Abstract
RNA-sequencing of the patient's bone marrow detected fusion transcripts in which the coding sequence of the FAM53B gene (from 10q26) was fused to a genomic sequence (from 19q13) that mapped upstream of the SLC7A10 locus. Reverse transcription-polymerase chain reaction together with Sanger sequencing verified the presence of this fusion transcript. The FAM53B fusion transcript is not expected to produce any chimeric protein. However, it may code for a truncated FAM53B protein consisting of the first 302 amino acids of FAM53B together with amino acids from the 19q13 sequence. Functionally, the truncated FAM53B would be similar to the protein encoded by the FAM53B sequence with accession no. BC031654.1 (FAM53B protein accession no. AAH31654.1). Furthermore, the truncated protein contains the entire conserved domain of the FAM53 protein family. The chromosome aberration t(10;19)(q26;q13) detected in this study was previously reported in a single case of ALL, in which it was also the sole karyotypic change. Both patients entered complete hematological and cytogenetic remission following treatment.
Keywords: acute lymphoblastic leukemia, chromosome translocation, RNA-sequencing, FAM53B gene, truncation
Introduction
Acute lymphoblastic leukemia (ALL) is a malignant disease of the bone marrow in which early lymphoid precursors proliferate and replace the normal hematopoietic cells (1). ALL is much more common in children than in older age groups (2).
In their leukemic bone marrow cells, ALL patients carry acquired genetic alterations that contribute to the increased proliferation, prolonged survival and/or impaired differentiation of the lymphoid hematopoietic progenitors (3–5). Many of these alterations are nonrandom numerical or structural chromosome aberrations that can be detected under the microscope (6,7). Often the aberrations have prognostic significance (4,6–8). For example, high hyperdiploidy (51–65 chromosomes) is associated with a favorable outcome in pre-B-ALL patients, whereas hypodiploidy (<44 chromosomes) is a marker of an adverse prognosis (4,6–8). The t(12;21)(p13;q22) translocation leading to an ETV6-RUNX1 fusion gene is another marker of standard risk disease with a high likelihood of cure, whereas the translocation t(4;11)(q21;q23), which generates the MLL-AF4 fusion and is common in infant ALL patients, is associated with a poor prognosis (4,6–8). To date, banding cytogenetics has had a pivotal role in disease classification and in the detection of new chromosomal rearrangements that can then be examined using molecular techniques to identify the important gene-level changes. Genome-wide profiling studies by means of microarray analysis and genome, RNA and exome sequencing have revealed many additional genetic rearrangements that are not detectable cytogenetically. In addition, these studies have identified multiple key cellular pathways that may be mutated in ALL (9).
High-throughput sequencing, in particular RNA-sequencing, has been shown to be an efficient tool for the detection of fusion genes in cancer (10). Many of the fusion genes detected to date correspond to chromosomal translocations that are often seen as the sole aberration by cytogenetic analysis, which adds to the likelihood that they represent primary tumorigenic events. The methodology is in principle simple: Extracted DNA or RNA from cancer cells is extensively sequenced, then the raw data are analyzed with one or more bioinformatics programs specifically designed to detect fusion genes (11). Numerous studies have used combinations of cytogenetics and RNA-sequencing to detect rearranged or fused genes that are associated with acquired cancer-specific chromosomal rearrangements (12).
In the present study, RNA-sequencing of an ALL sample with the t(10;19)(q26;q13) translocation was performed, in order to identify the genes affected by the translocation.
Materials and methods
Ethics statement
The present study was approved by the regional ethics committee (Regional Committees for Medical and Health Research Ethics, Oslo, Norway), and written informed consent was obtained from the patient to publish the case details. The ethics committee's approval included a review of the consent procedure. All patient information has been anonymized.
