Abstract
Colorectal cancer (CRC) is one of the most common types of malignant tumor in the world and occurs through a multi-step process resulting from the accumulation of genetic and epigenetic alterations of the genome. Although the molecular mechanisms of the pathogenesis of CRC remain unclear, the inactivation of tumor suppressor genes (TSGs) through promoter methylation serves an important role. Aberrant methylation is a well-defined marker of CRC. At present, the epigenetic silencing of protocadherin 10 (PCDH10) has been identified as an important TSG with key roles in colorectal carcinogenesis, invasion and metastasis as a frequent and early event. Advances in gene methylation detection in tumor tissues and body fluids have led to the development of non-invasive screening methods for CRC. The present study aimed to review the epigenetic alteration of PCDH10 in CRC development, and the potential of PCDH10 to be a non-invasive biomarker for CRC.
Keywords: DNA methylation, protocadherin 10, tumor suppressor gene, colorectal cancer, tumor biomarker
1. Introduction
Colorectal cancer (CRC) is one of the most common types of malignant tumor in the world (1). The development of CRC is a multi-step process resulting from the accumulation of genetic and epigenetic alterations of the genome (2). The loss of genomic stability and subsequent genetic alterations in tumor suppressor genes (TSG) and oncogenes may initiate carcinogenesis and tumor progression (3). A number of genetic aberrations have been identified in CRC, occurring in the APC, V-Ki-ras2 Kirsten rate sarcoma viral oncogene (KRAS) and TP53 genes (4,5). In addition to genetic changes, epigenetic deregulation including aberrant DNA methylation as a marker of cancer has been extensively studied in CRC (6). Epigenetic gene silencing is associated with the onset and progression of various types of cancer, and it is accepted that epigenetic alterations precede genetic changes during tumorigenesis (7). Aberrant methylation of cytosine-guanine (CpG) islands in TSG promoter regions has been previously proposed as a novel candidate cancer biomarker (8).
Protocadherins (PCDHs) are cadherin-associated receptors that serve an important role in the establishment and function of specific cell-cell connections and in tumor development (9,10). Previously, frequent epigenetic silencing of protocadherin 10 (PCDH10) was revealed in colorectal carcinogenesis. PCDH10 has been proposed to be a tumor suppressor gene involved in the processes of growth control, cell invasion and metastasis (11). The present study focuses on previous findings and aims to review the epigenetic alteration of PCDH10 and the possibility of PCDH10 methylation being a biomarker for CRC diagnosis.
2. Characteristics and biological functions of PCDH10
PCDHs constitute a major subfamily of the cadherin superfamily (12). The PCDH family may be divided largely into 2 groups, based on their genomic structure: Clustered PCDHs (PCDHα, δ and γ families) constituting gene clusters on a single chromosome and non-clustered PCDHs scattered over different chromosomes (13,14). The majority of non-clustered PCDHs are cell-adhesion molecules with 6 or 7 cadherin motifs in their extracellular domain and several cytoplasmic domains. Non-clustered PCDHs may be classified into 3 groups: δ1, δ2 and ε subgroups (15). All PCDHδ members contain highly conserved motifs (CM), CM1, 27 amino acids and CM2, 17 amino acids, in their cytoplasmic domains.
