Abstract
Kin17 DNA and RNA binding protein (Kin17) is a highly conserved protein that participates in DNA replication, DNA repair and cell cycle progression. Recently, the tumor-promoting function of Kin17 has been demonstrated and increasingly studied. In the present study, the role of Kin17 in the invasion and metastasis of non-small cells lung cancer (NSCLC) was investigated. Elevated Kin17 mRNA and protein expression was identified in a total of 97 NSCLC and benign lung lesion tissue specimens. Kin17 overexpression was significantly correlated with high tumor grade and lymph node metastasis, indicating poor patient prognosis. Scratch and Transwell assays demonstrated that the knockdown of KIN17 inhibited the ability of NSCLC cells to migrate and invade. Furthermore, reverse transcription-quantitative polymerase chain reaction and western blot analyses confirmed that knockdown of KIN17 decreased the expression of matrix metalloproteinase 7, epidermal growth factor receptor and v-myc avian myelocytomatosis viral oncogene homolog. The results of the present study indicate that Kin17 is markedly overexpressed in NSCLC tissues compared with benign lung lesion and peritumoral tissue. The upregulation of KIN17 may serve an important role in the metastasis of NSCLC cells. These results indicate that Kin17 is a novel diagnostic and prognostic biomarker of NSCLC, in addition to being a potential therapeutic target for the treatment of patients with NSCLC.
Keywords: Kin17 DNA and RNA binding protein, non-small cell lung cancer, expression, invasiveness
Introduction
Lung cancer is one of the most common types of cancer and the leading cause of cancer-associated mortality (1). It is estimated that worldwide, 1.8 million lung cancers are diagnosed annually (1). Non-small cell lung cancer (NSCLC) accounts for 80% of all lung cancer cases, with a 5-year survival rate of 16% (2). The poor prognosis of patients with NSCLC is primarily due to cancer metastasis and recurrence. Therefore, the identification of novel diagnostic markers and therapeutic targets is warranted for the early detection and inhibition of metastasis in patients with NSCLC.
Kin17 DNA and RNA binding protein (Kin17) is an evolutionarily conserved nuclear protein (3). KIN17 consists of three motifs, a zinc finger, a nuclear localization signal and a Kyprides-Onzonis-Woese motif that mediates transcription elongation (4). KIN17 is ubiquitously expressed in human tissues and has been demonstrated to regulate essential biological functions. (5). Kin17 is a member of the multiprotein DNA replication complex and associates with the origin of replication (6). Kin17 is also involved in DNA repair, whereby exposure of tissue to radiation has been demonstrated to upregulate its expression (7). Additionally, it has been suggested that Kin17 serves a role in transcription, due to its presence in the human spliceosome, as detected using proteomic analysis (4).
Emerging evidence has indicated the importance of Kin17 in the pathogenesis and progression of tumors. Previous studies have reported that KIN17 expression is elevated in colon cancer, hepatocellular carcinoma and breast cancer (8–10). In addition, previous studies have revealed that the upregulation of KIN17 is associated with proliferation, chemoresistance and radioresistance in tumors (5,11,12). However, the role of KIN17 in tumor invasion and metastasis remains unclear. In the current study, the association between KIN17 expression and the metastasis of NSCLC was investigated. In addition, KIN17 expression was silences to investigate its effect on the migration and metastasis of NSCLC cells.
Materials and methods
Patients and specimens
A total of 58 NSCLC samples (31 male, 27 female) and 39 benign pulmonary lesions samples (7 pneumonia, 11 tuberculosis, 12 bullae and 9 benign tumors) were obtained. Patients underwent surgery at Longgang Central Hospital (Shenzhen, China) between January 2010 and October 2013. The clinicopathological characteristics of NSCLC patients are summarized in Table I. The tumors were graded using a three-tier grading system based on cell differentiation and the percentage of cancer cells in the process of dividing (8,13). Tumors with higher grade showed lower cell differentiation, more aggressive and malignant in histological appearance. The present study was approved by the Institutional Review Board of Longgang Central Hospital and written informed consent was obtained from all participants.
Table I.