Case report
A 39-year-old male was transferred to Oslo University Hospital with a preliminary diagnosis of ALL. He presented with increasing fatigue and a reduced performance status. The clinical examination was unremarkable, except for the presence of an enlarged supraclavicular lymph node on the right side. Blood analysis revealed slight anemia (hemoglobin, 10.5 g/dl), moderate thrombocytopenia (thrombocytes, 55×109/l), moderate leukocytosis (leukocytes, 32.7×109/l) and elevated levels of lactate dehydrogenase, C-reactive protein and creatinine. Examination of a bone marrow aspirate and biopsy showed the complete replacement of normal hematopoiesis by blasts with a high nuclear to cytoplasmic ratio. The percentage of blasts in the blood was 65–75%, as determined by flow cytometric analysis. The immunophenotypic analysis confirmed the B-cell lineage differentiation of the lymphoblasts, which were positive for CD45, the B-cell markers, CD19, CD22 and cytoplasmic CD79a, human leukocyte antigen-antigen D related antigens, CD10 and terminal deoxynucleotidyl transferase. In addition, the B lymphoblasts showed abnormal expression of CD123 and CD58. The molecular genetic analysis was negative for BCR-ABL, TCF3-PBX1 and ETV6-RUNX1, but a clonal immunoglobulin kappa rearrangement was found.
The patient received therapy for ALL based on the Hammersmith 82 regimen (http://oncolex.org/Prosedyrer/TREATMENT/DrugTherapy/Leukemia_ALL?lg=print), which consists of an induction phase of 16 weeks followed by maintenance treatment for 3 years. The patient went into complete morphological remission 6 weeks following the initiation of induction therapy. A reassessment after 20 weeks of therapy confirmed continuous morphological remission and the absence of minimal residual disease (MRD) at a detection level of <0.01%, as assessed by flow cytometry. A G-banding analysis performed at the same time point revealed a normal karyotype. The patient then received a total of 13 maintenance cycles and finished treatment 3 years and 5 months following diagnosis. Bone marrow aspirates taken 24 months following diagnosis and after cessation of therapy revealed complete hematological remission and no MRD.
G-banding analysis
Bone marrow cells were cytogenetically investigated by standard methods at diagnosis and after 20 weeks of therapy. Chromosome preparations were made from metaphase cells after a 24-h culture. Peripheral blood T lymphocytes stimulated with phytohemagglutinin, M form (PHA-M; Thermo Fisher Scientific, Inc., Waltman, MA, USA) for 72 h were also examined. Chromosome preparations were G-banded using Leishman stain (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) and karyotyped according to the International System for Human Cytogenetic Nomenclature 2013 guidelines (13).
RNA-sequencing
Total RNA (3 µg) was extracted from the patient's bone marrow using the miRNeasy Mini kit (Qiagen GmbH, Hilden, Germany) at the time of diagnosis and subjected to high-throughput paired-end RNA-sequencing at the Norwegian Sequencing Centre, Oslo University Hospital. Detailed information about the procedure was described previously (14). The software deFuse (15) was used for detection of fusion transcripts.
Reverse transcription-polymerase chain reaction (RT-PCR) analysis and sequencing
For RT-PCR, 1 µg total RNA was reverse transcribed in a 20-µl reaction volume using the iScript™ Advanced cDNA Synthesis kit for RT-qPCR (Bio-Rad Laboratories AB, Oslo, Norway), according to the manufacturer's protocol. The 25-µl PCR volume consisted of 12.5 µl Premix Ex Taq™ DNA Polymerase Hot Start Version (Takara Bio Europe SAS, Saint-Germain-en-Laye, France), 1 µl cDNA and 0.4 µM of each of the forward and reverse primers. The PCR was run on a C-1000 Thermal cycler (Bio-Rad Laboratories AB) with an initial denaturation at 94°C for 30 sec, followed by 35 cycles at 98°C for 7 sec, 64°C for 30 sec and 72°C for 30 sec, and a final extension at 72°C for 5 min. For detection of the FAM53B fusion transcript, the following primer combination was used: Forward FAM53B-1149F1 (5′-ACACCTGGAGCCCTGACCTGCAC-3′) and reverse 19q13-Trans1-R1 (5′-AAGCCTCGCAGCTCTGAAGCCTG-3′). The primers were purchased from Thermo Fisher Scientific, Inc.