The human PCDH10 gene, termed OL-PCDH or KIAA1400, is located at 4q28.3 on the long arm of chromosome 4 and is a member of the subgroup of PCDHδ2 without phosphatase-1α (PP1α) binding domain (RRVTF, CM3) (16). Unlike other PCDH family members expressed predominantly in the nervous system, PCDH10 has been identified to exhibit widespread expression in almost all normal tissue (17,18) and contains 6 extracellular repeats, a transmembrane domain and a unique cytoplasmic domain (19), including CM2, homologous to a laminin-type epidermal growth factor (EGF)-like (LE) domain (17), as demonstrated in Fig. 1A. CM2 is also similar to the C2HC-type zinc-finger or zinc knuckle finger motif (20), and represents a functional interaction domain of PCDH10, which may mediate intracellular signal transduction. Previous studies have demonstrated that PCDH10 is involved in several important biological pathways in different types of tumor cells. Although PCDH10 appears to lack the β-catenin binding cytoplasmic site present in classical cadherins (21), it may affect the Wnt/β-catenin signaling pathway (22,23). It was revealed that PCDH10 induced apoptosis by inhibiting the nuclear factor (NF)-κB pathway in myeloma cells (24), or by interacting with human brain expressed X-linked 1 in imatinib-induced K562 cell apoptosis (25), indicating the proapoptotic and drug-resistance reversal role of PCDH10. PCDH10 exhibits cell-to-cell adhesion activity with a weak binding ability, suggesting that the cytoplasmic domain may not efficiently stabilize those interactions to facilitate adhesion or may negatively regulate their extracellular adhesions (15). The underlying mechanism is hypothesized to be distinct from that of classical cadherins. PCDH10 interacts with Nck-associated protein 1 (Nap1)/WAVE1, and the PCDH10/Nap1/WAVE1 complex affects actin assembly and subsequently regulates cell migration (26). However, how PCDH10/Nap1/WAVE1 complex controls actin assembly remains unknown. The molecules interacted with or regulated by PCDH10 are described in Table I.
Figure 1.
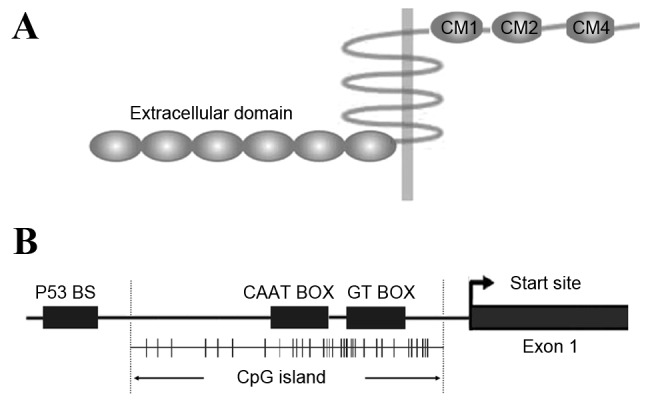
Characteristics of PCDH10. (A) Protein structure of PCDH10. PCDH10 possesses 6 extracellular domains, a transmembrane domain and 3 cytoplasmic domains: CM1, CM2 and CM4. (B) CpG island and promoter region of PCDH10. CpG sites are shown as short vertical lines. The transcription start site is indicated by a curved arrow. CAAT and GT BOX exist in the core promoter. A p53 binding site P53 BS is also located in the PCDH10 promoter region, which is responsive to p53 regulation. PCDH10, protocadherin 10; CM, cytoplasmic motif; p53, tumor protein 53; BS, binding site; CpG, cytosine-guanine site.
Table I.
Molecule/complex that interact with or are regulated by PCDH10.
| Molecule/complex | Functions | Ref |
|---|---|---|
| Nap1,WAVE1 | Actin assembly/cell migration | 22 |
| Wnt/β-catenin pathway | Cell cycle/apoptosis, carcinogenesis | 18,19 |
| CDKN1A, FGFR2, and HTATIP2 | Apoptosis, growth, invasion and metastasis | 24 |
| NF-kappaB pathway | Apoptosis | 20 |
| hBex1 | Drug resistance | 21 |
NAP1/Wave1, Nck-associated protein 1/WAVE1; CDKN1A, cyclin dependent kinase inhibitor 1A; HTATIP2, HIV-1 Tat interactive protein 2; NF-κB, nuclear factor-kappaB.
