Abstract
Gastrointestinal stromal tumors (GISTs) are mesenchymal tumors of the gastrointestinal tract. It is well known that activating mutations in the receptor tyrosine kinases KIT and platelet-derived growth factor receptor-α have essential roles in the pathogenesis of GISTs. The activation of these receptor protein kinases triggers multiple signaling pathways that promote cell proliferation and survival; however, the exact mechanism by which the activation of these kinases promotes the progression of GISTs remains uncertain. The aim of the present was to search for genes that are associated with the progression of GIST. The present study used reverse transcription-quantitative polymerase chain reaction to demonstrate that adenosine monophosphate deaminase 3 (AMPD3) was highly expressed in GISTs. Furthermore, transfection of GIST-T1 cells with KIT-specific small interfering RNA (siRNA) demonstrated that the expression of AMPD3 was dependent on KIT expression, while the depletion of AMPD3 in human GIST-T1 cells using AMPD3-specific siRNA resulted in the suppression of cell migration and invasion. In addition, AMPD3 depletion sensitized GIST-T1 cells to the tyrosine kinase inhibitor imatinib. The results of the present suggested that the combined inhibition of tyrosine kinases and AMPD3 may be effective for the treatment of GISTs.
Keywords: gastrointestinal stromal tumors, KIT, adenosine monophosphate deaminase 3, migration, invasion, imatinib
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors of the gastrointestinal tract, and although the exact incidence of GISTs worldwide is not known, the approximate annual incidence of GISTs in Japan is 1500–2000 cases. GISTs are believed to originate from the interstitial cells of Cajal (ICC) (1). The ICC are electrical pacemakers that regulate the motility of the gastrointestinal tract, and are located around the myenteric plexus and the muscularis propria (1). Approximately 85% of GISTs harbor activating mutations in the KIT receptor tyrosine kinase or platelet-derived growth factor receptor-α (PDGFRA) (2,3). Constitutive activation of these receptor tyrosine kinases induces multiple signaling pathways that are essential for the pathogenesis of GISTs (2,3). For example, oncogenic mutations of KIT promote the activation of various signaling cascades, including the Ras/mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K)/Akt and Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathways, which have been shown to be activated in various tumors (4,5). Imatinib, a potent inhibitor of KIT, is effective for the treatment of unresectable or metastatic GISTs. However, patients with metastatic GISTs eventually develop resistance to imatinib (6); thus, it is necessary to further define the molecular pathogenesis of GISTs, in order to develop alternative therapeutic strategies.
Adenosine monophosphate (AMP) deaminases (AMPDs) catalyze the hydrolytic deamination of AMP to inosine monophosphate, which is a critical step in nucleotide metabolism (7). There are three members of the AMPD family: AMPD1, AMPD2 and AMPD3 (8,9). AMPD1 is exclusively expressed in skeletal muscle, whereas AMPD2 and AMPD3 are ubiquitously expressed. Deficiencies in all three AMPDs have been reported in humans (7), and mutations in AMPD1 were associated with muscle weakness or pain in certain patients (10,11). AMPD2 deficiency is associated with pontocerebellar hypoplasia, which is a rare inherited and progressive neurodegenerative disorder (12). Conversely, individuals with a complete AMPD3 deficiency did not exhibit any significant disorders (13), although elevated levels of adenosine triphosphate (ATP) were detected in the cells of AMPD3-deficient mice (14). Accumulating evidence has suggested that metabolic alterations are associated with the progression of tumor formation (15–19). A recent report suggested that AMPD2 and AMPD3 may be potential targets for cancer treatment (20). The present study aimed to determine the expression and role of AMPD3 in GISTs.
Materials and methods
Patients
A total of 16 GIST samples and 6 normal gastrointestinal tract tissues samples were obtained from 16 patients with GIST who underwent surgery at Nagoya University Hospital (Nagoya, Japan) between September 2008 and April 2014. Informed consent was obtained from all patients. Ethical approval was obtained from the Nagoya University Graduate School of Medicine (Nagoya, Japan). Patients were histologically diagnosed. The clinical characteristics of the patients with GIST are shown in Table I.
Table I.
Clinical characteristics of patients with GISTs.
| Variable | Value |
|---|---|
| Gender | |
| Male | 6 |
| Female | 10 |
| Age (years) | 63 (46–81) |
| Anatomic site of GIST | |
| Stomach | 12 |
| Small intestine | 4 |
| c-KIT exon 11 mutation | |
| Deletion | 6 |
| Insertion | 5 |
| Point mutation | 5 |
| Malignancy | |
| Benign | 8 |
| Malignant | 8 |
| Imatinib treatment | |
| Yes/no | 4/9 |
| Unknown | 3 |
Data are presented as n or median (range). GISTs, gastrointestinal stromal tumors.