The PCR products (3 µl) were stained with GelRed (Biotium, Inc., Hayward, CA, USA), separated by 1.0% agarose gel electrophoresis and photographed. DNA gel electrophoresis was performed using lithium borate buffer (Faster Better Media LLC, Hunt Valley, MD, USA), according to a previous study (16). The remaining PCR products were purified using the Qiagen PCR purification kit (Qiagen GmbH) and directly sequenced using the dideoxy procedure with the ABI Prism BigDye Terminator v1.1 Cycle Sequencing kit (Thermo Fisher Scientific, Inc.) on the Applied Biosystems Model 3500 Genetic Analyzer sequencing system (Thermo Fisher Scientific, Inc.). The basic local alignment search tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for computer analysis of sequence data.
Results
Cytogenetics
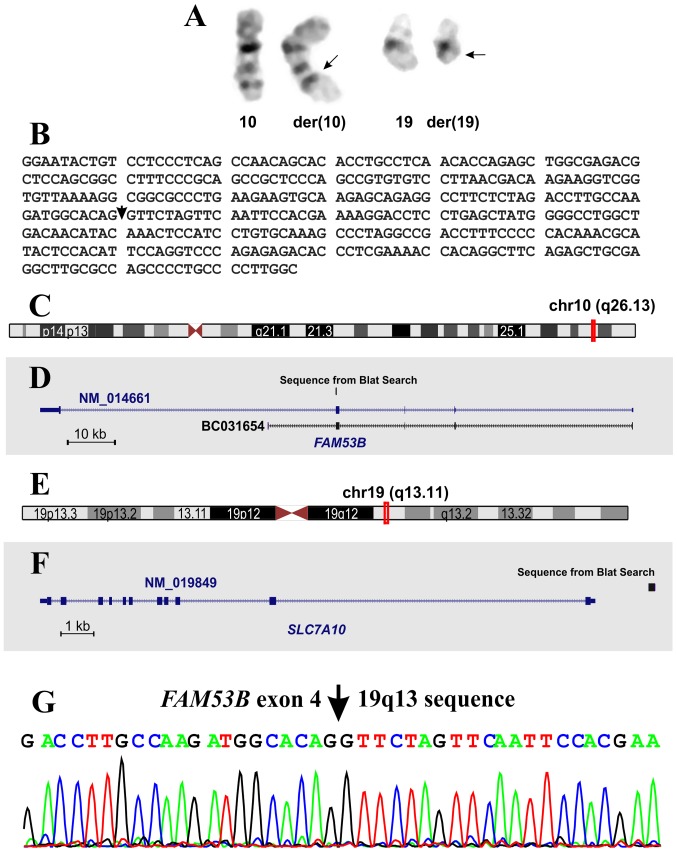
At the time of diagnosis, G-banding analysis of the bone marrow cells yielded a karyotype with a single clonal chromosome abnormality: 46,XY,t(10;19)(q26;q13)[15] (Fig. 1A). The G-banding analysis of PHA-M-stimulated peripheral blood mature T lymphocytes yielded a normal 46, XY karyotype. Therefore, the t(10;19)(q26;q13) translocation was found in the leukemic cells only. Notably, cytogenetic examination of the bone marrow cells 6 and 20 weeks following initiation of induction therapy showed a normal 46, XY karyotype (data not shown).
Figure 1.
Cytogenetic, RNA-sequencing and molecular genetic analysis of bone marrow from the ALL patient. (A) Partial karyotype showing the der(10) t(10;19)(q26;q13) and der (19) t(10;19)(q26;q13) abnormal chromosomes, together with the corresponding normal chromosome homologs. Arrows indicate the breakpoint positions. (B) The sequence obtained from the analysis of RNA-sequencing data using the deFuse program. The arrow indicates the junction point of FAM53B, with the sequence from 19q13. (C) Ideogram of chromosome 10 showing the mapping of FAM53B (red vertical line) on 10q26.13. (D) FAM53B gene with the sequences with accession numbers NM_014661 and BC031654. The sequence from the BLAT search is shown. (E) Ideogram of chromosome 19 showing the mapping of SLC7A10 (red vertical line) on 19q13.11. (F) The SLC7A10 gene with the sequence with accession number NM_019849. The result of the BLAT sequence from (B) is shown. (G) Partial sequence chromatogram of the amplified cDNA fragment showing the junction point of the FAM53B gene with the sequence from 19q13.
