Abstract
The present study aimed to investigate the association between the methylation status of the reversion-inducing cysteine-rich protein with kazal motifs (RECK) gene and its mRNA expression levels in patients with esophageal squamous cell carcinoma (ESCC). The methylation status of RECK was analyzed by methylation-specific polymerase chain reaction (PCR), and RECK mRNA expression levels were analyzed by quantitative PCR, in 310 paired ESCC tissues. The mean RECK methylation index (MI) was 0.65 in ESCCs and 0.49 in non-tumor samples. There was a significant association between RECK methylation and the American Joint Committee on Cancer stage and lymph node metastasis in ESCC (P<0.0001; P=0.001). The mRNA expression level of RECK was lower in ESCC tissues (mean-∆Cq=−4.66) compared with non-tumor tissues (mean-∆Cq=−2.79), and decreased RECK mRNA expression levels were associated with lymph node metastasis in ESCC. In addition, RECK mRNA levels were decreased in ESCC patients with hypermethylation of the RECK gene (∆MI >0.16; mean-∆∆Cq=−2.85) compared with those with hypomethylation of the RECK gene (∆MI ≤0.16; mean-∆∆Ct=−0.83), and there was a significant difference in the mRNA expression levels of RECK between those with N0–1 and N2–3 lymph node metastasis (P<0.0001). A significant correlation was observed between RECK mRNA expression levels, the MI of RECK and poor postoperative survival (P=0.0003; P<0.0001). The results of the present study suggested that promoter hypermethylation may be an important factor for loss of RECK mRNA expression and may be an indicator of poor survival in ESCC.
Keywords: reversion-inducing cysteine-rich protein with kazal motifs, esophageal squamous cell carcinoma, methylation
Introduction
The incidence of esophageal squamous cell carcinoma (ESCC) shows geographical variation and the highest rates have been reported in China (1). A number of risk factors have been reported for ESCC, including dietary factors, bad behavioral habits, socioeconomic status and genetics (1,2). Research has identified numerous molecular markers, particularly for the early stage of ESCC (1,2). Therefore, it is important to investigate molecular alterations in tumors and precancerous lesions for the early detection or classification of ESCC (1).
The vascular endothelial growth factor (VEGF)-A gene may be an important marker in the development and progression of ESCC in Chinese populations (3). The aberrant methylation of tumor related-genes, such as fragile histidine triad, E-cadherin and integrin-α4, has been reported to be an independent adverse prognostic factor in ESCC (4,5). Clinicopathological and molecular investigations of early squamous cell carcinoma in precancerous lesions and early-stage ESCCs have identified effective biomarkers to predict the risk of patients with dysplasia, the infiltration depth of tumors, and lymph node metastasis and vascular invasion (4–6).
Reversion-inducing cysteine-rich protein with kazal motifs (RECK) is a novel tumor suppressor gene that negatively regulates matrix metalloproteinases (MMPs). RECK is expressed in various normal human tissues, but is downregulated in several types of human tumors and has been positively correlated with the survival of patients with cancer (7–10). Furthermore, RECK is hypothesized to be involved in the maturation of blood vessels, and RECK and cluster of differentiation (CD) 34 (a known vessel marker) exhibited a strong positive correlation in glioma (11). In addition, there is a significant correlation between RECK gene expression and the formation of new blood vessels by VEGF (12), and the expression of RECK was associated with VEGF and CD105, which have an important role in esophageal carcinoma (13).
A previous study reported that hypoxia was able to induce RECK silenced by histone deacetylase (HDAC) 1 and the interaction of hypoxia-inducible factor (HIF)-1α with the reverse HRE2 site in the promoter, and that downregulation of RECK may be a therapeutic and preventive target for tumorigenesis (14). The inhibition of HDAC by small interfering (si)RNA-targeting RECK and trichostatin A successfully restored RECK expression under hypoxic conditions and inhibited the migration and invasion of cancer cells (15). It has been suggested that the binding activity of HIF-1α is important for the activation of the RECK promoter under hypoxic conditions (16). Furthermore, the results of a previous study supported the notion that RECK dsRNA formation in the promoter region has the potential to upregulate RECK gene expression (17).