Association between KIN17 expression and the clinicopathological characteristics of patients with non-small cell lung cancer.
| KIN17 expression, no. of patients (%) | ||||
|---|---|---|---|---|
| Clinicopathological characteristics | Low | High | No. of patients (%) | P-value |
| Age (years) | 50.3±7.9 | 52.2±8.7 | 0.381a | |
| Tumor size (mm) | 0.2122 | |||
| ≤29.4 | 15 (65.2) | 17 (48.6) | 32 (55.2) | |
| >29.4 | 8 (34.8) | 18 (51.4) | 26 (44.8) | |
| Lymph node metastasis | 0.0362 | |||
| No | 15 (65.2) | 13 (37.1) | 28 (48.3) | |
| Yes | 8 (34.8) | 22 (62.9) | 30 (51.7) | |
| Tumor grade | 0.0202 | |||
| 1 | 9 (39.1) | 4 (11.4) | 13 (22.4) | |
| 2 | 11 (47.8) | 18 (51.4) | 29 (50.0) | |
| 3 | 3 (13.1) | 13 (37.1) | 16 (27.6) | |
| NSE expression status | 0.9012 | |||
| Negative | 13 (68.4) | 18 (66.7) | 31 (67.4) | |
| Positive | 6 (31.6) | 9 (33.3) | 15 (32.6) | |
| TTF-1 expression status | 0.3802 | |||
| Negative | 17 (73.9) | 22 (62.9) | 39 (67.2) | |
| Positive | 6 (26.1) | 13 (37.1) | 19 (32.8) | |
| CEA expression status | 0.6342 | |||
| Negative | 13 (68.4) | 16 (61.5) | 29 (64.4) | |
| Positive | 6 (31.6) | 10 (38.5) | 16 (35.6) | |
Student's t-test
χ2 test. KIN17, KIN17 DNA and RNA binding protein. NSE, neuron-specific enolase; TTF-1, thyroid transcription factor 1; CEA, carcinoembryonic antigen.
Immunohistochemistry analysis
Specimens were fixed with 10% buffered formalin at room temperature for 24 h, embedded in paraffin wax and sectioned at a thickness of 4 µm. Subsequently, the sections were deparaffinized in xylene and rehydrated through by graded ethanol washes (absolute ethanol, 95, 80%, and distilled water), and quenched with 3% hydrogen peroxide to block endogenous peroxidase activity. Following a citrate buffer (0.01 M Sodium Citrate, pH 6.0) wash, 5% goat serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was applied to block non-specific binding at room temperature for 30 min. Sections were incubated overnight at 4°C with an anti-Kin17 primary antibody (dilution, 1:100; cat. no., sc-32769; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Subsequently, sections were incubated at room temperature for 45 min with horseradish peroxidase (HRP)-conjugated anti-mouse IgG secondary antibody (1:5,000; cat. no. K400511; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). The sections were visualized using a ChemMate™ DAKO EnVision™ Detection kit (Dako; Agilent Technologies, Inc.). The negative control was prepared in the same way, but excluding the primary antibody step. The stained slides were scored by two pathologists, who were unaware of the clinical diagnosis, as previously described (8). A staining score (intensity of staining × percentage of cells stained) of >100 was defined as high expression and a staining score of ≤100 was defined as low expression (8).
Cell culture
The NSCLC cell line A549 and human embryonic kidney (HEK 293T) cells were obtained from GeneChem Co., Ltd. (Shanghai, China) and maintained in a humidified atmosphere with 5% CO2 at 37°C. Cells were cultured in 75 cm2 culture flasks in Ham's F-12K medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified atmosphere with 5% CO2 at 37°C. HEK 293T cells were cultured in Opti-MEM I medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and was prepared for cDNAs synthesis using a reverse transcription kit (Promega Corporation, Madison, WI, USA). A total of 2 µg of total RNA was reverse transcribed in a total reaction volume of 20 µl containing: 1X RT buffer (50 mM Tris-HCl pH 8.3, 75 mM KCl and 3 mM MgCl2), 20 U RNase inhibitor (40 U/µl), 10 mM dithiothreitol, 50 mM deoxyribonucleoside triphosphates (dNTPs), 0.5 µg oligo-deoxythymidine and 200 U Moloney murine leukemia virus reverse transcriptase (200 U/µl, Promega Corporation). The mixture was incubated at 42°C for 60 min and at 70°C for 10 min. The cDNA was then used for PCR amplification with SYBR Premix Ex Taq (cat. no. DRR420A; Takara Bio, Inc., Otsu, Japan). The primers for KIN17 were as follows: forward: 5′-CCATGATTCCTTCATATTTGC-3′, reverse: 5′-GTAATACGGTTATCCACGCG-3′; GAP DH forward: 5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse: 5′-GGCTGTTGTCATACTTCTCATGG-3′. Subsequent to heating at 95°C for 15 min, PCR was performed in a thermal cycler (GeneAmp 2400; PE Applied Biosystems, Foster City, CA, USA) for 40 cycles, consisting of 95°C for 5 sec, 60°C for 30 sec, 72°C for 30 sec. All samples were run in triplicate and relative mRNA levels were calculated using the 2−ΔΔCt method (14) provided by the System software (Applied Biosystems). Data were normalized with GAPDH levels in the samples.