Analysis of RNA-sequencing
Using deFuse software to analyze the raw sequencing data, a fusion transcript (Fig. 1B) was found in which a coding sequence of the FAM53B gene from band 10q26 (Fig. 1C and D) was fused to a genomic sequence mapping upstream of the SLC7A10 locus (Fig. 1E and F). This corresponded well with the 19q13 breakpoint of the t(10;19)(q26;q13) found in the leukemic cells.
Molecular confirmation of the FAM53B fusions
RT-PCR using the FAM53B-1149F1/Trans1-R1 primers amplified a cDNA fragment (data not shown). Direct sequencing of the amplified fragment verified the FAM53B fusion detected with the deFuse program (Fig. 1G).
Discussion
The current study presents a case of precursor B-ALL in which the leukemic cells had acquired a t(10;19)(q26;q13) chromosome translocation as the sole cytogenetic abnormality. Molecular analysis of the translocation showed fusion of the FAM53B gene with an intergenic sequence from 19q13. Although the FAM53B fusion transcript is not expected to produce any chimeric protein, it may code for a truncated FAM53B protein consisting of the first 302 amino acids of FAM53B and amino acids from the sequence of 19q13. Functionally, the truncated FAM53B protein would be similar to the protein encoded by the FAM53B sequence with accession no. BC031654.1 (FAM53B protein accession no. AAH31654.1). The truncated FAM53B protein may be of biological importance, since it would contain the entire conserved domain of the FAM53 family of proteins (pfam15242: FAM53, http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=pfam15242). However, there is no evidence that this truncated protein is not degraded immediately or does not reach its target(s). Therefore, hemizygosity of the FAM53B gene, which acts as a tumor suppressor gene, may also be considered.
The FAM53 protein family members, which bind to a transcriptional regulator that modulates cell proliferation, are important in neural tube development, are found in eukaryotes and are typically between 303 and 413 amino acids in length (17,18). The FAM53B protein was reported to bind to 14-3-3 chaperones, as well as to the Ski-interacting proteins (SKIIP), which are part of the spliceosome and have an activator or a repressor role in transcription (18). Recently, FAM53B was shown to be involved in Wnt signal transduction and the regulation of nuclear localization of β-catenin (19). The Wnt signaling pathway has been implicated in the regulation of the proliferation, survival and differentiation of hematopoietic cells (20). Activation of the Wnt/β-catenin pathway was also shown to mediate growth and survival in B-cell progenitor acute lymphoblastic leukemia (21).
Chromosome translocations leading to the truncation of genes and the expression of aberrant truncated proteins in leukemias have been reported several times for the RUNX1 (22–30) and ETV6 (31) genes. Truncated RUNX1 protein was shown to reduce the transactivation capacity of core-binding factor (24). Furthermore, truncated RUNX1 protein resulting from the t(1;21)(p32;q22) chromosomal translocation was shown to impair the proliferation and differentiation of human hematopoietic progenitors (30). Transient expression of truncated forms of the ETV6 protein in zebrafish resulted in various hematopoietic effects, which acted dominantly over the wild-type ETV6 protein (32). Although the exact leukemogenic mechanism underlying truncated FAM53B is unknown, it may act via the Wnt/β-catenin pathway and the binding of 14-3-3 chaperones and SKIIP.
The chromosome aberration t(10;19)(q26;q13) was reported previously in a single case of ALL (33), where it was also the sole cytogenetic change and was detected in an adult patient. Both patients showed complete hematological and cytogenetic remission, indicating that this aberration is perhaps associated with a favorable prognosis. However, there is no evidence that these two translocations are identical at the molecular level, since no molecular analyses were performed in the first case.
Acknowledgements
The authors would like to thank Hege Kilen Andersen and Nina Øino for their excellent technical assistance. This study was supported by grants from the Norwegian Radium Hospital Foundation.