Cell lines
The human GIST cell line, GIST-T1, was purchased from Cosmo Bio, Co., Ltd. (Tokyo, Japan). GIST-T1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Wako Pure Chemical Industries, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS; Biowest, Nuaille, France), and penicillin and streptomycin (100 U/ml; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C with 5% CO2 for at least 1 month. SKOV3 (ovarian cancer) and HCT116 (colorectal cancer) cell lines were obtained from the American Type Culture Collection (Manassas VA USA), and KP4 (pancreatic cancer), TE1 (esophagus cancer), KASUMI1 (acute myeloid leukemia), MKN28 (stomach cancer) and QG90 (lung cancer) cell lines were obtained from RIKEN BioResource Center (Tsukuba, Japan). The cells were maintained in DMEM supplemented with 10% FBS and antibiotics.
Small interfering RNA (siRNA) transfection
siRNAs were obtained from Hokkaido System Science, Co., Ltd. (Sapporo, Japan). The sequences of the siRNAs targeting AMPD3 were 5′-CGGGACUUCUAUAACGUGAGA-3′ (siAMPD3-1) and 5′-CCGGAUGGCAUUCCGAUAUGA-3′ (siAMPD3-2). The sequences of the siRNAs used for KIT knockdown were 5′-GACGAGAUUAGGCUGUUAUGC-3′ (siKIT-1) and 5′-GCAUCACGGUGACUUCAAUUA-3′ (siKIT-2). The sequence of the control siRNA that targeted luciferase was 5′-CUUACGCUGAGUACUUCGATT-3′. GIST-T1 cells were transfected with 50 nM siRNAs using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was purified from the GIST and normal gastrointestinal tract tissue samples, the non-transfected cell lines and the siRNA-transfected cells using the RNeasy Mini kit (QIAGEN Benelux B. V., Venlo, The Netherlands). cDNA was synthesized using PrimeScript™ Reverse Transcriptase (Takara Bio Inc., Shiga, Japan). qPCR was performed on a LightCycler® Nano Instrument using a FastStart Essential DNA Green Master kit (Roche Diagnostics, Basel, Switzerland), according to the manufacturer's protocol. Thermal cycling conditions were 95°C for 10 sec and 60°C for 30 sec for 35 cycles. The relative mRNA expression levels were normalized to GAPDH using LightCyclerR® Nano Software 1.0 (Roche Life Science, Tokyo, Japan). The sequences of primers used to amplify each gene were as follows: GAPDH forward, 5′-AGGTGGAGGAGTGGGTGTCGCTGTT-3′ and reverse, 5′-CCGGGAAACTGTGGCGTGATGG-3′; AMPD3 forward, 5′-ACATCCTGGCTCTCATCACC-3′ and reverse, 5′-CAGCAGATGCTTTTGGTTCA-3′; AMPD2 forward, 5′-CGTAGTGCCCCGTATGAGTT-3′ and reverse, 5′-CGAGTCACTGTCCGTCTTCA-3′; and KIT forward, 5′-GAAGTCACCGTGATGCCAGC-3′ and reverse, 5′-CTCTGTCTGCATTGTTCTGTG-3′. RT-minus was used for negative control and three independent experiments were performed.
Western blotting
Total protein was extracted from the non-transfected and siRNA-transfected cell lines using laemmli sample buffer [20% glycerol, 135 mM Tris-HCl (pH 6.8), 4% SDS, 10% 2-Mercaptoethanol and 0.003% BPB]. The protein concentrations of the lysates were measured using the RC-DC Protein assay (Bio-Rad Laboratories, Hercules, CA). Proteins were separated by 0.01% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Following blocking with 0.5% skimmed milk in phosphate-buffered saline (PBS), membranes were incubated with anti-KIT (catalog no., 3392; rabbit polyclonal; 1,000 dilution; Cell Signaling Technology, Inc., Danvers, MA, USA) and anti-β-actin (catalog no., A1978; clone, AC15; mouse monoclonal; 1:5,000 dilution; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) antibodies overnight at 4°C. After washing with Tris-buffered saline plus Tween-20 [20 mM Tris (pH 7.5), 150 mM NaCl and 0.1% Tween 20], the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies, followed by 3,3-diaminobenzidine.