Western blot analysis
Total protein from cells or fresh tissue was used for western blot analysis. RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) containing a complete protease inhibitor cocktail Tablet (Roche Applied Science, Penzberg, Germany) was used to extract total proteins as described previously (8). In total, 10 µg of protein per lane was separated by 10% SDS-PAGE and transferred onto Immobilon®-P PVDF Transfer Membranes (EMD Millipore, Billerica, MA, USA). Following blocking with non-fat milk at room temperature for 30 min, the membranes were probed with anti-Kin17 (dilution, 1:500; cat. no. sc-32769; Santa Cruz Biotechnology, Inc.), anti-EGFR (dilution, 1:500; cat. no. sc-373746; Santa Cruz Biotechnology, Inc.), anti-MYC (dilution, 1:500; cat. no. sc-40; Santa Cruz Biotechnology, Inc.), anti-MMP7 (dilution, 1:500; cat. no. sc-58388; Santa Cruz Biotechnology, Inc.) and β-actin (dilution, 1:500; sc-47778; Santa Cruz Biotechnology, Inc.) monoclonal primary antibodies overnight at 4°C. HRP-conjugated secondary antibodies (dilution, 1:500; cat. no. sc-2318; Santa Cruz Biotechnology, Inc.) were used to develop immunoblots at room temperature for 1 h, which were processed using ECL enhanced chemiluminescence substrate (Thermo Fisher Scientific, Inc.). Images were captured using the ImageQuant RT ECL™ imager (GE Healthcare Life Sciences, Shanghai, China). Band intensities were quantified using ImageQuant TL image analysis software (version 7.0; GE Healthcare Life Sciences).
Lentiviral recombinant vector construction
Single-stranded oligonucleotides containing control small interfering RNA (siRNA; target sequence, 5′-TTCTCCGAACGTGTCACGT-3′) and 4 siRNA sequences targeting KIN17 gene (KD1, 5′-GTTGTGAAGATGATTGATT-3′; KD2, 5′-GCTACTATCGTCATTGAAA-3′; KD3, 5′-AATCTGCACTGGATGAAAT-3′; KD4, 5′-ACATTGTCTACAACGAATA-3′; GenBank accession no. NM_012311) were synthesized by GeneChem Co., Ltd. (Shanghai, China), and converted into double-stranded DNA via annealing. Then, the DNA was inserted into the lentiviral GV248 vector (GeneChem Co., Ltd.) following double enzyme digestion by HpaI and XhoI enzymes as previously described (8). The recombinant vectors were confirmed through DNA sequencing prior to transfection into HEK 293T cells with two helper plasmids (pHelper 1 carrying gag/pol/rev gene and pHelper 2 carrying VSV-G gene, GeneChem Co., Ltd.) to package virus particles using Lipofectamine® 2000 Transfection reagent (Thermo Fisher Scientific, Inc.).