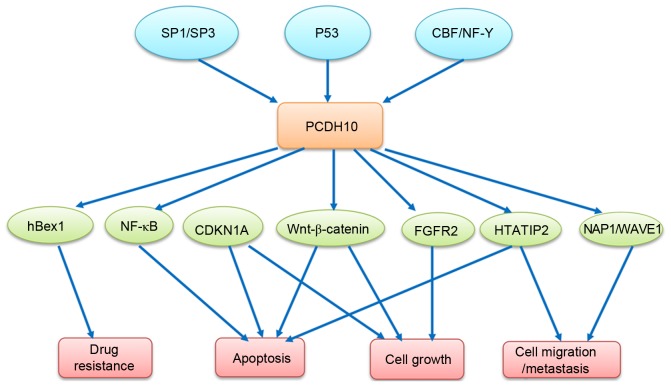
The minimal promoter region of PCDH10 is identified as the segment between nucleotides −144 and −99, containing a CAAT box and a GT box, which are required for promoter activity, and a putative transcription factor binding site for AP-4. Specificity protein (Sp)1/Sp3 and core binding factor (CBF)/NF-Y transcription factors are crucial to the basal expression of PCDH10 (27). Previously, PCDH10 was revealed to be a transcriptional target of tumor protein (p)53 and that the levels of PCDH10 expression may be induced by wild type p53 but not mutant p53 in a number of human cancer cell lines (28), suggesting that PCDH10 may be involved in the signaling pathway of p53 regulation, as illustrated in Fig. 1B. Yu et al (29) identified cyclin dependent kinase inhibitor 1A (CDKN1A), FGFR2 and HIV-1 Tat interactive protein 2 (HTATIP2) as the upregulated target genes in a PCDH10 re-expressing gastric cancer cell line by complementary DNA microarray analysis, and hypothesized the molecular events mediated by PCDH10, including induction of apoptosis, control of cell growth and inhibition of invasion and metastasis. The signaling pathways involved in PCDH10 regulation are illustrated in Fig. 2. The comprehensive functions of PCDH10 require additional investigation.
Figure 2.
Molecular events for induction of cell apoptosis, reversal of drug resistance, suppression of cell growth and migration/invasion mediated by PCDH10. Sp1/Sp3, specificty protein 1/3; p53, tumor protein 53; CBF/NF-Y, core binding factor/nuclear transcription factor Y; NF-κB, nuclear factor-kappa B; CDKN1A, cyclin dependent kinase inhibitor 1A; HTATIP2, HIV-1 Tat interactive protein 2; NAP1/Wave1, Nck-associated protein 1/WAVE1.
3. Role of PCDH10 as a functional TSG in CRC
Previously, an association between PCDH10 and tumor development has been proposed. PCDH10 downregulation is involved into several types of cancer, including gastric, hepatocellular, colorectal, breast, cervical, lung, nasopharyngeal, esophageal, pancreatic and bladder cancer (30–35). Functional studies have also identified PCDH10 as a potential novel tumor suppressor gene in multiple types of cancer (18,32,33). The present study demonstrated that PCDH10 may suppress cell proliferation and invasion in colorectal cancer RKO cells. Flow cytometry analysis of PCDH10-re-expressed RKO cells revealed a significant decrease in the number of cells in the G2/M phase, and an increase in the number of cells in the G0/G1 phase, without inducing apoptosis (11). The ectopic expression of PCDH10 significantly reduced the colony formation efficiencies of the HCT116 cell line in monolayer culture and soft-agar assays in vitro (18) and suppressed tumorigenesis and liver metastasis in vivo, resulting in prolonged mice survival (36). Thus, PCDH10 may be an effective therapeutic target for the treatment of CRC and the mechanism requires attention.