Gene promoter hypermethylation has been associated with the silencing of tumor-related genes, which is considered the most important epigenetic disruption in numerous tumors (18,19). In a previous study, hypermethylation of the RECK gene was partially reversed by epigallocatechin gallate treatment, and the mRNA expression level of RECK was significantly enhanced in oral squamous cell carcinoma cell lines (20). RAS was able to increase the binding of DNA methyltransferase (DNMT) 3B to the RECK promoter and induce promoter methylation, while treatment with 5′-azacytidine and DNMT3B-targeting siRNA restored RECK expression and potently suppressed cell invasion (21). These results suggested that RECK methylation may be an important regulatory mechanism of RECK expression in ESCC. However, the molecular mechanism underlying this downregulation and its biological significance in ESCC remain unclear. Furthermore, no studies have investigated RECK methylation and RECK mRNA expression in ESCC. The present study aimed to investigate the relationship between RECK mRNA expression and promoter methylation in ESCC.
Materials and methods
Patients and tissue samples
Tumor and non-tumor samples were obtained by surgical resection from 310 ESCC patients between May 2001 and June 2014. These specimens were collected from Changzhou Cancer Hospital and Nanyang Center Hospital in China. The matched non-tumor tissues were obtained from >3 cm away from the tumors and were confirmed to be tumor-free by microscopic examination. The tissues were maintained at −196°C until processing for RNA/DNA extraction. The patients included 180 men and 130 women, and ranged in age from 36–82 years (mean age, 52.47±12.43 years). In total, 88 cases were well-differentiated, 142 were moderately-differentiated and 80 were poorly-differentiated. The present study was approved by the committee on Human Experimentation of Soochow University (Changzhou, China). Written informed consent was obtained from all patients.
RECK promoter methylation analysis
Tissue DNA was isolated using the Commercial QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany). Bisulfite modification of the DNA was performed using the EpiSeeker DNA Purification and Modification kit (Abcam, Cambridge, MA, USA) as described previously (22), and RECK methylation was measured by methylation-specific polymerase chain reaction (PCR) (19). Unmethylated RECK primers were as follows: Forward, 5′-TAAAGAGTTTTGGTATGGGGTATGT-3′ and reverse, 5′-CTCCAAACCACAAAATACTCAAA-3′. Methylated RECK primers were: Forward, 5′-AATAAAGAGTTTTGGTACGGGGTAC-3′ and reverse, 5′-AAAACCGCGAAATACTCGAA-3′. Modified DNA samples (2 µl) were used for PCR amplification. PCR was performed in a thermal cycler for 40 cycles. The conditions for PCR were denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for 30 sec. The PCR products were separated on 2% agarose gels and visualized by ethidium bromide staining. The methylation index (MI) of RECK was calculated using the following formula: MI = 100 × methylated reaction / (unmethylated reaction + methylated reaction) (23). ΔMI was defined as MIESCC - MINon-tumor.
RECK mRNA quantitative analysis
Total RNA was isolated from 310 ESCC and adjacent normal tissues using TRIzol reagent (Takara Biotechnology Co., Ltd., Dalian, China). First-strand cDNA was synthesized from 2 µg total RNA using PrimeScript™ RT Reagent kit (Takara Biotechnology Co., Ltd.). RECK mRNA primers were as follows: Forward, 5′-CCTCAGTGAGCACAGTTCAGA-3′ and reverse, 5′-GCAGCACACACACTGCTGTA-3′ (19). Quantitative PCR (qPCR) was performed using 20 µl SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) on the Mx3000P QPCR System (Agilent Technologies, Inc., Santa Clara, CA, USA). qPCR was performed for 40 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 1 min. β-actin mRNA was amplified from the same cDNA samples as an internal control. All results were normalized to β-actin. The relative RECK expression level was determined using the comparative Cq method (24), using average Cq values for RECK and β-actin.
Statistical analysis
The clinical data, MI and mRNA expression levels were analyzed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). Comparisons were performed using Pearson's χ2 test, or unpaired or paired t-tests. All P-values are two-sided, and a P-value of <0.05 was considered statistically significant. Survival curves were based on Kaplan-Meier estimates.
Results
MI of the RECK gene in tissues
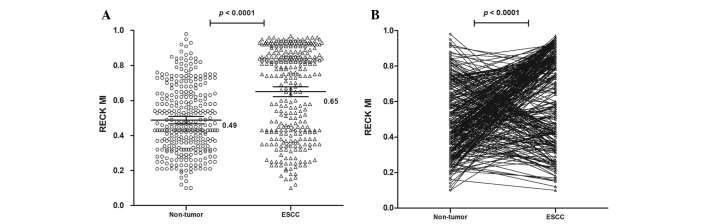
The methylation status of the RECK gene was analyzed in 310 ESCC and non-tumor tissues. The methylated PCR product contains methylated and/or unmethylated PCR products (25). It was observed that the mean MI of the RECK gene was 0.65 [95% confidence interval (CI)=0.62 to 0.68] in ESCC and 0.49 (95% CI=0.47 to 0.51) in non-tumor samples, which was significantly different (P<0.0001; Fig. 1). These results suggest that the RECK promoter is hypermethylated in ESCC compared with non-tumor samples.