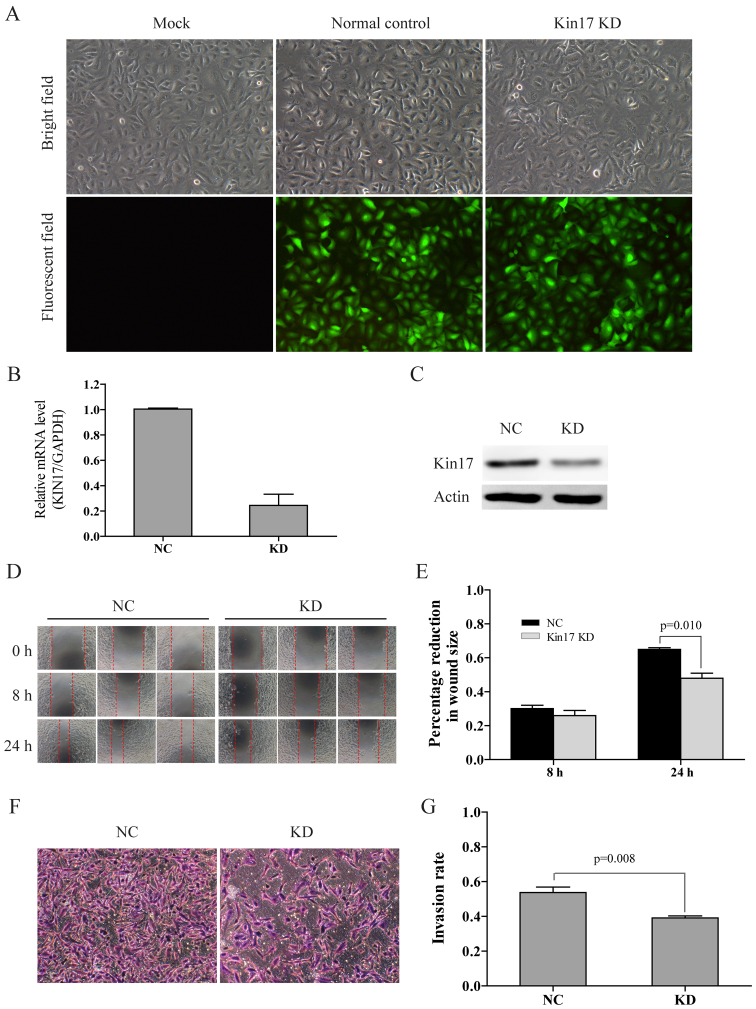
Following purification and titration, the harvested virus particles were used to infect the A549 cells as described by Kou (9). KIN17 mRNA levels in total RNA extracted from the infected cells was determined through RT-qPCR as described above. Results demonstrated that KIN17 expression was markedly decreased following transfection with the recombinant vectors containing KIN17-targeting siRNA sequences. Recombinant vectors with the KD4 siRNA sequence were chosen for use in subsequent experiments, as they demonstrated the highest knockdown efficiency. A549 cells transfected with recombinant vectors carrying the KD4 sequence (A549KIN17 KD cells) and cells transfected with the control vector (A549NC cells) were cultured with puromycin until ~90% of cells contained the fluorescent vector (Fig. 2A). Stable transfection was maintained via continued culture in Ham's F-12K medium with 10% FBS and puromycin. A549 cells without transfection with vector (A549MOCK cells) were used as blank control.
Figure 2.
Knockdown of KIN17 by lentiviral recombinant vector-mediated siRNA transfection inhibited the invasiveness of A549 cells. (A) Morphological features and fluorescence-indicated transfection of A549Mock, A549NC and A549KIN17 KD cells (×100). (B) mRNA and (C) protein levels of KIN17 in A549NC and A549KIN17 KD cells were decreased following KIN17 knockdown, identified by reverse transcription-quantitative polymerase chain reaction analysis and western blotting, respectively. Scratch assay (D) representative images (×100) and (E) quantification. Matrigel assay (F) representative images (×100) and (G) quantification. KIN17, KIN17 DNA and RNA binding protein; A549KIN17 KD, A549 cells transfected with recombinant lentiviral vectors carrying the KD4 siRNA sequence targeting KIN17 gene; A549NC, A549 cells transfected with the control vector; A549MOCK, A549 cells without transfection of vector.
Scratch assay
A549 cells (3×104 cells/well) were seeded into a 96-well plate and cultured for 24 h. Cells in the individual wells were scratched with a sterile pipette tip, prior to the addition of Ham's F-12 medium without serum. Cell migration was measured using a fluorescence microscope (CKX41, Olympus Corporation, Tokyo, Japan) with ImageJ software (v1.48u, National Institutes of Health, Bethesda, MD, USA).