References
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Katz AJ, Chia VM, Schoonen WM, Kelsh MA. Acute lymphoblastic leukemia: An assessment of international incidence, survival, and disease burden. Cancer Causes Control. 2015;26:1627–1642. doi: 10.1007/s10552-015-0657-6. [DOI] [PubMed] [Google Scholar]
- 3.Harrison CJ, Foroni L. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Rev Clin Exp Hematol. 2002;6:91–113. doi: 10.1046/j.1468-0734.2002.00069.x. 200–202. [DOI] [PubMed] [Google Scholar]
- 4.Hunger SP, Mullighan CG. Redefining ALL classification: Toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125:3977–3987. doi: 10.1182/blood-2015-02-580043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullighan CG. The molecular genetic makeup of acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program. 2012;2012:389–396. doi: 10.1182/asheducation-2012.1.389. [DOI] [PubMed] [Google Scholar]
- 6.Heim S, Mitelman F, editors. Cancer Cytogenetics: Chromosomal and Molecular Genetic Abberations of Tumor Cells. 4th. Wiley-Blackwell; Hoboken, NJ: 2015. [DOI] [Google Scholar]
- 7.Johansson B, Mertens F, Mitelman F. Clinical and biological importance of cytogenetic abnormalities in childhood and adult acute lymphoblastic leukemia. Ann Med. 2004;36:492–503. doi: 10.1080/07853890410018808. [DOI] [PubMed] [Google Scholar]
- 8.Mrózek K, Harper DP, Aplan PD. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Hematol Oncol Clin North Am. 2009;23:991–1010. doi: 10.1016/j.hoc.2009.07.001. v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: Insights and treatment implications. Nat Rev Clin Oncol. 2015;12:344–357. doi: 10.1038/nrclinonc.2015.38. [DOI] [PubMed] [Google Scholar]
- 10.Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340:192–200. doi: 10.1016/j.canlet.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Xia J, Jia P, Pao W, Zhao Z. Application of next generation sequencing to human gene fusion detection: Computational tools, features and perspectives. Brief Bioinform. 2013;14:506–519. doi: 10.1093/bib/bbs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panagopoulos I, Thorsen J, Gorunova L, Micci F, Heim S. Sequential combination of karyotyping and RNA-sequencing in the search for cancer-specific fusion genes. Int J Biochem Cell Biol. 2014;53:462–465. doi: 10.1016/j.biocel.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Schaffer LG, McGowan-Jordan J, Schmid M, editors. ISCN 2013: An International System for Human Cytogenetic Nomenclature. Karger; Basel: 2013. [DOI] [Google Scholar]
- 14.Panagopoulos I, Torkildsen S, Gorunova L, Tierens A, Tjonnfjord GE, Heim S. Comparison between karyotyping- FISH-reverse transcription PCR and RNA-sequencing-fusion gene identification programs in the detection of KAT6A-CREBBP in acute myeloid leukemia. PLoS One. 2014;9:e96570. doi: 10.1371/journal.pone.0096570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Moussavi A Heravi, Senz J, Melnyk N, et al. deFuse: An algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singhal H, Ren YR, Kern SE. Improved DNA electrophoresis in conditions favoring polyborates and lewis acid complexation. PLoS One. 2010;5:e11318. doi: 10.1371/journal.pone.0011318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jun L, Balboni AL, Laitman JT, Bergemann AD. Isolation of DNTNP, which encodes a potential nuclear protein that is expressed in the developing, dorsal neural tube. Dev Dyn. 2002;224:116–123. doi: 10.1002/dvdy.10090. [DOI] [PubMed] [Google Scholar]
- 18.Thermes V, Candal E, Alunni A, Serin G, Bourrat F, Joly JS. Medaka simplet (FAM53B) belongs to a family of novel vertebrate genes controlling cell proliferation. Development. 2006;133:1881–1890. doi: 10.1242/dev.02350. [DOI] [PubMed] [Google Scholar]