Imatinib sensitivity assay
GIST-T1 cells were transfected with siRNAs, and 48 h later, the cells were treated with different concentrations of imatinib for 48 h. A cell proliferation assay was performed and the ratio of viable cells to control cells (not treated with imatinib) was calculated. Imatinib was obtained from Sigma-Aldrich (Merck Millipore).
Cell proliferation assay
The siRNA-transfected cells were plated onto a 96-well plate at a density of 1,000 cells/well. The day of transfection was set as day 0, and the number of viable cells on days 1, 2, 3 and 4 were assessed using the Cell Counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan).
EdU incorporation assay
EdU incorporation assays were performed using the Click-iT® Plus EdU Alexa Fluor® 594 Imaging kit (Thermo Fisher Scientific, Inc.). The siRNA-transfected cells were incubated with EdU for 24 h and fixed with 4% paraformaldehyde. The cells were permeabilized using 0.5% Triton X-100, and stained with a reaction cocktail (0.5% Triton X-100 in PBS) and Hoechst stain, according to the manufacturer's protocol. Images of the cells were captured using a fluorescence microscope, and the percentage of EdU-positive cells was evaluated.
Invasion assay
The invasion assay was performed using 8-µm pore filters inserted into 24-well Boyden Chambers (Corning Incorporated, Corning, NY, USA). The filter was pre-coated with Matrigel (BD Bioscience, San Jose, CA, USA). GIST-T1 cells (1.5×105) were seeded into the upper chamber and allowed to invade the lower surface of the filter. After 18 h, the cells were fixed with 100% ethanol and stained with 0.5% crystal violet. The number of cells in five randomly selected fields was counted, and three independent experiments were performed.
Migration assay
Cell migration was determined using a wound healing assay and a Boyden chamber assay. For the wound healing assay, confluent monolayers of siRNA-transfected cells were scratched with a 200 µl sterile pipette tip and imaged 24 h later. The distance between the leading edges of the wound was measured in five randomly selected fields, and three independent experiments were performed. To assess cell migration using the Boyden chamber (8 µm pore size and 6.5 mm membrane diameter), siRNA-transfected cells were seeded onto the upper chamber and allowed to migrate to the lower surface of the filter that had been pre-coated with fibronectin. After 12 h, the cells were fixed in 100% ethanol and 0.5% crystal violet. The number of cells in five randomly selected fields was counted, and three independent experiments were performed.
Statistical analysis
Data are expressed as the mean ± standard error. An unpaired t-test was performed to evaluate P-values. The correlation of KIT expression with AMPD3 expression was analyzed by Pearson's correlation analysis using Analyse-it 3.0 (Analyse-it Software, Ltd., Leeds, UK). A difference was considered statistically significant when P<0.05.
Results
AMPD3 is highly expressed in GISTs and is positively correlated with KIT
To obtain further insight into the molecular basis of GISTs, genes whose expression levels were upregulated in GISTs were searched for using the Oncomine database (21). This search suggested that AMPD3 is upregulated in GISTs. The present study examined the expression levels of AMPD1, AMPD2 and AMPD3 in 16 GIST specimens and 6 normal gastrointestinal tissues (2 gastric and 4 colon tissues) using RT-qPCR. As is shown in Fig. 1A, the mRNA expression levels of AMPD3 were significantly increased in the GIST specimens, as compared with the normal tissue specimens (P=0.0002). In addition, there was a significant correlation between KIT and AMPD3 expression levels in the GIST samples (r=0.687; P=0.0047; Fig. 1B). The expression levels of AMPD1 and AMPD2 in the GIST samples were ~10-times lower than that of AMPD3. Similar to AMPD3, increased expression of AMPD2 in GIST specimens was observed by RT-qPCR (P=0.0045; Fig. 1C), whereas AMPD1 expression was not increased in GIST specimens (data not shown). AMPD2 expression was also significantly correlated with KIT expression (r=0.449; P=0.0806; Fig. 1D), although to a lesser extent than AMPD3 and KIT. The present study assessed whether AMPD3 expression was correlated with the malignancy of GISTs; however, there was no significant difference in AMPD3 expression between benign and malignant GISTs (P=0.455; Fig. 1E).
Figure 1.
