Abstract
Lung cancer is the most frequent cause of mortality in cancer patients; non-small-cell lung cancer (NSCLC) accounts for ~80% of lung cancer cases. MicroRNAs (miRNAs) have been revealed to perform an important role in cancer development and progression. Based on a custom miRNA microarray analysis of patients with NSCLC, miRNA-615-3p (miR-615-3p) downregulation was identified in NSCLC tissues compared with normal lung tissues, which suggested that miR-615-3p acted as a tumor suppressor in lung cancer. The overexpression of miR-615-3p was then validated using 40 pairs of NSCLC and adjacent normal tissue samples using a TaqMan reverse transcription-quantitative polymerase chain reaction assay. In order to investigate the tumor suppressor function of miR-615-3p, the ectopic expression of miR-615-3p in the NSCLC A549, H1299 and H1650 cell lines was established. The results revealed that overexpressed miR-615-3p markedly inhibited cell proliferation and colony formation in the 3 NSCLC cell lines compared with the cells overexpressing the negative control sequence (NC). Additional investigation revealed that miR-615-3p overexpression significantly induced apoptosis and cell cycle arrest at the G1 phase in the A549, H1299 and H1650 cell lines compared with the cells overexpressing NC. Finally, ectopic expression of miR-615-3p was found to repress the cell migration and invasion of the 3 lung cancer cell lines. The results of the present study demonstrate, for the first time, that miR-615-3p functions as a tumor suppressor in NSCLC, and may be a novel potential molecular therapeutic target for patients with NSCLC.
Keywords: miR-615-3p, non-small cell lung cancer, RT-qPCR, microarray, biological role
Introduction
Lung cancer is the leading cause of cancer-associated mortality in the world (1), and ~80% of lung cancer cases are non-small-cell lung cancer (NSCLC) (2). Patients with NSCLC possess an extremely poor prognosis, partly due to the lack of early diagnostic biomarkers and prognostic indicators. The identification of microRNAs (miRNAs) has opened a new avenue for developing highly reliable diagnostic and prognostic biomarkers to facilitate the early diagnosis of NSCLC and predict the clinical outcomes of patients (3,4). At present, numerous single miRNAs, including let-7a-2 (5), miRNA-146b (6), miRNA-155 (5,6), miRNA-31 (7), miRNA-374a (8), miRNA-451 (9) and miRNA-21 (10,11) have been reported to be associated with the tumorigenesis and progression of lung cancer.
The expression of 1,849 miRNAs has previously been profiled in 397 patients with NSCLC (Pu et al, unpublished data). The results showed that the expression level of miRNA-615-3p (miR-615-3p) was notably downregulated in NSCLC tissues compared with the corresponding normal lung tissues, which suggested that miR-615-3p may act as a tumor suppressor in NSCLC development and progression. Subsequent to confirming the downregulation of miR-615-3p in NSCLC tissues, the biological role of miR-615-3p in NSCLC cells was assessed. To the best of our knowledge, the present study is the first to report that miR-615-3p functions as a tumor suppressor in NSCLC.
Materials and methods
Patients, tissue specimens and cell culture
In total, 40 matched pairs of NSCLC and normal lung tissues were randomly selected from 397 patients diagnosed with NSCLC that had not been treated with radiotherapy or chemotherapy prior to undergoing surgical resection at the Sun Yat-Sen University Cancer Center (Guangzhou, China) between 2004 and 2009. The miRNA expression in 397 NSCLC and 151 matched normal control tissue samples had been assessed using a custom miRNA microarray (Pu et al, unpublished data). All patients with NSCLC were staged according to the 7th edition of the American Joint Committee on Cancer tumor-node-metastasis staging system for lung cancer (12).
The fresh tissues were immediately immersed in RNAlater (Ambion, Inc., Austin, TX, USA) subsequent to surgical resection, stored at 4°C for 24 h to allow thorough penetration of the RNase inhibitor, and then frozen at −80°C in a freezer (Sanyo Electric Co., Ltd., Moriguchi, Osaka, Japan) until use. The present study was reviewed and approved by the Ethical Committee of Sun Yat-Sen University Cancer Center. Written informed consent was obtained from each patient.
A549, H1299 and H1650 cells were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in Gibco RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% Gibco fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 atmosphere.