Matrigel invasion assay
Matrigel-coated Transwell cell culture chambers (8 µm pore size) were used to determine the invasion abilities of the cells. A549 cells were trypsinized, suspended in starvation medium and placed in the upper Transwell chambers (5×105 cells/well). The lower chambers contained 750 µl Ham's F-12K medium containing 30% FBS (Gibco; Thermo Fisher Scientific, Inc.). The plate was placed in a humidified atmosphere with 5% CO2 at 37°C for 24 h. Cells on the upper surface of the Matrigel membrane were removed and invasive cells on the bottom surface were stained with Giemsa for 30 min and counted using a light microscope (CKX41, Olympus Corporation). The number of cells placed into the upper chamber was divided by the number of cells from the bottom surface of Matrigel membrane to get the invasion rate.
Statistical analysis
Immunohistochemistry results about Kin17 expression were analyzed by c2 test. Data analysis of the scratch assay and Transwell invasion assay were performed by the Student's t-test. Baseline clinicopathological characteristics of the patients with NSCLC (for example age and lymph nodes metastasis), in addition to biological variables of the tumors (for example tumor size, tumor grade, NSE expression, TTF-1 expression and CEA expression) were compared using the student's t-test (continuous variables) or χ2 test (categorical variables). All statistical tests and corresponding P-values reported were two-tailed. P<0.05 was considered to indicate a statistically significant difference. SPSS software (version 16.0; SPSS, Inc., Chicago, IL, USA) was used to perform all statistical analyses.
Results
Kin17 expression in NSCLC and cells
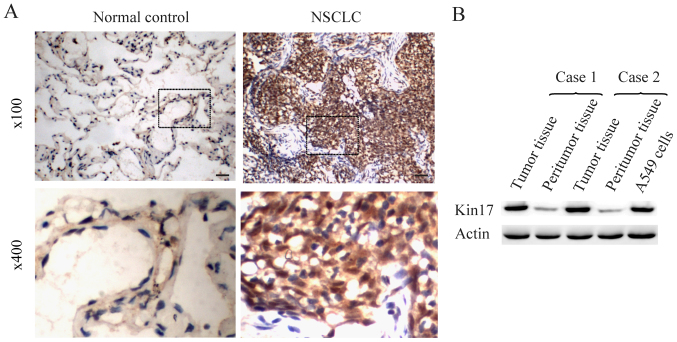
To investigate the function of Kin17 in NSCLC, Kin17 protein expression in NSCLC and benign pulmonary lesion tissue was examined. A total of 65 samples of NSCLC and 40 benign pulmonary lesion samples were investigated using immunohistochemistry analysis. The results obtained using the scoring method indicated that the mean expression of Kin17 in the NSCLC sample group was markedly increased compared with the benign pulmonary lesion sample group (normal control; Fig. 1A). Furthermore, Kin17 protein expression in the samples and A549 cells was detected through western blot analysis. Kin17 protein expression was markedly higher in the NSCLC tumor samples compared with their adjacent healthy tissues (Fig. 1B). In addition, Kin17 protein was overexpressed in A549 cells compared with epithelial cells in peritumoral lung tissue (Fig. 1B).
Figure 1.
Kin17 protein expression in NSCLC tissues and A549 cells. (A) Immunohistochemistry staining for Kin17 protein in NSCLC samples and benign pulmonary lesion samples. 100x, Scale bars, 30 µm. (B) Western blot analysis of Kin17 protein expression in NSCLC tissues, peritumoral tissues and A549 cells. NSCLC, non-small cell lung cancer; Kin17, Kin17 DNA and RNA binding protein.
Association between Kin17 protein expression and the clinicopathological characteristics of patients with NSCLC
The clinicopathological characteristics of 58 patients with NSCLC were reviewed and their association with Kin17 protein expression was examined. High Kin17 expression was significantly associated with lymph nodes metastasis (P=0.036) and high tumor grade (grade 2 or 3, P=0.020; Table I). However, Kin17 expression was not associated with age, tumor size or expression status of the following tumor biomarkers: Neuronal enriched enolase, thyroid transcription factor 1 and carcinoembryonic antigen (Table I). The significant correlation between Kin17 expression and lymph node metastasis suggests that Kin17 may be associated with the invasion and metastasis of NSCLC.
