCLC, non-small cell lung cancer.
Discussion
The clinical significance of CTCs is well known and has been described previously (4,15). Monitoring the CTC count of patients during treatment may aid in establishing guidelines for surrogate endpoints of anticancer treatments. For example, CTCs are recommended by the American Society of Clinical Oncology as an acceptable breast cancer marker (16). The significance of CTCs has not been well established for lung cancer; however, several studies have reported that the detection of CTCs may be a prognostic factor for lung cancer subsequent to surgical and chemotherapeutic treatment (17–19). Since NSCLC lacks validated biomarkers, a radiological response is recognized as a surrogate for survival or recurrence (20). CTC count may be an additional prognostic factor for survival, recurrence and response to treatment.
We previously evaluated a platform for the recovery of various cancer cell lines transfused into whole blood (13), and subsequently performed a clinical validation study of 82 patients with NSCLC. To avoid chemotherapeutic interference with CTC quantification, blood was drawn from patients prior to surgery or chemotherapy. The results revealed that the mean CTC count for patients with stage IV cancer was significantly higher than that of patients with cancer of lower stages. Chen et al (19) reported that CTC count in lung cancer correlates with pathological TNM stage, consistent with the results of the present study.
Of 82 patients undergoing treatment for NSCLC, 53 were diagnosed with stage I or II cancer, among which 33 patients (62.3%) were identified to have ≥1 CTC per 5-ml sample. The CTC status of the patients with early-stage lung cancer will continue to be monitored. Patients with CTC positivity may be candidates for postoperative chemotherapy if additional investigation reveals that ≥1 CTC correlates with recurrence or poor survival. If so, CTC count may be a clinicopathological parameter for liquid biopsy.
To identify changes in CTC count in individual patients, blood was drawn at different time points and subjected to CTC enrichment using the size-based filtration platform. The results showed that CTC count did not change with time, suggesting that the platform produces consistent CTC counts.
A number of reports on CTC capture using ISET technology have been published since the initial report in 2000 by Vona et al (7). ISET may have an advantage over the CellSearch method for enriching rare CTCs in the blood, as the ISET enrichment principle is based on size rather than specific tumor antigens. Although the specificity and sensitivity of ISET are low, the retrieved cells are viable and negatively selected; therefore, ISET may be combined with other approaches for CTC detection and identification.
The present study had four main limitations that require attention. Firstly, CTCs that did not express EpCAM may have been excluded. EpCAM expression on CTCs has been debated, as cancer cells that undergo EMT lose EpCAM expression on the cell surface (12,21). Using flow cytometry, we have observed that the lung cancer PC-9 and H358 cell lines express EpCAM (unpublished data). In the present study, immunofluorescent staining of the the collected cells was performed using anti-EpCAM and anti-CD45 mAbs, and the EpCAM-positive and CD45-negative cells were counted, excluding CD45-positive cells as leukocytes. To more accurately count CTCs, a cocktail of antibodies targeting a broad spectrum of tumor antigens is required.
The second limitation of the present study was that the size-based filtration platform was unable to detect CTCs in patients with early-stage cancer. Negligible CTC counts from patients with early-stage NSCLC suggest that only a small number of CTCs migrate from the primary tumor site to the bloodstream. Numerous studies have suggested that current CTC detection technologies may only be applied to patients with advanced cancer (4,22). Studies using the CellSearch platform found that the CTC count of patients with early-stage cancer is error-prone and too low to determine tumor prognosis (23–25). Thus, based on the present results, we propose that the CellSearch platform be used to monitor only the CTC counts of patients with advanced cancer.
Thirdly, CTCs may be lost during processing steps such as cell transfer and centrifugation. The size-based filtration platform worked well with blood transfused with cancer cell lines of a uniform size (13). However, cancer cells are typically heterogeneous in size and deformable, which may allow them to pass through the pores of the filters used in the size-based filtration platform.
Finally, additional evidence is required to determine whether EpCAM-positive cells are actually CTCs. Captured cells must be additionally characterized using assays targeting specific mRNAs or genomic DNA such as quantitative polymerase chain reaction (PCR), chromosomal fluorescence in situ hybridization assays and next-generation sequencing (26,27).
CTC detection and identification may be used as an early prognostic tool and for personalized therapeutic applications. The majority of patients with pulmonary nodules must undergo fine-needle biopsies to be diagnosed by cytological examination. More convenient and precise diagnostic markers for cancer progression are required. In addition, early detection and diagnosis platforms with higher sensitivity and specificity should be developed. More evidence is also required to conclude that CTC quantification and identification can be used to accurately determine cancer status.
With the increased importance of CTCs, numerous studies have performed DNA sequencing from single CTCs to characterize mutational status (26,28). Maheswaran and Haber (28) reported capturing CTCs from the blood of patients with NSCLC using microfluidic device microposts coated with antibodies against epithelial cells. The authors performed EGFR mutational analysis using allele-specific PCR amplification. Lohr et al (26) reported on an integrated process for the isolation, validation and whole-exome sequencing of CTCs in patients with prostate cancer. Genomic analysis of CTCs showed that mutations in CTCs resemble mutations in primary tumors and metastases. These data affect the potential clinical utility of CTCs as part of a liquid biopsy. In additional studies, next-generation sequencing will be required to improve the understanding of CTC biology in the blood of patients with lung cancer and of the mechanisms of metastasis.
In conclusion, the present study evaluated a novel, size-based filtration platform for CTC quantification. CTCs in the blood of patients with lung cancer were enriched using the size-based filtration platform and immunofluorescently stained with DAPI, CD45 and EpCAM. The CTC count in patients with stage IV cancer was significantly higher than that of patients with stages I–III cancer. These results suggest that the novel platform may be a useful tool for determining lung cancer prognosis.