Figure 1.
RECK promoter hypermethylation was observed in ESCC patients. (A) The mean MI of the RECK promoter in ESCC and corresponding non-tumor tissues. (B) The RECK MI showed a decreasing trend in non-tumor tissues compared with ESCC tissues. Statistical analyses were performed using the paired t-test. RECK, reversion-inducing cysteine-rich protein with kazal motif; MI, methylation index; ESCC, esophageal squamous cell carcinoma.
RECK methylation and clinicopathological features of ESCC
Associations between the RECK methylation level and the clinical characteristics of ESCC patients, including age, gender, American Joint Committee on Cancer (AJCC) stage, differentiation and others, were analyzed (Table I). A cutoff value of 0.16 was set for ΔMI and the patients were classified according to the mean MI of RECK in ESCC and non-tumor tissues. There was a significant difference in the AJCC stage (P<0.0001) and the stage of lymph node metastasis (P=0.001) between cases where ΔMI ≤0.16 and ΔMI >0.16. These results suggest that RECK hypermethylation may occur more frequently in ESCC patients with advanced tumors and lymph node metastasis.
Table I.
Correlation of clinicopathological variables with RECK MI in esophageal squamous cell carcinoma.
| Variable | n | ΔMI ≤0.16 (n=151) | ΔMI >0.16 (n=159) | Odds ratio (95% CI) | P-valuea |
|---|---|---|---|---|---|
| Gender | 0.092 | ||||
| Male | 180 | 95 | 85 | 1.48 (0.94–2.33) | |
| Female | 130 | 56 | 74 | ||
| Age (years) | 0.965 | ||||
| ≥60 | 170 | 83 | 87 | 1.01 (0.65–1.58) | |
| <60 | 140 | 68 | 72 | ||
| Size (cm) | 0.387 | ||||
| <3 | 172 | 80 | 92 | 0.82 (0.52–1.29) | |
| ≥3 | 138 | 71 | 67 | ||
| Tobacco | 0.062 | ||||
| Yes | 168 | 90 | 78 | 1.53 (0.98–2.40) | |
| No | 142 | 61 | 81 | ||
| Alcohol | 0.056 | ||||
| Yes | 147 | 80 | 67 | 1.55 (0.99–2.42) | |
| No | 163 | 71 | 92 | ||
| Depth of invasion | 0.079 | ||||
| T1–2 | 224 | 108 | 116 | 0.93 (0.57–1.53) | |
| T3–4 | 86 | 43 | 43 | ||
| AJCC stage | <0.0001b | ||||
| I–II | 125 | 78 | 47 | 2.55 (1.60–4.06) | |
| III–IV | 185 | 73 | 112 | ||
| Lymph node metastasis | 0.001b | ||||
| N0–1 | 148 | 87 | 61 | 2.18 (1.39–3.44) | |
| N2–3 | 162 | 64 | 98 | ||
| Distant metastasis | 0.513 | ||||
| M0 | 251 | 120 | 131 | 0.83 (0.47–1.46) | |
| M1 | 59 | 31 | 28 | ||
| Differentiation | 0.806 | ||||
| G1 | 88 | 41 | 47 | ||
| G2 | 142 | 72 | 70 | ||
| G3 | 80 | 38 | 42 |
Comparisons were performed using Pearson's χ2 test.
P<0.05. RECK, reversion-inducing cysteine-rich protein with kazal motif; MI, methylation index; CI, confidence interval; AJCC, American Joint Committee on Cancer.
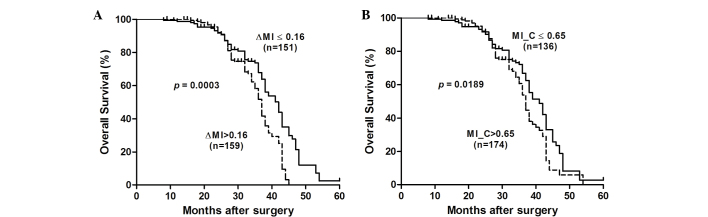
Subsequently, associations between the RECK MI and postoperative outcomes were assessed using the Kaplan-Meier method and log-rank test (Fig. 2). The results suggested that the median cumulative survival time was significantly shorter for ESCC patients with ΔMI >0.16 compared with those with ΔMI ≤0.16 [37 vs. 42 months; log-rank P=0.0003; hazard ratio (HR)=1.896 (95% CI=1.34 to 2.68)]. Furthermore, it was observed that ESCC patients with an MI of >0.65 had a significantly shorter median cumulative survival time compared with those with an MI of ≤0.65 [37 vs. 41 months; log-rank P=0.0189; HR=1.484 (95% CI=1.07 to 2.06)]. These results suggest that the RECK MI may be a good prognostic marker in ESCC.