AMPD3 is highly expressed in GISTs. (A) Box plots show the relative mRNA expression levels of KIT and AMPD3 in normal and GIST specimens. Horizontal lines in the boxes indicate the medians and the boxes extend from the 25th to the 75th percentiles. (B) Pearson correlation analysis detected a positive correlation between the mRNA expression levels of AMPD3 and KIT in GIST specimens (r=0.687; P=0.0047). (C) Box plots show the relative mRNA expression level of AMPD2 in normal and GIST specimens. (D) Pearson correlation analysis detected a positive correlation between the mRNA expression levels of AMPD2 and KIT in GIST specimens (r=0.449; P=0.0806). (E) Box plots show the distribution of AMPD3 expression in benign (n=8) and malignant (n=8) GIST specimens. AMPD, adenosine monophosphate deaminase; GISTs, gastrointestinal stromal tumors.
AMPD3 and KIT regulate the expression of each other's protein
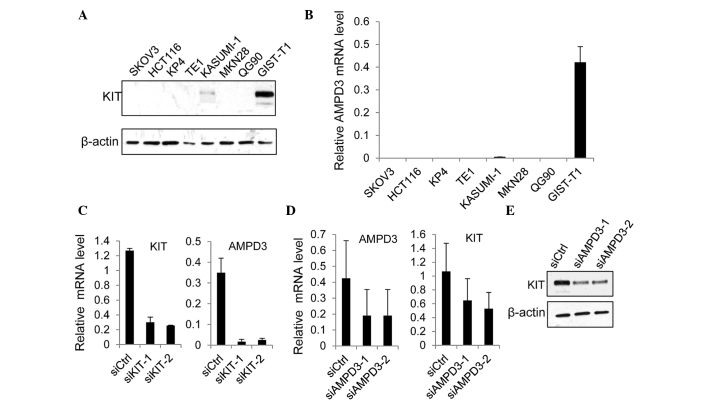
The present study examined whether AMPD3 expression was regulated by KIT in GIST-T1 cells, which were originally established using GISTs from a Japanese woman (22). The GIST-T1 cell line contains heterozygous deletion mutations at 57 bases in exon 11 of the KIT gene (22). Western blot analysis demonstrated that the protein expression levels of KIT were markedly increased in GIST-T1 cells, as compared with the other cancer cell lines examined (Fig. 2A). Notably, the level of AMPD3 mRNA was significantly increased in GIST-T1 cells, as compared with the other cancer cell lines (Fig. 2B). The expression of AMPD2 was also upregulated in GIST-T1 cells, as compared with other cell lines, although its expression was ~10-times lower than AMPD3 expression (data not shown).
Figure 2.
AMPD3 and KIT regulate the expression of each other. (A) The protein expression of KIT in various cancer cell lines was examined by western blotting. (B) The mRNA expression levels of AMPD3 in the different cell lines were evaluated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). (C) GIST-T1 cells were transfected with siCtrl, siKIT-1 or siKIT-2. After 72 h, the mRNA expression levels of KIT and AMPD3 were measured by RT-qPCR. (D) The mRNA expression levels of AMPD3 and KIT in GIST-T1 cells transfected with siCtrl, siAMPD3-1 or siAMPD3-2 were evaluated by RT-qPCR. (E) The protein expression of KIT in AMPD3-depleted GIST-T1 cells was confirmed by western blotting. AMPD, adenosine monophosphate deaminase; siCtrl, control-small interfering RNA (siRNA); siKIT, siRNA targeting KIT; siAMPD3, siRNA targeting AMPD3.
Subsequently, the effect of KIT depletion on AMPD3 expression was assessed. The expression of KIT was suppressed using two different siRNAs, and the level of AMPD3 mRNA was determined using RT-qPCR. Both KIT-specific siRNAs markedly reduced the KIT mRNA level (Fig. 2C). In addition, the level of AMPD3 mRNA was significantly reduced by KIT knockdown (Fig. 2C). Furthermore, whether AMPD3 knockdown using two AMPD3-specific siRNAs was able to affect the level of KIT mRNA in GIST-T1 cells was evaluated using RT-qPCR. As is shown in Fig. 2D and E, AMPD3 knockdown reduced KIT expression at the mRNA and protein level. Notably, depletion of AMPD3 did not affect the level of AMPD2 mRNA (data not shown). These results indicate that KIT and AMPD3 regulate the expression of each other's transcript.