KIN17 knockdown suppresses the migration and metastasis of NSCLC A549 cells
To investigate the role of Kin17 in NSCLC, lentiviral-mediated transfection of siRNA targeting KIN17 mRNA was used to knock down KIN17 expression in A549 cells (Fig. 2A). RT-qPCR and western blot assays confirmed that the mRNA and protein levels of KIN17 in A549KIN17 KD cells were markedly decreased compared with levels in A549NC cells (Fig. 2B and C).
Scratch and Transwell assays were conducted in order to investigate the effect of a KIN17 knockdown on the metastasis and invasion abilities of A549 cells, respectively. In the scratch assay, A549KIN17 KD cells demonstrated a significant reduction in migration 24 h following the scratch compared with A549NC cells (P=0.01; Fig. 2D and E). Furthermore, the number of invasive A549KIN17 KD cells was significantly lower compared with the A549NC cells (P=0.01; Fig. 2F and G).
KIN17 knockdown effects matrix metallopeptidase 7 (MMP7), epidermal growth factor receptor (EGFR) and v-myc avian myelocytomatosis viral oncogene homolog (MYC) protein expression
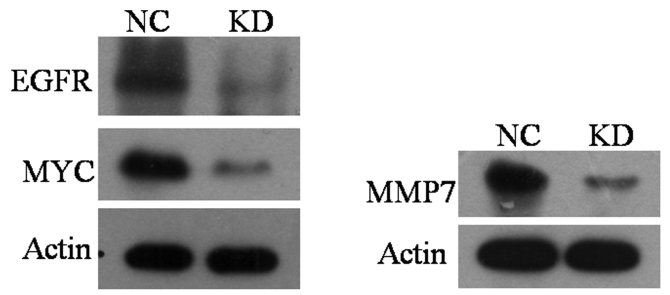
Ectopic activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase (MEK/ERK) signaling pathway has been associated with the invasion and metastasis of various types of tumor. Our previous study of breast cancer revealed that silencing KIN17 may effect this signaling pathway (8). In order to explore the role of KIN17 in the MEK/ERK signaling pathway in NSCLC, influencing of KIN17 knockdown on the expression levels of EGFR, MMP7 and MYC, 3 key components of the MEK/ERK signaling pathway, were investigated in A549 cell in the present study. Western blot analysis revealed that A549KIN17 KD cells expressed markedly lower levels of EGFR, MMP7 and MYC compared with A549NC cells (Fig. 3). These findings suggest that KIN17 expression is associated with the expression of EGFR, MMP7 and MYC.
Figure 3.
Effect of KIN17 knockdown on the expression of EGFR, Myc and MMP7. KIN17 via western blot analysis, KIN17 DNA and RNA binding protein; MMP7, matrix metallopeptidase 7; EGFR, epidermal growth factor receptor; Myc, v-myc avian myelocytomatosis viral oncogene homolog; NC, negative control; KD, knock down.
Discussion
The expression level of KIN17 in the majority of healthy tissues is low, with the exception of the testis, ovary, muscle and heart (5). At present, the expression level of KIN17 in human tumors remains unclear. Recently, studies performed by Yu et al (10) and Kou et al (9) revealed that KIN17 gene expression is upregulated in colorectal cancer and hepatocellular carcinoma, respectively. In addition, a previous study demonstrated that KIN17 was markedly overexpressed in breast cancer (8). In the present study, the expression of KIN17 was demonstrated to be increased in NSCLC tissue compared with benign pulmonary lesion and peritumoral tissues. Further analysis revealed that upregulation of KIN17 was significantly correlated with advanced tumor grade and lymph node metastasis. Similar results have been reported by Yu et al (10). The results of the present study suggest that patients with elevated KIN17 expression have a poorer prognosis, and that KIN17 is a potential diagnostic and prognostic biomarker for NSCLC.