Figure 2.
RECK methylation is associated with a poor prognosis in patients with ESCC. (A) Kaplan-Meier analysis of survival after surgery according to the RECK MI. Hypermethylation of RECK (∆MI ≥0.16) was significantly correlated with a poor overall postoperative survival in ESCC patients. (B) The MI of RECK in tumor tissues was significantly correlated with a poor overall survival after surgery. ∆MI, MIESCC-MInon-tumor; MI_C, methylation index of RECK in tumor tissues; RECK, reversion-inducing cysteine-rich protein with kazal motif; ESCC, esophageal squamous cell carcinoma.
RECK mRNA expression level in tissues
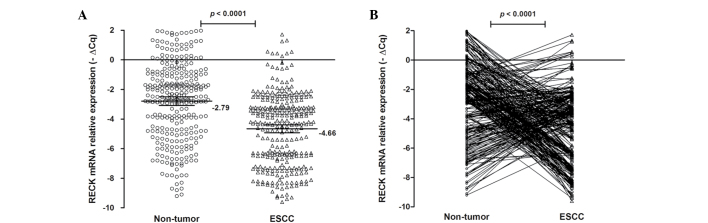
qPCR was performed to quantify the relative mRNA expression levels of RECK in 310 ESCC and matched non-tumor tissues. The RECK mRNA expression levels are shown in Fig. 3. The mean-∆Cq RECK mRNA expression level was −4.66 (95% CI=−4.92 to −4.39) in ESCC tissues and −2.79 (95% CI=−3.08 to −2.50) in non-tumor tissues. There was a significant difference between ESCC tissues and matching non-tumor tissues (P<0.0001; Fig. 3). Furthermore, the RECK mRNA expression level was increased (−∆∆Cq >0) in 130 patients (41.94%), but decreased (−∆∆Cq ≤0) in 150 patients (58.06%). These results suggest that RECK mRNA expression may be an important factor for ESCC.
Figure 3.
mRNA expression of RECK in ESCC and non-tumor tissues. (A) The mRNA expression level of RECK in ESCC and non-tumor samples was determined by reverse transcription-quantitative polymerase chain reaction. Data are shown as the mean-∆Cq and 95% confidence interval. The RECK mRNA expression level in ESCC was lower compared with the matched non-tumor tissues (P<0.0001). (B) There was a decreased tendency for RECK expression in non-tumor tissues compared with ESCC tissues. Statistical analyses were performed using the paired t-test. RECK, reversion-inducing cysteine-rich protein with kazal motif; ESCC, esophageal squamous cell carcinoma.
RECK mRNA is associated with the clinical features of ESCC
The relationship between the mRNA expression level of RECK and clinical factors was evaluated for 310 tumor tissues. These analyses are summarized in Table II. There was a significant difference in the mRNA expression levels of RECK between ESCC patients with N0–1 and N2–3 stages of lymph node metastasis. A lower mRNA expression level of RECK (−ΔΔCq ≤0) was observed in 67/148 (43.24%) ESCC patients with N0–1 lymph node metastasis and 113/162 (69.75%) ESCC patients with N2–3 lymph node metastasis (P<0.0001).
Table II.
Correlation of clinicopathological variables with RECK mRNA expression in esophageal squamous cell carcinoma.