Depletion of AMPD3 reduces the proliferation, migration and invasion of GIST cells
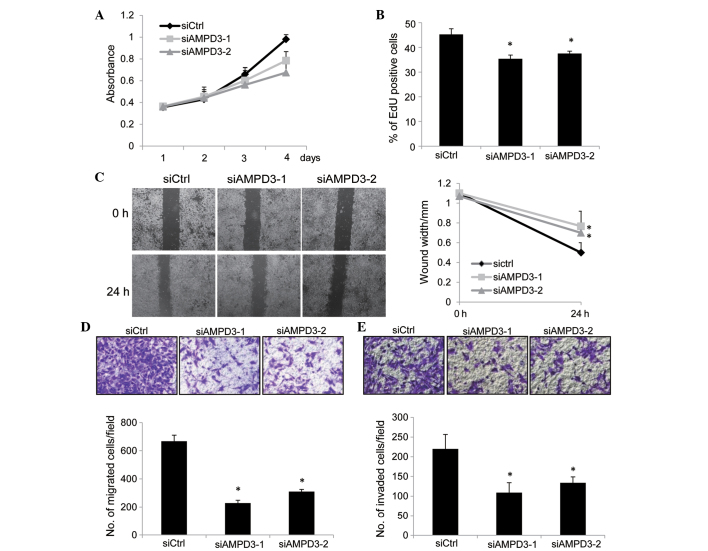
To determine the role of AMPD3 in the malignant characteristics of GIST-T1 cells, the effect of AMPD3 knockdown on cell proliferation was assessed using CCK-8 assays. GIST-T1 cells were transfected with siRNAs, and the number of cells was evaluated at various time points. As is shown in Fig. 3A, the proliferation of AMPD3-knockdown cells was reduced compared with the control-siRNA-transfected cells. To examine whether the suppression of cell proliferation was mediated by the inhibition of cell cycle progression or by the induction of apoptosis, EdU incorporation and TUNEL assays (data not shown) were performed. EdU is a thymidine analog and is incorporated into newly synthesized DNA during the S phase. As is shown in Fig. 3B, depletion of AMPD3 significantly reduced the ratio of EdU-positive cells (siAMPD3-1, P=0.01; siAMPD3-2, P=0.02), whereas apoptosis was not induced by AMPD3 knockdown (data not shown). These results indicate that AMPD3 is associated with the progression of the GIST cell cycle.
Figure 3.
AMPD3 depletion suppressed the proliferation, migration and invasion of gastrointestinal stromal tumor (GIST) cells. (A) GIST-T1 cells were transfected with small interfering RNAs (siRNAs), and the number of viable cells was evaluated using the Cell Counting kit-8 assay. (B) GIST-T1 cells were transfected with siRNAs and, after 48 h, cell proliferation was assessed using the EdU incorporation assay. The graph shows the percentage of EdU-positive cells. Three independent experiments were performed, and the data are presented as the mean ± standard deviation (SD) (*P<0.05). (C) Confluent monolayers of siRNA-transfected cells were scratched, and the distances between the leading edges were measured at 0 and 24 h. Representative images of the migrated cells are shown. The graph shows the average distance of the wound edge at the indicated time point (*P<0.05). (D) siRNA-transfected GIST-T1 cells were subjected to a migration assay. Representative images of the migrated cells are shown (0.5% crystal violet stain), and the graph indicates the average number of migrated cells per field. Three independent experiments were performed, and the data are shown as the mean ± SD (*P<0.05). (E) siRNA-transfected GIST-T1 cells were subjected to a cell invasion assay. Representative images of the invaded cells are shown (0.5% crystal violet stain), and the graph indicates the average number of invaded cells per field. Three independent experiments were performed and data are shown as the mean ± SD (*P<0.05). Original magnification, ×40. AMPD, adenosine monophosphate deaminase 3; siCtrl, control-siRNA; siAMPD3, siRNA targeting AMPD3.
The migration of AMPD3-knockdown cells was evaluated by performing a scratch assay. Confluent monolayers of siRNA-transfected GIST-T1 cells were scratched, and the migration of the cells into the scratch was observed after 24 h. The migration of AMPD3-knockdown cells was delayed compared with the control-siRNA-transfected cells (siAMPD3-1, P=0.008; siAMPD3-2, P=0.002; Fig. 3C). A modified Boyden chamber assay was performed to further confirm this result. The siRNA-transfected cells were placed on the upper surface of the filter and allowed to migrate to the bottom surface, which was coated with fibronectin. Cells that migrated to the bottom surface were counted to evaluate cell migration. Notably, the migration of AMPD3-depleted cells was suppressed compared with the control-siRNA-transfected cells (siAMPD3-1, P=0.008; siAMPD3-2, P=0.002; Fig. 3D). Finally, the invasion of AMPD3-depleted GIST-T1 cells was examined using a Matrigel-coated Boyden chamber. As is shown in Fig. 3E, AMPD3 suppression significantly delayed the invasion of GIST-T1 cells (siAMPD3-1, P=0.0004; siAMDP3-2, P=0.0007).