4. Aberrant promoter methylation and inactivation of PCDH10 in CRC
DNA methylation represents one of the most studied types of epigenetic marker in CRC (37). The methylation of CpG islands in the gene promoter region may induce chromatin conformational modifications and inhibit the access of the transcriptional machinery, thus altering gene expression levels. The downregulation of cadherins or their functional alterations have been frequently observed in human malignancies (38). Several studies have demonstrated the aberrant methylation of the PCDH10 promoter region in a wide spectrum of types of cancer by various techniques, such as methylation-specific polymerase chain reaction (MSP PCR) and MSP quantitative (q) PCR and high-resolution melting (HRM) analysis (35). Promoter methylation and transcriptional silencing of PCDH10 were detected in all analyzed colorectal carcinoma cell lines, but not in immortalized normal epithelial cell lines (11). Ying et al (18) demonstrated that the transcriptional silencing of PCDH10 may be reversed by pharmacologic demethylation with 5-aza-2′-deoxycytidine or genetic demethylation with double knockout of DNA methyltransferase (DNMT)1 and DNMT3B in the colorectal cancer HCT116 cell line, suggesting a direct epigenetic mechanism (18). The aberrant methylation of the PCDH10 promoter was also observed in 43–85% of colorectal cancer tissues in several studies, indicating that PCDH10 methylation is a frequent event in colorectal carcinogenesis (11,35,39). In the present study, PCDH10 methylation was detected in colorectal cancer samples, but not in paired adjacent colorectal tissues, in accordance with the study by Yu et al (35), suggesting that the aberrant promoter methylation of PCDH10 is specific to colorectal cancer cells.
As recognized previously, candidate TSGs may be inactivated through epigenetic inactivation, biallelic genetic inactivation or the two (7). Notably, the allelic loss of PCDH10 was revealed in 53/171 types of colorectal carcinoma and the expression of the PCDH10 gene was silenced or markedly downregulated in 41 of the aforementioned 53 types of colorectal carcinoma compared with their matched normal mucosa, indicating genetic deletion may be an additional mechanism for PCDH10 inactivation in CRC (36), although Ying et al (18) did not detect any homozygous deletion of the PCDH10 gene in CRC cell lines. This suggests that the epigenetic inactivation and genetic alteration of PCDH10 serve important roles in the development of CRC. Additional study is required to assess whether other types of genetic inactivation, including point mutations, are involved in colorectal tumorigenesis.
5. Non-invasive biomarker for CRC diagnosis
CRC prognosis remains poor, and the 5-year survival rates are ~90% for patients with early stage disease but decrease to <10% in patients with distant metastases (40). The early detection of CRC through screening programs that detect mucosal changes that are predictive of colorectal tumors may reduce the incidence and mortality rates of this disease (41,42). However, novel biomarkers that are absent in the healthy population and present in patients with CRC need to be identified, particularly those that may be detected at the early development stage of the disease and applied in screening tests. Since methylated genes that are present in tumor tissues may be identified in serum/plasma, stool or ascitic fluid, epigenetic biomarkers represent a well-suited biological material for a non-invasive screening method for CRC diagnosis (43). Using the MSP qPCR method, Danese et al (44) demonstrated that the PCDH10 promoter methylation was detected in 94.0% of surgically resected tumors and in 62.7% of plasma samples, with a high concordance rate between tissues and plasma samples of 66.6% in total, and 76.3% in patients with early (stage I/II) CRC (45), demonstrating a significant correlation between PCDH10 methylation in cell-free DNA and tumor tissue. Since methylated alleles may be detected with a high degree of sensitivity, there is great scope in using methylation as a potential early detection system for CRC. Stool DNA is another well-suited biological material for CRC diagnosis. The methylated PCDH10 promoter sequence in stool as a biomarker may be investigated in additional studies. With respect to CRC diagnosis, fecal occult blood tests (FOBT) are a commonly used non-invasive screening procedure, which reduce CRC-associated mortality by 20% when performed every 2 years (46). Detecting PCDH10 methylation in plasma and/or stool, combined with FOBT, may be a good diagnostic regimen to improve the sensitivity and effectiveness. Additional case-control studies are required to address this issue.