Total RNA extraction
Total RNA was extracted from the NSCLC tissues and the adjacent normal lung tissues using Invitrogen TRIzol reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The RNA concentration was measured using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.), and the quality of all RNA samples was monitored by electrophoresis on 1.5% denaturing agarose gels.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT was performed on the 40 paired tissue samples (including tumor tissues and paired peri-tumor tissues) using 10 ng of total RNA, 1X-miRNA specific RT primers, 100 µM nucleoside triphosphates (Thermo Fisher Scientific, Inc.,), 3.33 U/µl MultiScribe™ Reverse Transcriptase (Thermo Fisher Scientific, Inc.), 1X RT buffer and 1.33 U/µl RNase inhibitor (Thermo Fisher Scientific, Inc.), in a final volume of 15 µl. The reaction was conducted at 16°C for 30 min, followed by 30 min at 42°C and 5 min at 85°C. No negative control was performed. Small nuclear RNA U6 was used as a normalization control. Each experiment was repeated three times. The qPCR reaction was performed using a 20-µl volume containing 1.33 µl complementary DNA (produced during the RT step), 1X TaqMan® Small RNA Assay solution (which includes specific primers and probes; Applied Biosystems; Thermo Fisher Scientific, Inc.) and 1X Universal PCR Master Mix II (which does not contain uracil-N glycoslyase; Thermo Fisher Scientific, Inc.). RT-qPCR was performed using an Applied Biosystems PRISM 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, Inc.) with the following conditions: 50°C for 2 min; 95°C for 10 min; and 45 cycles of 95°C for 15 sec and 60°C for 60 sec. Small nuclear RNA U6 was used as a normalization control. The 2−ΔΔCq method was used to represent the relative expression of miRNA (13). The primer sequences were as follows: mir-615-3p, forward 5′-CTGCCTTTCACCTTGGAGAC-3′ and reverse 5′-CGTTTCCTGGGGATGAGATA-3′; and U6 forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Transient transfection of lung cancer cells with oligonucleotides
For the transfection assay, the oligonucleotides of miR-615-3p (forward 5′-UCCGAGCCUGGGUCUCCCUCUU-3′ and reverse 5′-GAGGGAGACCCAGGCUCGGAUU-3′), and the negative control sequence (NC) for miR-615-3p (forward 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse 5′-ACGUGACACGUUCGGAGAATT-3′) were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The NSCLC cells were transfected with 100 nM miRNA-615-3p or NC using Invitrogen Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Lentivirus infection
An OmicsLink miRNA expression plasmid (Shanghai GenePharma Co., Ltd.), which contains a hsa-miR-615-3p sequence or a scramble sequence driven by a cytomegalovirus promoter and followed by an enhanced green fluorescent protein reporter, was packed into lentiviral particles to infect A549 cells. The cells were then selected with puromycin (Weijia; Thermo Fisher Scientific, Inc., Guangzhou, China). The expression level of miR-615-3p in A549 cells was measured by RT-qPCR subsequent to 1 week of treatment with puromycin and then frozen down in aliquots for further use.
Methyl thiazolyl tetrazolium (MTT) assay
An MTT assay was used to measure the viability of cells transfected with the oligonucleotides. Indicated cells were seeded onto a 96-well plate 24 h subsequent to transfection, at a density of 500 cells per well, and the cells were cultured for 7 days. Every 24 h, 20 µl 0.5 mg/ml MTT (Weijia; Thermo Fisher Scientific, Inc.) was added to the cells. Following incubation at 37°C for 4 h, the supernatant was removed and 150 µl dimethyl sulfoxide (DMSO; Weijia; Thermo Fisher Scientific, Inc.) was added to each well. The cells were gently agitated for 10 min, and the absorbance of the solution in each well was measured at 490 nm with an Evolution 201/220/260 UV–Visible Spectrophotometer (Thermo Fisher Scientific, Inc.). The absorbance for each well was calculated as follows: Absorbance = ODtreated - ODblank.
Colony formation assay
Following transfection for 24 h, indicated cells were seeded into each well of 6-well plate at a density of 500 cells per well. The surviving colonies, which were classified as containing >50 cells per colony, were counted using crystal violet staining (Weijia; Thermo Fisher Scientific, Inc.) subsequent to 2 weeks of culture.