AMPD3 depletion increases GIST-T1 cell sensitivity to imatinib
The ability of AMPD3 depletion to affect the sensitivity of GIST-T1 cells to imatinib was evaluated. The siRNA-transfected GIST-T1 cells were treated with various concentrations of imatinib, and cell growth was assessed. As shown in Fig. 4, AMPD3-siRNA-transfected cells were more sensitive to imatinib than the control-siRNA-transfected cells.
Figure 4.
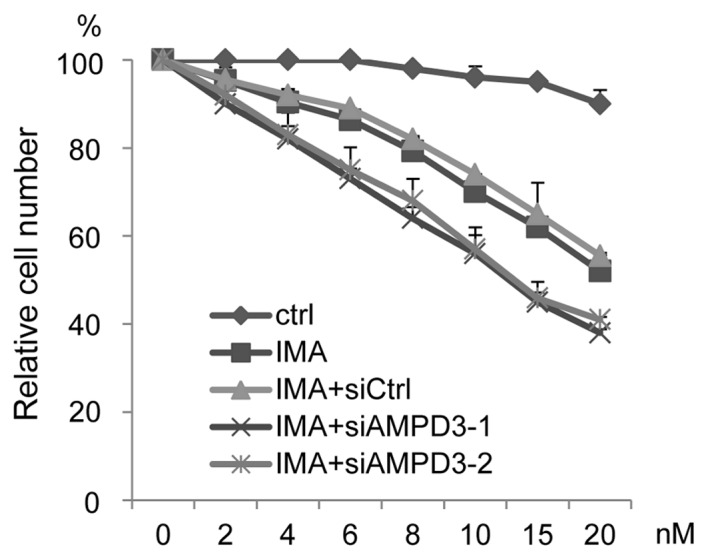
Depletion of AMPD3 sensitizes GIST-T1 cells to imatinib. The siRNA-transfected GIST-T1 cells were cultured in the presence of various concentrations of imatinib, and the proliferation ratio was evaluated. Data are presented as the percent growth of cells treated without imatinib (IMA; imatinib).
Discussion
Previous studies have demonstrated that activating mutations in the receptor tyrosine kinases KIT and PDGFRA are critical for the pathogenesis of GISTs (23–25). Recent studies have shown the important function of ETS variant 1 (ETV1), an ETS family transcriptional factor, in the progression of GISTs, and suggested that targeting both KIT and ETV1 may be effective for the treatment of this type of tumor (26,27). KIT is known to activate multiple pathways, including the RAS/MAPK and PI3K/AKT signaling pathways (28,29); thus, activated KIT may promote the expression of various cancer-associated genes. The present study demonstrated that AMPD3 was significantly upregulated in GIST tissue samples, as compared with normal tissue samples, and that AMPD3 expression was correlated with KIT expression. In addition, KIT depletion was shown to suppress the expression of AMPD3 in GIST-T1 cells at the mRNA level. These results suggested that KIT regulates the expression of AMPD3 in GISTs. Notably, it was also demonstrated that AMPD3 knockdown suppressed KIT expression in GIST-T1 cells. Therefore, KIT and AMPD3 may form a positive feedback loop to promote their expression and the progression of GISTs.
AMPD3 is ubiquitously expressed, and mice with a total AMPD3 deficiency showed increased levels of ATP in their cells (14), indicating that AMPD3 contributes to the energy balance in cells. In the present study, the depletion of AMPD3 in GIST-T1 cells suppressed the cell proliferation, migration and invasion. These results indicated that AMPD3 is associated with the malignant characteristics of GIST-T1 cells. AMPD3 depletion reduced KIT expression; thus, the suppression of malignant characteristics by AMPD3 may be partly mediated by KIT suppression. AMPD3 may also promote cancer progression by contributing to the energy-dependent activation of cancer-associated pathways. For example, AMP-activated protein kinase (AMPK) is known to suppress the progression of various tumors (30–32). Therefore, increases in the levels of AMP as a result of AMPD3 knockdown may promote the activation of AMPK, which can subsequently inhibit anabolic pathways that are essential for cancer cell growth and survival. AMPD3 depletion may promote the activation of protein kinases, such as AMPK, and suppress cancer cell migration and invasion.