Acknowledgements
The present study was supported by the Converging Research Center Program funded by the Ministry of Science, ICT and Future Planning (grant no. 2013K000278).
References
- 1.GBD 2015 Mortality and Causes of Death Collaborators: Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the global burden of disease study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mello RA, Liu DJ, Aguiar PN, Tadokoro H. EGFR and EML4-ALK updated therapies in non-small cell lung cancer. Recent Pat Anticancer Drug Discov. 2016;11:393–400. doi: 10.2174/1574892811666160803090944. [DOI] [PubMed] [Google Scholar]
- 3.Rothschild SI. Targeted therapies in non-small cell lung cancer-beyond EGFR and ALK. Cancers (Basel) 2015;7:930–949. doi: 10.3390/cancers7020816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 5.Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, Ward TH, Backen A, Clack G, Hughes A, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol. 2012;7:306–315. doi: 10.1097/JTO.0b013e31823c5c16. [DOI] [PubMed] [Google Scholar]
- 6.Hofman V, Bonnetaud C, Ilie MI, Vielh P, Vignaud JM, Fléjou JF, Lantuejoul S, Piaton E, Mourad N, Butori C, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res. 2011;17:827–835. doi: 10.1158/1078-0432.CCR-10-0445. [DOI] [PubMed] [Google Scholar]
- 7.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K, Capron F, Franco D, Pazzagli M, Vekemans M, et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofman V, Ilie MI, Long E, Selva E, Bonnetaud C, Molina T, Vénissac N, Mouroux J, Vielh P, Hofman P. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: Comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int J Cnacer. 2011;129:1651–1660. doi: 10.1002/ijc.25819. [DOI] [PubMed] [Google Scholar]
- 12.Gorges TM, Stein A, Quidde J, Hauch S, Röck K, Riethdorf S, Joosse SA, Pantel K. Improved detection of circulating tumor cells in metastatic colorectal cancer by the combination of the CellSearch® System and the AdnaTest®. PLoS One. 2016;11:e0155126. doi: 10.1371/journal.pone.0155126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim EH, Lee JK, Kim BC, Rhim SH, Kim JW, Kim KH, Jung SM, Park PS, Park HC, Lee J, Jeon BH. Enrichment of cancer cells from whole blood using a microfabricated porous filter. Anal Biochem. 2013;440:114–116. doi: 10.1016/j.ab.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC cancer staging manual. 7th edition. Springer; New York, NY: 2010. [Google Scholar]
- 15.Braun S, Naume B. Circulating and disseminated tumor cells. J Clin Oncol. 2005;23:1623–1626. doi: 10.1200/JCO.2005.10.073. [DOI] [PubMed] [Google Scholar]
- 16.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, RC Jr; American Society of Clinical Oncology Bast. American society of clinical oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 17.Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 18.Zhu WF, Li J, Yu LC, Wu Y, Tang XP, Hu YM, Chen YC. Prognostic value of EpCAM/MUC1 mRNA-positive cells in non-small cell lung cancer patients. Tumour Biol. 2014;35:1211–1219. doi: 10.1007/s13277-013-1162-8. [DOI] [PubMed] [Google Scholar]
- 19.Chen Q, Ge F, Cui W, Wang F, Yang Z, Guo Y, Li L, Bremner RM, Lin PP. Lung cancer circulating tumor cells isolated by the EpCAM-independent enrichment strategy correlate with Cytokeratin 19-derived CYFRA21-1 and pathological staging. Clin Chim Acta. 2013;419:57–61. doi: 10.1016/j.cca.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Berghmans T, Pasleau F, Paesmans M, Bonduelle Y, Cadranel J, Cs Toth I, Garcia C, Giner V, Holbrechts S, Lafitte JJ, et al. Surrogate markers predicting overall survival for lung cancer: ELCWP recommendations. Eur Respir J. 2012;39:9–28. doi: 10.1183/09031936.00190310. [DOI] [PubMed] [Google Scholar]
- 21.Konigsberg R, Obermayr E, Bises G, Pfeiler G, Gneist M, Wrba F, de Santis M, Zeillinger R, Hudec M, Dittrich C. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011;50:700–710. doi: 10.3109/0284186X.2010.549151. [DOI] [PubMed] [Google Scholar]
- 22.Truini A, Alama A, Dal Bello MG, Coco S, Vanni I, Rijavec E, Genova C, Barletta G, Biello F, Grossi F. Clinical applications of circulating tumor cells in lung cancer patients by CellSearch system. Front Oncol. 2014;4:242. doi: 10.3389/fonc.2014.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis JW, Nakanishi H, Kumar VS, Bhadkamkar VA, McCormack R, Fritsche HA, Handy B, Gornet T, Babaian RJ. Circulating tumor cells in peripheral blood samples from patients with increased serum prostate specific antigen: Initial results in early prostate cancer. J Urol. 2008;179:2187–2191, 2191. doi: 10.1016/j.juro.2008.01.102. [DOI] [PubMed] [Google Scholar]
- 24.Hong B, Zu Y. Detecting circulating tumor cells: Current challenges and new trends. Theranostics. 2013;3:377–394. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SJ, Masago A, Tamaki Y, Akazawa K, Tsukamoto F, Sato J, Ozawa T, Tsujino Y, Noguchi S. A novel approach using telomerase-specific replication-selective adenovirus for detection of circulating tumor cells in breast cancer patients. Breast Cancer Res Treat. 2011;128:765–773. doi: 10.1007/s10549-011-1603-2. [DOI] [PubMed] [Google Scholar]
- 26.Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS, et al. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010;5:e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maheswaran S, Haber DA. Circulating tumor cells: A window into cancer biology and metastasis. Curr Opin Genet Dev. 2010;20:96–99. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]