The invasion and metastasis of lung cancer cells significantly affects the prognosis of patients. Therefore, the identification of potential therapeutic targets for the inhibition of metastasis in NSCLC is important. Through statistical analysis of clinical data, an association between KIN17 expression and metastasis of lymph nodes was demonstrated in the present study. To further determine the role of KIN17 in the invasion and metastasis of NSCLC, siRNA was used to knock down KIN17 expression in A549 cells, and scratch and Transwell assays were used to determine the effects of this. The results of these experiments demonstrated that downregulation of KIN17 reduced the invasion and migration of A549 cells, suggesting that KIN17 serves an important role in the invasion and metastasis of NSCLC.
The underlying molecular mechanism through which KIN17 promotes the malignant features of tumors remains unclear. Altered activation of the MEK/ERK signaling pathway is associated with the stimulation tumor invasion and metastasis (15). The effect of KIN17 on this signaling pathway has previously been investigated in breast cancer cells (8). This revealed that silencing of KIN17 decreased EGF-stimulated cell proliferation and phosphorylation of ERK in breast cancer cells. Therefore, it was hypothesized that the role of KIN17 in promoting the invasion and metastasis of NSCLC is mediated through the MEK/ERK signaling pathway. Egfr, Mmp7 and Myc are important components of the MEK/ERK signaling pathway, and the genes encoding these proteins are frequently mutated and overexpressed in various types of human cancer, including in NSCLC.
The MEK/ERK signaling pathway is one of the most important downstream signaling pathways that EGFR serves a role in (16). The phosphorylation of ERK leads to the activation of multiple transcription factors, including activating protein-1 (AP-1) and Myc (17). MMP7 is an important target gene of AP-1. MMP7 is a member of the MMP family, which performs important roles in tumor invasion and metastasis (18). The association between MMP7 and the progression and metastasis of lung cancer is due to its ability to degrade the extracellular matrix and cause the accumulation of β-catenin (19). Ectopic expression of Myc, a multifunctional transcription factor, results in the altered expression of a variety of genes, including those that are associated with the development and progression of tumors (20). Myc has also been identified as a metastasis-associated gene in NSCLC. A previous study revealed that overexpression of nuclear Myc led to early macrometastasis of NSCLC to the liver and lymph nodes in mice models (21). This suggests that the MEK/ERK signaling pathway is a mechanism through which KIN17 promotes NSCLC progression and metastasis. Further studies on the association between KIN17 and the MEK/ERK signaling pathway are warranted.
In conclusion, the present study demonstrated that the expression of KIN17 protein and mRNA is markedly elevated in NSCLC. Upregulation of KIN17 was identified to be associated with lymph node metastasis, indicating poor patient prognosis. In addition, silencing of KIN17 inhibited NSCLC cell invasion and metastasis. These findings indicate that KIN17 is a novel diagnostic and prognostic biomarker of NSCLC, in addition to a potential therapeutic target for the treatment of patients with NSCLC.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81201831), the Guangdong Planning Project for Science and Technology (grant no. 2014A020212190) and the Science and Technology Planning Project of Zhangjiang (grant no. 2012C3104014).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Tieulent J Lortet, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Schabath MB, Thompson ZJ, Gray JE. Temporal trends in demographics and overall survival of non-small-cell lung cancer patients at Moffitt Cancer Center from 1986 to 2008. Cancer Control. 2014;21:51–56. doi: 10.1177/107327481402100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cloutier P, Lavallée-Adam M, Faubert D, Blanchette M, Coulombe B. Methylation of the DNA/RNA-binding protein KIN17 by METTL22 affects its association with chromatin. J Proteomics. 2014;100:115–124. doi: 10.1016/j.jprot.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Angulo JF, Mauffirey P, Pinon-Lataillade G, Miccoli L, Biard DS. Putative roles of KIN17, a mammalian protein binding curved DNA, in transcription. Springer; pp. 75–89. [Google Scholar]