In summary, we have shown that the expression levels of KIT and AMPD3 were correlated in GIST-T1 cells, and that the two proteins likely form a positive feedback loop. In addition, it was demonstrated that AMPD3 depletion suppressed the migration and invasion of GIST-T1 cells. Although imatinib is effective in GIST treatment, the drug cannot completely eradicate the tumor. The results of the present study suggested that the combined inhibition of KIT and AMPD3 may be effective for the treatment of GIST.
Acknowledgements
The authors would like to thank the members of the Division of Cancer Biology, Nagoya University Graduate School of Medicine for their helpful discussions and technical assistance. This research was funded by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant no. 24790689), and supported by the Naito Foundation.
References
- 1.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 2.Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA, et al. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- 3.Corless CL, Heinrich MC. Molecular pathobiology of gastrointestinal stromal sarcomas. Annu Rev Pathol. 2008;3:557–586. doi: 10.1146/annurev.pathmechdis.3.121806.151538. [DOI] [PubMed] [Google Scholar]
- 4.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: Origin and molecular oncology. Nat Rev Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 5.Duensing A, Medeiros F, McConarty B, Joseph NE, Panigrahy D, Singer S, Fletcher CD, Demetri GD, Fletcher JA. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumors (GISTs) Oncogene. 2004;23:3999–4006. doi: 10.1038/sj.onc.1207525. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 7.Morisaki T, Sabina RL, Holmes EW. Adenylate deaminase. A multigene family in humans and rats. J Biol Chem. 1990;265:11482–11486. [PubMed] [Google Scholar]
- 8.Mahnke DK, Sabina RL. Calcium activates erythrocyte AMP deaminase [isoform E (AMPD3)] through a protein-protein interaction between calmodulin and the N-terminal domain of the AMPD3 polypeptide. Biochemistry. 2005;44:5551–5559. doi: 10.1021/bi048121p. [DOI] [PubMed] [Google Scholar]
- 9.Mahnke-Zizelman DK, Sabina RL. Cloning of human AMP deaminase isoform E cDNAs. Evidence for a third AMPD gene exhibiting alternatively spliced 5′-exons. J Biol Chem. 1992;267:20866–20877. [PubMed] [Google Scholar]
- 10.Coley W, Rayavarapu S, Pandey GS, Sabina RL, Van der Meulen JH, Ampong B, Wortmann RL, Rawat R, Nagaraju K. The molecular basis of skeletal muscle weakness in a mouse model of inflammatory myopathy. Arthritis Rheum. 2012;64:3750–3759. doi: 10.1002/art.34625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Adel BA, Tarnopolsky MA. Metabolic myopathies: Update 2009. J Clin Neuromuscul Dis. 2009;10:97–121. doi: 10.1097/CND.0b013e3181903126. [DOI] [PubMed] [Google Scholar]
- 12.Akizu N, Cantagrel V, Schroth J, Cai N, Vaux K, McCloskey D, Naviaux RK, Van Vleet J, Fenstermaker AG, Silhavy JL, et al. AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell. 2013;154:505–517. doi: 10.1016/j.cell.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada Y, Goto H, Wakamatsu N, Ogasawara N. A rare case of complete human erythrocyte AMP deaminase deficiency due to two novel missense mutations in AMPD3. Hum Mutat. 2001;17:78. doi: 10.1002/1098-1004(2001)17:1<78::AID-HUMU21>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 14.Cheng J, Morisaki H, Toyama K, Ikawa M, Okabe M, Morisaki T. AMPD3-deficient mice exhibit increased erythrocyte ATP levels but anemia not improved due to PK deficiency. Genes Cells. 2012;17:913–922. doi: 10.1111/gtc.12006. [DOI] [PubMed] [Google Scholar]