Cell cycle analysis
The cells were collected at 48 h subsequent to transfection with the indicated RNAs (miR-615-3p mimics and NC), washed twice with ice-cold phosphate-buffered saline (PBS; Weijia; Thermo Fisher Scientific, Inc.), and fixed in ice-cold 75% ethanol (Guangzhou Chemical Reagent Factory, Guangzhou, China) at −20°C for 1 h. The cells were re-suspended in 400 µl ice-cold PBS subsequent to washing twice with PBS, and then incubated with 50 µg/ml RNase (BestBio, Shanghai, China) in a 37°C water bath for 30 min. Subsequently, the cells were incubated with propidium iodide (PI; BestBio) at 4°C for 30–60 min and analyzed by flow cytometry (CytoFLEX; Beckman Coulter, Inc., Brea, CA, USA).
Apoptosis analysis
In total, 48 h subsequent to transfection, the cells were washed twice with ice-cold PBS and re-suspended in 400 µl of 1X binding buffer (BestBio). Subsequent to incubation of the cells with Annexin V-fluorescein isothiocyanate (BestBio) at 4°C for 15 min, PI was added and the cells continued to be incubated at 4°C for an additional 5 min. Finally, the cells were analyzed by flow cytometry (CytoFLEX; Beckman Coulter, Inc.).
Invasion and migration assays
Cell migration was examined using 24-well Transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA), according to the manufacturer's protocol. In total, 2×105 A549, H1299 or H1650 cells were seeded into the upper well of the chamber in RPMI-1640 medium without fetal bovine serum. RPMI-1640 medium containing 10% fetal bovine serum was added to the lower well to stimulate invasion. Following incubation for 18 h, the cells remaining on at the upper well were removed while the bottom cells were fixed with 100% methanol (Guangzhou Chemical Reagent Factory) and then stained with 0.1% crystal violet. The number of migrated cells was calculated with an inverted microscope (IX83; Olympus Corporation, Tokyo, Japan) using 5 randomly-selected fields of view at a ×10 magnification. Cell invasion ability was analyzed using the same procedure subsequent to coating the upper well of the chambers with Matrigel (BD Biosciences), and up to 2×106 A549, H1299 or H1650 cells were seeded.
Statistical analysis
Subsequent to digitization of the microarray images, the signal intensity from probes was processed with local background subtraction, averaging the duplicated probe intensities, and then quantile normalization was performed. The microarray data have been deposited in the Gene Expression Omnibus Public Database at the National Center for Biotechnology Information (GSE53882). Biometric Research Branch-ArrayTools, version 3.8.0 (available from http://linus.nci.nih.gov/BRB-ArrayTools.html) was used for microarray data analysis.
Pearson's correlation coefficient was used to analyze the association between the microarray and RT-qPCR data. The 2−ΔΔCq method was used to measure the miRNA expression level detected by RT-qPCR. Two-tailed, unpaired Student's t-test was used to compare the differences between experimental and control groups in in vitro assays, and was conducted using SPSS software, version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Expression level of miR-615-3p was downregulated in NSCLC tissues
The differential microRNA expression profiling performed on 397 NSCLC tumor tissues and 151 adjacent normal lung tissues using a custom microarray revealed that miR-615-3p was downregulated in NSCLC tissues compared with normal lung tissues. To validate the downregulation of miR-615-3p in NSCLC, RT-qPCR was conducted using 40 matched pairs of tumor and normal tissues, which were randomly selected from the 151 NSCLC patients for which there were tumor and matched normal lung samples available. The results revealed that the expression level of miR-615-3p was significantly downregulated in NSCLC tissues compared with normal lung tissues (P<0.001; Fig. 1), which was strongly associated with the microarray result (Pu et al, unpublished data). This result indicates that miR-615-3p may act as a tumor suppressor in NSCLC.
Figure 1.