Whether PCDH10 may be a prognostic biomarker has also been investigated. However, a significant association has not been revealed between PCDH10 methylation status and tumor node metastasis staging, DFS or OS in CRC (35). A possible reason for this is that PCDH10 promoter methylation is an early event in CRC carcinogenesis, and is hypothesized to be the cause, rather than the outcome, of carcinogenesis. Additionally, the different extent, full or partial, of promoter methylation may affect the gene expression level and function. Thus, it is important to determine the presence and extent of gene methylation. Jao et al (36) reported that genetic and/or epigenetic alterations contribute to the PCDH10 inactivation that occurs in the majority of CRC tumors, and the genetic aberration of PCDH10 was demonstrated to be significantly associated with tumor progression and distant metastasis, indicating it may be an independent predictor of poor survival rates for patients with CRC. Heitzer et al (47) found that the promoter methylation statuses of PCDH10, SPARC and UCHL1 may be used as prognostic and predictive molecular markers for patients with CRC, and facilitate treatment decisions for stage II colorectal cancer. These results suggest that genetic changes or epigenetic aberrations of PCDH10, combined with other biomarkers, may exhibit prognostic value for CRC.
6. Conclusion
Aberrant methylation of the PCDH10 promoter, causing gene inactivation, is a frequent and early event in CRC. PCDH10 has been identified as an important TSG, with key roles in colorectal carcinogenesis, invasion and metastasis. PCDH10 methylation status may be a valuable non-invasive biomarker for the diagnosis of CRC.
Acknowledgements
The present study was supported by Health Program of Hangzhou (grant no. 20140633B26), National Natural Science Foundation of China (grant nos. 81000945, 81071619, 81101477) and the Zhejiang Provincial Natural Science Foundation of China (grant no. Y2110018).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21314. [DOI] [PubMed] [Google Scholar]
- 2.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bach SP, Renehan AG, Potten CS. Stem cells: The intestinal stem cell as a paradigm. Carcinogenesis. 2000;21:469–476. doi: 10.1093/carcin/21.3.469. [DOI] [PubMed] [Google Scholar]
- 4.Cho KR, Vogelstein B. Genetic alterations in the adenoma-carcinoma sequence. Cancer. 1992;70(6 Suppl):S1727–S1731. doi: 10.1002/1097-0142(19920915)70:4+<1727::AID-CNCR2820701613>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 5.Zoratto F, Rossi L, Verrico M, Papa A, Basso E, Zullo A, Tomao L, Romiti A, Lo Russo G, Tomao S. Focus on genetic and epigenetic events of colorectal cancer pathogenesis: Implications for molecular diagnosis. Tumour Biol. 2014;35:6195–6206. doi: 10.1007/s13277-014-1845-9. [DOI] [PubMed] [Google Scholar]
- 6.Wong JJ, Hawkins NJ, Ward RL. Colorectal cancer: A model for epigenetic tumorigenesis. Gut. 2007;56:140–148. doi: 10.1136/gut.2005.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 8.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Chen MW, Terry S, Vacherot F, Chopin DK, Bemis DL, Kitajewski J, Benson MC, Guo Y, Buttyan R. A human- and male-specific protocadherin that acts through the wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Res. 2005;65:5263–5271. doi: 10.1158/0008-5472.CAN-05-0162. [DOI] [PubMed] [Google Scholar]