- 15.Desman G, Waintraub C, Zippin JH. Investigation of cAMP microdomains as a path to novel cancer diagnostics. Biochim Biophys Acta. 2014;1842:2636–2645. doi: 10.1016/j.bbadis.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu QF, Liu Y, Fan Y, Hua SN, Qu HY, Dong SW, Li RL, Zhao MY, Zhen Y, Yu XL, et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathway. J Hematol Oncol. 2015;8:22. doi: 10.1186/s13045-015-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantano F, Santoni M, Procopio G, Rizzo M, Iacovelli R, Porta C, Conti A, Lugini A, Milella M, Galli L, et al. The changes of lipid metabolism in advanced renal cell carcinoma patients treated with everolimus: A new pharmacodynamic marker? PLoS One. 2015;10:e0120427. doi: 10.1371/journal.pone.0120427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Wang JP, Santen RJ, Yue W. Adenosine monophosphate activated protein kinase (AMPK), a mediator of estradiol-induced apoptosis in long-term estrogen deprived breast cancer cells. Apoptosis. 2015;20:821–830. doi: 10.1007/s10495-015-1111-7. [DOI] [PubMed] [Google Scholar]
- 19.Shieh JM, Chen YC, Lin YC, Lin JN, Chen WC, Chen YY, Ho CT, Way TD. Demethoxycurcumin inhibits energy metabolic and oncogenic signaling pathways through AMPK activation in triple-negative breast cancer cells. J Agric Food Chem. 2013;61:6366–6375. doi: 10.1021/jf4012455. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Wang Y, Shang D, Yu F, Liu W, Zhang Y, Feng C, Wang Q, Xu Y, Liu Y, et al. Characterizing and optimizing human anticancer drug targets based on topological properties in the context of biological pathways. J Biomed Inform. 2015;54:132–140. doi: 10.1016/j.jbi.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taguchi T, Sonobe H, Toyonaga S, Yamasaki I, Shuin T, Takano A, Araki K, Akimaru K, Yuri K. Conventional and molecular cytogenetic characterization of a new human cell line, GIST-T1, established from gastrointestinal stromal tumor. Lab Invest. 2002;82:663–665. doi: 10.1038/labinvest.3780461. [DOI] [PubMed] [Google Scholar]
- 23.Buleje J, Acosta Ó, Guevara-Fujita M, Enriquez Y, Taxa L, Machicado E, Lizaraso-Caparó F, Fujita R. Mutational profile of KIT and PDGFRA genes in gastrointestinal stromal tumors in Peruvian samples. Rev Esp Enferm Dig. 2015;107:72–78. [PubMed] [Google Scholar]
- 24.Comandone A, Boglione A. The importance of mutational status in prognosis and therapy of GIST. Recenti Prog Med. 2015;106:17–22. doi: 10.1701/1740.18950. (In Italian) [DOI] [PubMed] [Google Scholar]
- 25.Joensuu H, Rutkowski P, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Braconi C, Bordoni A, Magnusson MK, et al. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J Clin Oncol. 2015;33:634–642. doi: 10.1200/JCO.2014.57.4970. [DOI] [PubMed] [Google Scholar]
- 26.Ran L, Sirota I, Cao Z, Murphy D, Chen Y, Shukla S, Xie Y, Kaufmann MC, Gao D, Zhu S, et al. Combined inhibition of MAP kinase and KIT signaling synergistically destabilizes ETV1 and suppresses GIST tumor growth. Cancer Discov. 2015;5:304–315. doi: 10.1158/2159-8290.CD-14-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Gu ML, Zhou XX, Ma H, Yao HP, Ji F. Altered expression of ETV1 and its contribution to tumorigenic phenotypes in gastrointestinal stromal tumors. Oncol Rep. 2014;32:927–934. doi: 10.3892/or.2014.3281. [DOI] [PubMed] [Google Scholar]
- 28.Liang J, Wu YL, Chen BJ, Zhang W, Tanaka Y, Sugiyama H. The C-kit receptor-mediated signal transduction and tumor-related diseases. Int J Biol Sci. 2013;9:435–443. doi: 10.7150/ijbs.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu K. Stem cell factor (SCF)-kit mediated phosphatidylinositol 3 (PI3) kinase signaling during mammalian oocyte growth and early follicular development. Front Biosci. 2006;11:126–135. doi: 10.2741/1785. [DOI] [PubMed] [Google Scholar]
- 30.Kim A, Im M, Ma JY. Ethanol extract of Remotiflori radix induces endoplasmic reticulum stress-mediated cell death through AMPK/mTOR signaling in human prostate cancer cells. Sci Rep. 2015;5:8394. doi: 10.1038/srep08394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuster S, Penke M, Gorski T, Gebhardt R, Weiss TS, Kiess W, Garten A. FK866-induced NAMPT inhibition activates AMPK and downregulates mTOR signaling in hepatocarcinoma cells. Biochem Biophys Res Commun. 2015;458:334–340. doi: 10.1016/j.bbrc.2015.01.111. [DOI] [PubMed] [Google Scholar]
- 32.Popovics P, Frigo DE, Schally AV, Rick FG. Targeting the 5′-AMP-activated protein kinase and related metabolic pathways for the treatment of prostate cancer. Expert Opin Ther Targets. 2015;19:617–632. doi: 10.1517/14728222.2015.1005603. [DOI] [PubMed] [Google Scholar]