| Variable | n | −ΔΔCqRECK >0 (n=130) | −ΔΔCqRECK ≤0 (n=180) | Odds ratio (95% CI) | P-valuea |
|---|---|---|---|---|---|
| Gender | 0.292 | ||||
| Male | 180 | 80 | 100 | 1.28 (0.81–2.03) | |
| Female | 130 | 50 | 80 | ||
| Age (years) | 0.221 | ||||
| ≥60 | 170 | 66 | 104 | 0.75 (0.48–1.19) | |
| <60 | 140 | 64 | 76 | ||
| Size (cm) | 0.012 | ||||
| <3 | 172 | 83 | 89 | 1.81 (1.14–2.87) | |
| ≥3 | 138 | 47 | 91 | ||
| Tobacco | 0.571 | ||||
| Yes | 168 | 68 | 100 | 0.88 (0.59–1.38) | |
| No | 142 | 62 | 80 | ||
| Alcohol | 1.000 | ||||
| Yes | 147 | 65 | 82 | 1.0 (0.64–1.56) | |
| No | 163 | 65 | 98 | ||
| Depth of invasion | 0.296 | ||||
| T1–2 | 224 | 98 | 126 | 1.31 (0.79–2.19) | |
| T3–4 | 86 | 32 | 54 | ||
| AJCC stage | 0.545 | ||||
| I–II | 125 | 55 | 70 | 1.15 (0.73–1.82) | |
| III–IV | 185 | 75 | 110 | ||
| Lymph node metastasis | <0.0001b | ||||
| N0–1 | 148 | 85 | 67 | 3.19 (1.99–5.10) | |
| N2–3 | 162 | 45 | 113 | ||
| Distant metastasis | 0.421 | ||||
| M0 | 251 | 108 | 143 | 1.27 (0.71–2.28) | |
| M1 | 59 | 22 | 37 | ||
| Differentiation | 0.533 | ||||
| G1 | 88 | 36 | 52 | ||
| G2 | 142 | 64 | 78 | ||
| G3 | 80 | 30 | 50 |
Comparisons were performed using Pearson's χ2 test.
P<0.05. RECK, reversion-inducing cysteine-rich protein with kazal motif; MI, methylation index; CI, confidence interval; AJCC, American Joint Committee on Cancer.
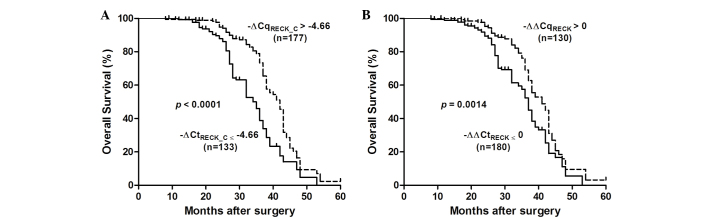
The association between RECK mRNA expression and survival in patients with ESCC was assessed using the Kaplan-Meier method and log rank test (Fig. 4). Decreased RECK mRNA expression (−∆∆Cq <0) was significantly correlated with a poor overall survival [37 vs. 41 months; P=0.0014; HR=0.586 (95% CI=0.42 to 0.81)]. In addition, ESCC patients with -∆CqRECK ≤-4.66 had a shorter median cumulative survival time compared with those with -∆CqRECK >-4.66 [35 vs. 42 months; log-rank P<0.0001; HR=0.379 (95% CI=0.26 to 0.55)]. These results suggest that RECK mRNA silencing may have an important role in the poor survival of patients with ESCC.
Figure 4.
RECK mRNA expression is associated with a poor prognosis in patients with ESCC. (A) Kaplan-Meier analysis of survival after surgery according to the RECK mRNA expression level. The decreased expression of RECK (−∆∆Cq ≤0) was significantly correlated with a poor overall postoperative survival in ESCC patients. (B) The decreased expression of RECK (−∆Cq ≤-4.66) in tumor tissues was significantly correlated with a poor overall postoperative survival. -∆∆Cq, -(∆CqESCC - ∆CqNon-tumor); -∆Cq, -(CqESCC - Cqβ-actin); RECK, reversion-inducing cysteine-rich protein with kazal motif; ESCC, esophageal squamous cell carcinoma.
Decreased RECK mRNA expression and RECK hypermethylation in ESCC
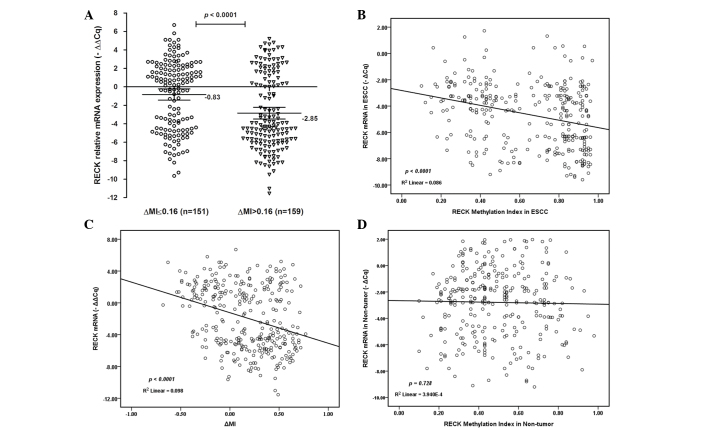
Subsequently, the association between the MI and mRNA expression of RECK was investigated (Fig. 5). Notably, the mRNA expression levels of RECK were lower in ESCC tissues with ΔMI >0.16 [mean-∆∆Cq =−2.85; 95% CI=−3.48 to 2.21) compared with ESCC tissues with ΔMI ≤0.16 [mean-∆∆Cq =−0.83; 95% CI=−1.43 to −0.23; P<0.0001; Fig. 5A]. There was a decreasing tendency for RECK mRNA expression from promoter hypermethylation to hypomethylation in ESCC tissues. A decreased RECK mRNA expression level was significantly associated with the demethylation status of RECK in ESCC patients (R2=0.086; P<0.0001; Fig. 5B; R2=0.098; P<0.0001; Fig. 5C). However, there was no significant association between the demethylation status of RECK and mRNA expression in non-tumor tissues (R2=0.00039; P=0.728; Fig. 5D). These results suggest that promoter hypermethylation may be an important factor for RECK silencing in ESCC.