- 10.Waha A, Güntner S, Huang TH, Yan PS, Arslan B, Pietsch T, Wiestler OD, Waha A. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005;7:193–199. doi: 10.1593/neo.04490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong X, Zhu Y, Mao J, Zhang J, Zheng S. Frequent epigenetic silencing of PCDH10 by methylation in human colorectal cancer. J Cancer Res Clin Oncol. 2013;139:485–490. doi: 10.1007/s00432-012-1353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St John T, Taketani S, Suzuki S. Protocadherins: A large family of cadherin-related molecules in central nervous system. EMBO J. 1993;12:2249–2256. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morishita H, Yagi T. Protocadherin family: Diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/S0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 15.Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adh Migr. 2011;5:97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanhalst K, Kools P, Staes K, van Roy F, Redies C. delta-Protocadherins: A gene family expressed differentially in the mouse brain. Cell Mol Life Sci. 2005;62:1247–1259. doi: 10.1007/s00018-005-5021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolverton T, Lalande M. Identification and characterization of three members of a novel subclass of protocadherins. Genomics. 2001;76:66–72. doi: 10.1006/geno.2001.6592. [DOI] [PubMed] [Google Scholar]
- 18.Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, Putti T, Murray P, Chan AT, Tao Q. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070–1080. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 19.Hirano S, Yan Q, Suzuki ST. Expression of a novel protocadherin, OL-protocadherin, in a subset of functional systems of the developing mouse brain. J Neurosci. 1999;19:995–1005. doi: 10.1523/JNEUROSCI.19-03-00995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews JM, Kowalski K, Liew CK, Sharpe BK, Fox AH, Crossley M, MacKay JP. A class of zinc fingers involved in protein-protein interactions biophysical characterization of CCHC fingers from fog and U-shaped. Eur J Biochem. 2000;267:1030–1038. doi: 10.1046/j.1432-1327.2000.01095.x. [DOI] [PubMed] [Google Scholar]
- 21.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–562. doi: 10.1016/S0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, Yang Y, Trovik J, Sun K, Zhou L, Jiang P, Lau TS, Hoivik EA, Salvesen HB, Sun H, Wang H. A novel wnt regulatory axis in endometrioid endometrial cancer. Cancer Res. 2014;74:5103–5117. doi: 10.1158/0008-5472.CAN-14-0427. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Yang Z, Yuan H, Li Z, Li Y, Liu Q, Chen J. PCDH10 inhibits cell proliferation of multiple myeloma via the negative regulation of the Wnt/β-catenin/BCL-9 signaling pathway. Oncol Rep. 2015;34:747–754. doi: 10.3892/or.2015.4056. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Yang Z, Peng X, Li Y, Liu Q, Chen J. Nuclear factor-κB is involved in the protocadherin-10-mediated pro-apoptotic effect in multiple myeloma. Mol Med Rep. 2014;10:832–838. doi: 10.3892/mmr.2014.2285. [DOI] [PubMed] [Google Scholar]
- 25.Ding K, Su Y, Pang L, Lu Q, Wang Z, Zhang S, Zheng S, Mao J, Zhu Y. Inhibition of apoptosis by downregulation of hBex1, a novel mechanism, contributes to the chemoresistance of Bcr/Abl+ leukemic cells. Carcinogenesis. 2009;30:35–42. doi: 10.1093/carcin/bgn251. [DOI] [PubMed] [Google Scholar]
- 26.Nakao S, Platek A, Hirano S, Takeichi M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J Cell Biol. 2008;182:395–410. doi: 10.1083/jcb.200802069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Xie J, Li W, Tang A, Li X, Jiang Z, Han Y, Ye J, Jing J, Gui Y, Cai Z. Identification and characterization of human PCDH10 gene promoter. Gene. 2011;475:49–56. doi: 10.1016/j.gene.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Shi D, Murty VV, Gu W. PCDH10, a novel p53 transcriptional target in regulating cell migration. Cell Cycle. 2015;14:857–866. doi: 10.1080/15384101.2015.1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM, et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136:640–651.e1. doi: 10.1053/j.gastro.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 30.Fang S, Huang SF, Cao J, Wen YA, Zhang LP, Ren GS. Silencing of PCDH10 in hepatocellular carcinoma via de novo DNA methylation independent of HBV infection or HBX expression. Clin Exp Med. 2013;13:127–134. doi: 10.1007/s10238-012-0182-9. [DOI] [PubMed] [Google Scholar]