- 19.Kizil C, Kuchler B, Yan JJ, Özhan G, Moro E, Argenton F, Brand M, Weidinger G, Antos CL. Simplet/Fam53b is required for Wnt signal transduction by regulating β-catenin nuclear localization. Development. 2014;141:3529–3539. doi: 10.1242/dev.108415. [DOI] [PubMed] [Google Scholar]
- 20.Lento W, Congdon K, Voermans C, Kritzik M, Reya T. Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb Perspect Biol. 2013;5:a008011. doi: 10.1101/cshperspect.a008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. 2007;138:338–348. doi: 10.1111/j.1365-2141.2007.06667.x. [DOI] [PubMed] [Google Scholar]
- 22.Nucifora G, Birn DJ, Espinosa R, III, Erickson P, LeBeau MM, Roulston D, McKeithan TW, Drabkin H, Rowley JD. Involvement of the AML1 gene in the t(3;21) in therapy-related leukemia and in chronic myeloid leukemia in blast crisis. Blood. 1993;81:2728–2734. [PubMed] [Google Scholar]
- 23.Zent CS, Mathieu C, Claxton DF, Zhang DE, Tenen DG, Rowley JD, Nucifora G. The chimeric genes AML1/MDS1 and AML1/EAP inhibit AML1B activation at the CSF1R promoter, but only AML1/MDS1 has tumor-promoter properties. Proc Natl Acad Sci USA. 1996;93:1044–1048. doi: 10.1073/pnas.93.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hromas R, Busse T, Carroll A, Mack D, Shopnick R, Zhang DE, Nakshatri H, Richkind K. Fusion AML1 transcript in a radiation-associated leukemia results in a truncated inhibitory AML1 protein. Blood. 2001;97:2168–2170. doi: 10.1182/blood.V97.7.2168. [DOI] [PubMed] [Google Scholar]
- 25.Ramsey H, Zhang DE, Richkind K, Burcoglu-O'Ral A, Hromas R. Fusion of AML1/Runx1 to copine VIII, a novel member of the copine family, in an aggressive acute myelogenous leukemia with t(12;21) translocation. Leukemia. 2003;17:1665–1666. doi: 10.1038/sj.leu.2403048. [DOI] [PubMed] [Google Scholar]
- 26.Mikhail FM, Coignet L, Hatem N, Mourad ZI, Farawela HM, El Kaffash DM, Farahat N, Nucifora G. A novel gene, FGA7, is fused to RUNX1/AML1 in a t(4;21)(q28;q22) in a patient with T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2004;39:110–118. doi: 10.1002/gcc.10302. [DOI] [PubMed] [Google Scholar]
- 27.Agerstam H, Lilljebjörn H, Lassen C, Swedin A, Richter J, Vandenberghe P, Johansson B, Fioretos T. Fusion gene-mediated truncation of RUNX1 as a potential mechanism underlying disease progression in the 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer. 2007;46:635–643. doi: 10.1002/gcc.20442. [DOI] [PubMed] [Google Scholar]
- 28.Giguère A, Hébert J. CLCA2, a novel RUNX1 partner gene in a therapy-related leukemia with t(1;21)(p22;q22) Cancer Genet Cytogenet. 2010;202:94–100. doi: 10.1016/j.cancergencyto.2010.07.116. [DOI] [PubMed] [Google Scholar]
- 29.Giguère A, Hébert J. Identification of a novel fusion gene involving RUNX1 and the antisense strand of SV2B in a BCR-ABL1-positive acute leukemia. Genes Chromosomes Cancer. 2013;52:1114–1122. doi: 10.1002/gcc.22105. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Perales S, Torres-Ruiz R, Suela J, Acquadro F, Martin MC, Yebra E, Ramirez JC, Alvarez S, Cigudosa JC. Truncated RUNX1 protein generated by a novel t(1;21)(p32;q22) chromosomal translocation impairs the proliferation and differentiation of human hematopoietic progenitors. Oncogene. 2016;35:125–134. doi: 10.1038/onc.2015.70. [DOI] [PubMed] [Google Scholar]
- 31.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: A review. Leuk Res. 2012;36:945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Rasighaemi P, Liongue C, Onnebo SM, Ward AC. Functional analysis of truncated forms of ETV6. Br J Haematol. 2015;171:658–662. doi: 10.1111/bjh.13428. [DOI] [PubMed] [Google Scholar]
- 33.De Braekeleer M, Poon MC, Russell J, Lin CC. A case of acute lymphoblastic leukemia with t(10;19)(q26;q13) Cancer Genet Cytogenet. 1985;16:369–372. doi: 10.1016/0165-4608(85)90247-X. [DOI] [PubMed] [Google Scholar]