Figure 5.
RECK mRNA expression and promoter methylation in patients with ESCC. (A) RECK mRNA expression was lower in ESCC patients with ∆MI >0.16 compared with those with ∆MI ≤0.16. Statistical analyses were performed using the unpaired t-test. (B) The scatter plot shows that the mRNA expression level of RECK (−∆Cq) was significantly associated with the MI in ESCC tissues. (C) The scatter plot shows that RECK mRNA expression (−∆∆Cq) was significantly associated with ∆MI in ESCC patients. (D) The scatter plot shows that the mRNA expression level of RECK (−∆Cq) was not significantly associated with the MI in non-tumor tissues. Statistical analyses were performed using Pearson's correlation coefficient test. -∆∆Cq, -(∆CqESCC - ∆CqNon-tumor); ∆MI, MIESCC - MINon-tumor; MI, methylation index; RECK, reversion-inducing cysteine-rich protein with kazal motif; ESCC, esophageal squamous cell carcinoma.
Discussion
RECK, as a tumor-related gene, has an important role in regulating the invasion and metastasis of tumor cells. RECK is widely expressed in normal tissues, but is significantly reduced in tumor tissues (26). In a previous study, the RECK expression level was inversely correlated with the MMP9 expression level in nasopharyngeal carcinoma (27). Furthermore, there was a significant association between the positive expression of RECK and that of MMP2 in adenoid cystic carcinoma (28). These findings suggest that RECK has an important role in regulating the proliferation and migration of normal epithelial cells and carcinoma cells (29). Therefore, understanding RECK expression is useful for delineating the molecular basis of malignancies.
A previous study detected RECK methylation in 27.5% of adjacent normal mucosa samples and 47.5% of gastric cancer samples (30). Non-small cell lung cancer (NSCLC) patients with RECK methylation exhibited lower RECK mRNA expression compared with patients with promoter hypomethylation (31). RECK methylation was detected in 63.6% (35/55) of lung cancer specimens, and the methyltransferase inhibitor 5′-azacytidine was shown to upregulate RECK expression and reduce the invasive ability of NSCLC cells (17). In the present study, the mean MI of the RECK promoter was 0.65 in ESCC and 0.49 in non-tumor samples. Furthermore, there was a significant association between the MI of RECK and AJCC stage and lymph node metastasis in ESCC patients, with ESCC patients with RECK hypomethylation tending to show better survival.
The present study demonstrated that RECK mRNA expression was lower in ESCC tissues compared with non-tumor tissues. In addition, patients with lymph node metastasis showed a lower RECK mRNA expression level, and ESCC patients with high RECK mRNA expression showed better survival. Therefore, RECK silencing may have an important role in the pathogenesis of ESCC. In the present study, the mRNA expression level of RECK was lower in ESCC patients with hypermethylation (∆MI >0.16; mean-∆∆Cq =−2.85) compared with those with hypomethylation (∆MI ≤0.16; mean-∆∆Cq=−0.83), and there was a decreased tendency for RECK mRNA expression in ESCC patients with promoter hypermethylation (P<0.0001). These results suggested that hypermethylation of RECK gene may lead to RECK silencing in ESCC, and RECK expression could be regulated by DNA methylation in ESCC.
In conclusion, RECK methylation was frequently observed in ESCC and was associated with the downregulation of its mRNA expression, which was significantly correlated with a poor survival in ESCC. Further studies are required to elucidate the detailed mechanism of promoter methylation and RECK mRNA expression in ESCC.
Acknowledgements
This study was supported by the Natural Science Foundation of Jiangsu (grant nos. BL2013012 and BE2016656), the Changzhou Sci&Tech Program, China (grant nos. CE20155043 and CJ20159023), the High-level Health Talents of Changzhou City grant (nos. 2016CZLJ021 and 2016CZLJ009) and the Health Talents Project for Jiangsu, China (grant no. RC2016038).