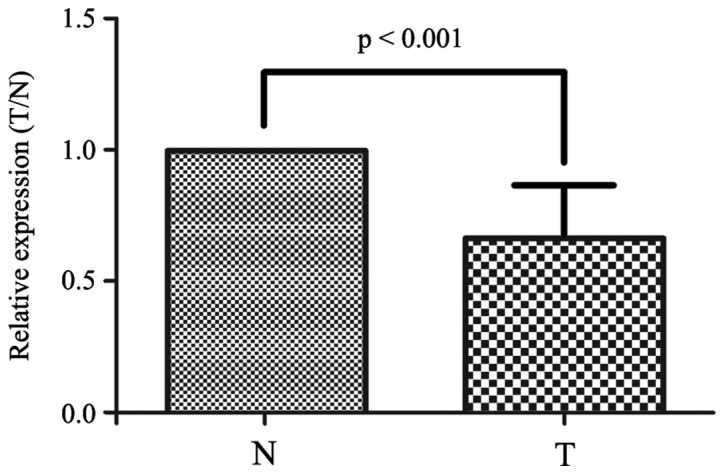
miR-615-3p is downregulated in non-small cell lung cancer tissues. Comparison of the expression ratio of miR-615-3p between the 40 paired T and N samples detected by reverse transcription-quantitative polymerase chain reaction. miR-615-3p, microRNA-615-3p; T, non-small cell lung cancer tissue; N, adjacent normal tissue.
Overexpression of miR-615-3p inhibits the growth of NSCLC cells
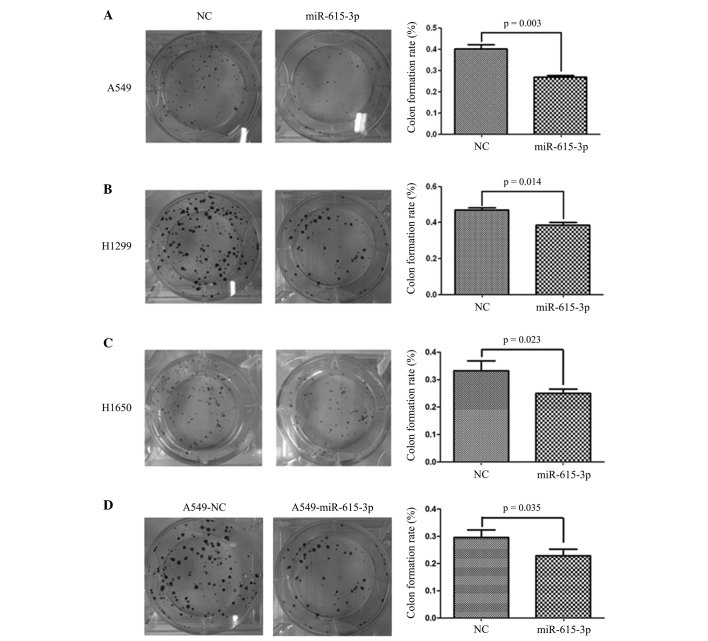
To demonstrate the tumor suppressor function of miR-615-3p in vitro, the lung cancer A549, H1299 and H1650 cell lines were transiently transfected with the miR-615-3p mimic or NC, and the overexpression of miR-615-3p in the 3 cell lines was verified by RT-qPCR (data not shown). An MTT assay was used to assess the effect of upregulated miR-615-3p on the proliferation of A549, H1299 and H1650 cells. It was found that 72 h subsequent to transfection, overexpression of miR-615-3p in the A549, H1299 or H1650 cells resulted in significant inhibition of cell proliferation compared with the proliferation of cells transfected with NC (P<0.001; Fig. 2). To confirm the vital role of miR-615-3p in NSCLC cell growth, the A549 cells with stable expression of miR-615-3p, which had been established by transfecting the cells with lentivirus containing miR-615-3p mimics, were confirmed to demonstrate a high expression level by RT-qPCR (data not shown). The MTT assay clearly showed that the inhibition of cell proliferation was also notable in the A549 cells with stable overexpression of miR-615-3p (P<0.001; Fig. 2). Furthermore, a plate colony formation assay was performed. Ectopic expression of miR-615-3p was also found to significantly reduce the colony formation ability of the A549, H1299 and H1650 cells (all P<0.05; Fig. 3). These observations support the hypothesis that miR-615-3p acts as a tumor suppressor in NSCLC cells.
Figure 2.