- 5.Kannouche P, Mauffrey P, Pinon-Lataillade G, Mattei MG, Sarasin A, Daya-Grosjean L, Angulo JF. Molecular cloning and characterization of the human KIN17 cDNA encoding a component of the UVC response that is conserved among metazoans. Carcinogenesis. 2000;21:1701–1710. doi: 10.1093/carcin/21.9.1701. [DOI] [PubMed] [Google Scholar]
- 6.Miccoli L, Frouin I, Novac O, Di Paola D, Harper F, Zannis-Hadjopoulos M, Maga G, Biard DS, Angulo JF. The human stress-activated protein KIN17 belongs to the multiprotein DNA replication complex and associates in vivo with mammalian replication origins. Mol Cell Biol. 2005;25:3814–3830. doi: 10.1128/MCB.25.9.3814-3830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay BC. Post-transcriptional regulation of DNA damage-responsive gene expression. Antioxid Redox Sign. 2014;20:640–654. doi: 10.1089/ars.2013.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng T, Gao H, Yu P, He H, Ouyang X, Deng L, Zhang Y. Upregulation of KIN17 is essential for proliferation of breast cancer. PLoS One. 2011;6:e25343. doi: 10.1371/journal.pone.0025343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kou WZ, Xu SL, Wang Y, Wang LW, Wang L, Chai XY, Hua QL. Expression of Kin17 promotes the proliferation of hepatocellular carcinoma cells in vitro and in vivo. Oncol Lett. 2014;8:1190–1194. doi: 10.3892/ol.2014.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu M, Zhang Z, Yu H, Xue C, Yuan K, Miao M, Shi H. KIN enhances stem cell-like properties to promote chemoresistance in colorectal carcinoma. Biochem Bioph Res Commun. 2014;448:63–69. doi: 10.1016/j.bbrc.2014.04.057. [DOI] [PubMed] [Google Scholar]
- 11.Despras E, Miccoli L, Créminon C, Rouillard D, Angulo JF, Biard DS. Depletion of KIN17, a human DNA replication protein, increases the radiosensitivity of RKO cells. Radiat Res. 2003;159:748–758. doi: 10.1667/0033-7587(2003)159[0748:DOKAHD]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Deng L, Zhang H, Zeng T, Wang H, Zhang Y. Secretory KIN17 is correlated with chemoresistance in oral squamous cell carcinoma. J Anal Oncol. 2014;3:18–25. [Google Scholar]
- 13.Kadota K, Suzuki K, Kachala SS, Zabor EC, Sima CS, Moreira AL, Yoshizawa A, Riely GJ, Rusch VW, Adusumilli PS, Travis WD. A grading system combining architectural features and mitotic count predicts recurrence in stage I lung adenocarcinoma. Mod Pathol. 2012;25:1117–1127. doi: 10.1038/modpathol.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Johnson GL, Stuhlmiller TJ, Angus SP, Zawistowski JS, Graves LM. Molecular pathways: Adaptive kinome reprogramming in response to targeted inhibition of the BRAF-MEK-ERK pathway in cancer. Clin Cancer Res. 2014;20:2516–2522. doi: 10.1158/1078-0432.CCR-13-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Y, He H, Liu H, Zhang C, Zhao W, Shao R. Phosphorylation of myofibrillogenesis regulator-1 activates the MAPK signaling pathway and induces proliferation and migration in human breast cancer MCF7 cells. FEBS Lett. 2014;588:2903–2910. doi: 10.1016/j.febslet.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Sizemore ST, Sizemore GM, Booth CN, Thompson CL, Silverman P, Bebek G, Abdul-Karim FW, Avril S, Keri RA. Hypomethylation of the MMP7 promoter and increased expression of MMP7 distinguishes the basal-like breast cancer subtype from other triple-negative tumors. Breast Cancer Res Treat. 2014;146:25–40. doi: 10.1007/s10549-014-2989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Luo J, Ni J, Tang L, Zhang HP, Zhang L, Xu JF, Zheng D. MMP-7 is upregulated by COX-2 and promotes proliferation and invasion of lung adenocarcinoma cells. Eur J Histochem. 2014;58:2262. doi: 10.4081/ejh.2014.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kress TR, Sabò A, Amati B. MYC: Connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 21.Rapp UR, Korn C, Ceteci F, Karreman C, Luetkenhaus K, Serafin V, Zanucco E, Castro I, Potapenko T. MYC is a metastasis gene for non-small-cell lung cancer. PLoS One. 2009;4:e6029. doi: 10.1371/journal.pone.0006029. [DOI] [PMC free article] [PubMed] [Google Scholar]