References
- 1.Shang L, Wang M. Molecular alterations and clinical relevance in esophageal squamous cell carcinoma. Front Med. 2013;7:401–410. doi: 10.1007/s11684-013-0286-y. [DOI] [PubMed] [Google Scholar]
- 2.Aghcheli K, Marjani HA, Nasrollahzadeh D, Islami F, Shakeri R, Sotoudeh M, Abedi-Ardekani B, Ghavamnasiri MR, Razaei E, Khalilipour E, et al. Prognostic factors for esophageal squamous cell carcinoma-a population-based study in Golestan Province, Iran, a high incidence area. PloS one. 2011;6:e22152. doi: 10.1371/journal.pone.0022152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golozar A, Beaty TH, Gravitt PE, Ruczinski I, Qiao YL, Fan JH, Ding T, Tang ZZ, Etemadi A, Hu N, et al. Oesophageal squamous cell carcinoma in high-risk Chinese populations: Possible role for vascular epithelial growth factor A. Eur J Cancer. 2014;50:2855–2865. doi: 10.1016/j.ejca.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee EJ, Lee BB, Kim JW, Shim YM, Hoseok I, Han J, Cho EY, Park J, Kim DH. Aberrant methylation of fragile histidine triad gene is associated with poor prognosis in early stage esophageal squamous cell carcinoma. Eur J Cancer. 2006;42:972–980. doi: 10.1016/j.ejca.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Lee EJ, Lee BB, Han J, Cho EY, Shim YM, Park J, Kim DH. CpG island hypermethylation of E-cadherin (CDH1) and integrin alpha4 is associated with recurrence of early stage esophageal squamous cell carcinoma. Int J Cancer. 2008;123:2073–2079. doi: 10.1002/ijc.23598. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu M, Zaninotto G, Nagata K, Graham DY, Lauwers GY. Esophageal squamous cell carcinoma with special reference to its early stage. Best Pract Res Clin Gastroenterol. 2013;27:171–186. doi: 10.1016/j.bpg.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Tseng SH. The potential of RECK inducers as antitumor agents for glioma. Anticancer Res. 2012;32:2991–2998. [PubMed] [Google Scholar]
- 8.Clark JC, Thomas DM, Choong PF, Dass CR. RECK-a newly discovered inhibitor of metastasis with prognostic significance in multiple forms of cancer. Cancer Metastasis Rev. 2007;26:675–683. doi: 10.1007/s10555-007-9093-8. [DOI] [PubMed] [Google Scholar]
- 9.Masui T, Doi R, Koshiba T, Fujimoto K, Tsuji S, Nakajima S, Koizumi M, Toyoda E, Tulachan S, Ito D, et al. RECK expression in pancreatic cancer: Its correlation with lower invasiveness and better prognosis. Clin Cancer Res. 2003;9:1779–1784. [PubMed] [Google Scholar]
- 10.Mao X, Liu L, Zhang B, Zhang D. Reversion-inducing cysteine-rich protein with Kazal motifs gene expression and its clinical significance in peripheral T-cell lymphoma. Oncology Lett. 2013;5:1867–1871. doi: 10.3892/ol.2013.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahmah NN, Sakai K, Sano K, Hongo K. Expression of RECK in endothelial cells of glioma: Comparison with CD34 and VEGF expressions. J Neurooncol. 2012;107:559–564. doi: 10.1007/s11060-011-0778-z. [DOI] [PubMed] [Google Scholar]
- 12.Alexius-Lindgren M, Andersson E, Lindstedt I, Engstrom W. The RECK gene and biological malignancy-its significance in angiogenesis and inhibition of matrix metalloproteinases. Anticancer Res. 2014;34:3867–3873. [PubMed] [Google Scholar]
- 13.Li SL, Gao DL, Zhao ZH, Liu ZW, Zhao QM, Yu JX, Chen KS, Zhang YH. Correlation of matrix metalloproteinase suppressor genes RECK, VEGF and CD105 with angiogenesis and biological behavior in esophageal squamous cell carcinoma. World J Gastroenterol. 2007;13:6076–6081. doi: 10.3748/wjg.13.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KJ, Lee KY, Lee YM. Downregulation of a tumor suppressor RECK by hypoxia through recruitment of HDAC1 and HIF-1alpha to reverse HRE site in the promoter. Biochim Biophys Acta. 2010;1803:608–616. doi: 10.1016/j.bbamcr.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Jeon HW, Lee YM. Inhibition of histone deacetylase attenuates hypoxia-induced migration and invasion of cancer cells via the restoration of RECK expression. Mol Cancer Ther. 2010;9:1361–1370. doi: 10.1158/1535-7163.MCT-09-0717. [DOI] [PubMed] [Google Scholar]