- 31.Lin YL, Li ZG, He ZK, Guan TY, Ma JG. Clinical and prognostic significance of protocadherin-10 (PCDH10) promoter methylation in bladder cancer. J Int Med Res. 2012;40:2117–2123. doi: 10.1177/030006051204000609. [DOI] [PubMed] [Google Scholar]
- 32.Narayan G, Scotto L, Neelakantan V, Kottoor SH, Wong AH, Loke SL, Mansukhani M, Pothuri B, Wright JD, Kaufmann AM, et al. Protocadherin PCDH10, involved in tumor progression, is a frequent and early target of promoter hypermethylation in cervical cancer. Genes Chromosomes Cancer. 2009;48:983–992. doi: 10.1002/gcc.20703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang X, Yin X, Xiang T, Li H, Li F, Chen L, Ren G. Protocadherin 10 is frequently downregulated by promoter methylation and functions as a tumor suppressor gene in non-small cell lung cancer. Cancer Biomark. 2012;12:11–19. doi: 10.3233/CBM-2012-00280. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Xie PG, Lin YL, Ma JG, Li WP. Aberrant methylation of PCDH10 predicts worse biochemical recurrence-free survival in patients with prostate cancer after radical prostatectomy. Med Sci Monit. 2014;20:1363–1368. doi: 10.12659/MSM.891241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu B, Yang H, Zhang C, Wu Q, Shao Y, Zhang J, Guan M, Wan J, Zhang W. High-resolution melting analysis of PCDH10 methylation levels in gastric, colorectal and pancreatic cancers. Neoplasma. 2010;57:247–252. doi: 10.4149/neo_2010_03_247. [DOI] [PubMed] [Google Scholar]
- 36.Jao TM, Tsai MH, Lio HY, Weng WT, Chen CC, Tzeng ST, Chang CY, Lai YC, Yen SJ, Yu SL, Yang YC. Protocadherin 10 suppresses tumorigenesis and metastasis in colorectal cancer and its genetic loss predicts adverse prognosis. Int J Cancer. 2014;135:2593–2603. doi: 10.1002/ijc.28899. [DOI] [PubMed] [Google Scholar]
- 37.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pignatelli M. Integrins, cadherins, and catenins: Molecular cross-talk in cancer cells. J Pathol. 1998;186:1–2. doi: 10.1002/(SICI)1096-9896(199809)186:1<1::AID-PATH135>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 39.Silva TD, Vidigal VM, Felipe AV, DE Lima JM, Neto RA, Saad SS, Forones NM. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncol Lett. 2013;6:1687–1692. doi: 10.3892/ol.2013.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toribara NW, Sleisenger MH. Screening for colorectal cancer. N Engl J Med. 1995;332:861–867. doi: 10.1056/NEJM199503303321306. [DOI] [PubMed] [Google Scholar]
- 41.Pignone MP, Lewis CL. Using quality improvement techniques to increase colon cancer screening. Am J Med. 2009;122:419–420. doi: 10.1016/j.amjmed.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winawer SJ. The multidisciplinary management of gastrointestinal cancer. Colorectal cancer screening. Best Pract Res Clin Gastroenterol. 2007;21:1031–1048. doi: 10.1016/j.bpg.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10:123–132. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL, Gusella M, Pasini F, Guidi GC, Lippi G. Epigenetic alteration: New insights moving from tissue to plasma- the example of PCDH10 promoter methylation in colorectal cancer. Br J Cancer. 2013;109:807–813. doi: 10.1038/bjc.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC cancer staging manual. 7th edition. New York, NY: Springer; 2010. [Google Scholar]
- 46.Atkin W. Options for screening for colorectal cancer. Scandin J Gastroenterol. 2009;38:13–16. doi: 10.1080/00855910310001421. [DOI] [PubMed] [Google Scholar]
- 47.Heitzer E, Artl M, Filipits M, Resel M, Graf R, Weißenbacher B, Lax S, Gnant M, Wrba F, Greil R, et al. Differential survival trends of stage II colorectal cancer patients relate to promoter methylation status of PCDH10, SPARC, and UCHL1. Mod Pathol. 2014;27:906–915. doi: 10.1038/modpathol.2013.204. [DOI] [PubMed] [Google Scholar]