- 16.Jeon HW, Lee KJ, Lee SH, Kim WH, Lee YM. Attenuated expression and function of the RECK tumor suppressor under hypoxic conditions is mediated by the MAPK signaling pathways. Arch Pharm Res. 2011;34:137–145. doi: 10.1007/s12272-011-0116-1. [DOI] [PubMed] [Google Scholar]
- 17.Chang HC, Cho CY, Hung WC. Downregulation of RECK by promoter methylation correlates with lymph node metastasis in non-small cell lung cancer. Cancer Sci. 2007;98:169–173. doi: 10.1111/j.1349-7006.2006.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, Cao L. DNA methylation, its mediators and genome integrity. Int J Biol Sci. 2015;11:604–617. doi: 10.7150/ijbs.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Ling Y, Zhang C, Xu Y, Gao L, Li R, Zhu J, Fan L, Wei L. The silencing of RECK gene is associated with promoter hypermethylation and poor survival in hepatocellular carcinoma. Int J Biol Sci. 2012;8:451–458. doi: 10.7150/ijbs.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato K, Long NK, Makita H, Toida M, Yamashita T, Hatakeyama D, Hara A, Mori H, Shibata T. Effects of green tea polyphenol on methylation status of RECK gene and cancer cell invasion in oral squamous cell carcinoma cells. Br J Cancer. 2008;99:647–654. doi: 10.1038/sj.bjc.6604521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang HC, Cho CY, Hung WC. Silencing of the metastasis suppressor RECK by RAS oncogene is mediated by DNA methyltransferase 3b-induced promoter methylation. Cancer Res. 2006;66:8413–8420. doi: 10.1158/0008-5472.CAN-06-0685. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Xu Y, Zhao J, Fan L, Jiang G, Li R, Ling Y, Wu M, Wei L. Elevated expression of the stem cell marker CD133 associated with Line-1 demethylation in hepatocellular carcinoma. Ann Surg Oncol. 2011;18:2373–2380. doi: 10.1245/s10434-011-1599-1. [DOI] [PubMed] [Google Scholar]
- 23.Bais AJ, Gardner AE, McKenzie OL, Callen DF, Sutherland GR, Kremmidiotis G. Aberrant CBFA2T3B gene promoter methylation in breast tumors. Mol Cancer. 2004;3:22. doi: 10.1186/1476-4598-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Guo X, Zhang L, Lu Z, Ma N, Cheng Y, Shen F, Zhang B, Wu M, Wei L. Methylation-related silencing of p14ARF gene correlates with telomerase activity and mRNA expression of human telomerase reverse transcriptase in hepatocellular carcinoma. J Surg Oncol. 2008;98:462–468. doi: 10.1002/jso.21131. [DOI] [PubMed] [Google Scholar]
- 26.Noda M, Takahashi C, Matsuzaki T, Kitayama H. What we learn from transformation suppressor genes: Lessons from RECK. Future Oncol. 2010;6:1105–1116. doi: 10.2217/fon.10.80. [DOI] [PubMed] [Google Scholar]
- 27.Zhou DN, Deng YF, Li RH, Yin P, Ye CS. Concurrent alterations of RAGE, RECK, and MMP9 protein expression are relevant to Epstein-Barr virus infection, metastasis and survival in nasopharyngeal carcinoma. Int J Clin Exp Pathol. 2014;7:3245–3254. [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Huang S, Jiang L, Zhang S, Li W, Chen Z, Zhang D. Expression of RECK and MMP-2 in salivary adenoid cystic carcinoma: Correlation with tumor progression and patient prognosis. Oncology Lett. 2014;7:1549–1555. doi: 10.3892/ol.2014.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuki K, Yoshida Y, Inagaki R, Hiai H, Noda M. E-cadherin-downregulation and RECK-upregulation are coupled in the non-malignant epithelial cell line MCF10A but not in multiple carcinoma-derived cell lines. Sci Rep. 2014;4:4568. doi: 10.1038/srep04568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du YY, Dai DQ, Yang Z. Role of RECK methylation in gastric cancer and its clinical significance. World J Gastroenterol. 2010;16:904–908. doi: 10.3748/wjg.v16.i7.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesta M, Kulda V, Topolcan O, Safranek J, Vrzalova J, Cerny R, Holubec L. Significance of methylation status and the expression of RECK mRNA in lung tissue of patients with NSCLC. Anticancer Res. 2009;29:4535–4539. [PubMed] [Google Scholar]