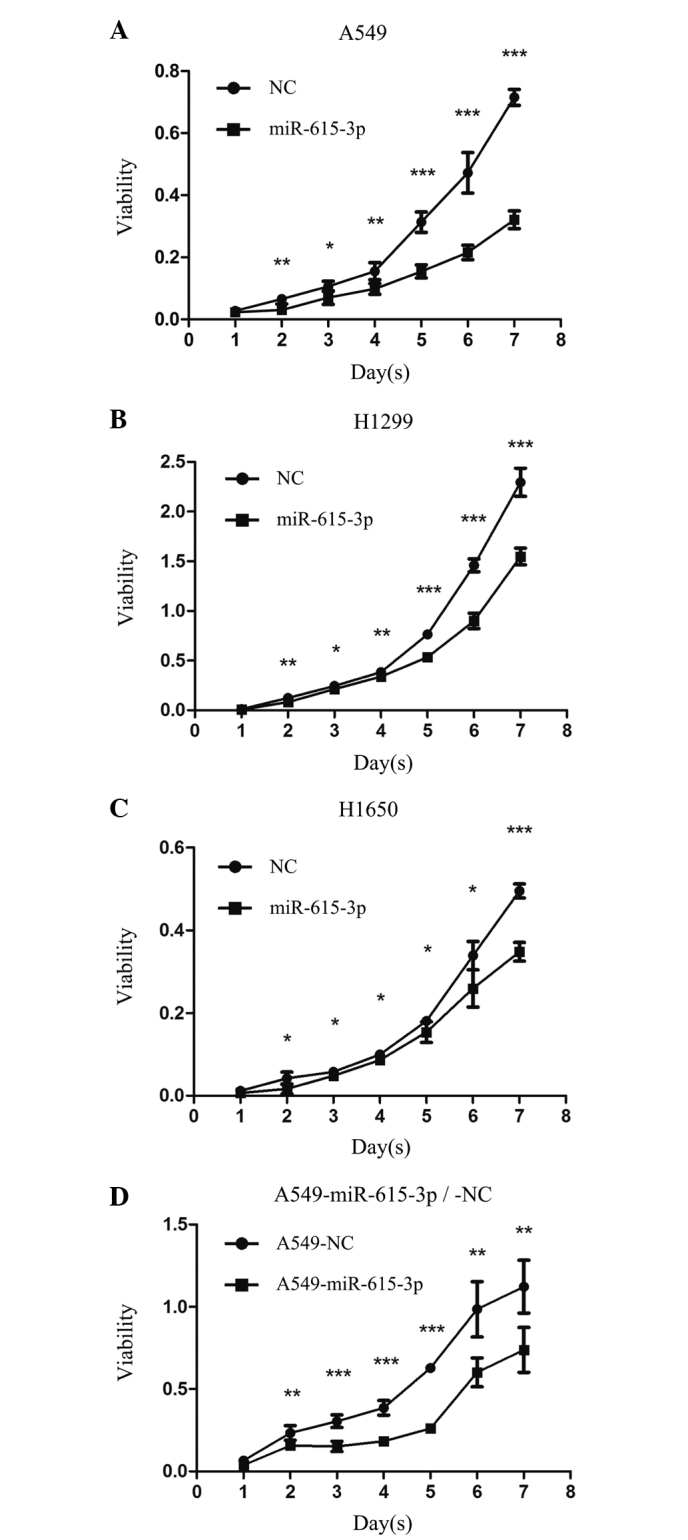
Overexpression of miR-615-3p inhibited the proliferation of NSCLC cells in vitro. (A) A549, (B) H1299 and (C) H1650 cells were transiently transfected with miR-615-3p or NC and the cell viabilities were measured by an MTT assay. (D) The viability of A549 cells with stable overexpression of miR-615-3p or NC was also measured by an MTT assay. The growth curves showed significantly different cell proliferations between the NSCLC cells with miR-615-3p and NC. *P<0.01, **P<0.001, ***P<0.0001. NSCLC, non-small cell lung cancer; miR-615-3p, microRNA-615-3p; NC, negative control sequence; MTT, methyl thiazolyl tetrazolium.
Figure 3.
Overexpression of miR-615-3p inhibited the formation of NSCLC cell colonies in vitro. (A) A549, (B) H1299 and (C) H1650 cells were transiently transfected with miR-615-3p or NC and a colony formation assay was performed. (D) A549 cells stably transfected with miR-615-3p and NC, termed A549-miR-615-3p and A549-NC, respectively, were also assessed by colony formation assay. The number of colonies of NSCLC cells with miR-615-3p overexpression was markedly reduced compared with the number of colonies of NSCLC cells overexpressing NC. NSCLC, non-small cell lung cancer; miR-615-3p, microRNA-615-3p; NC, negative control sequence.
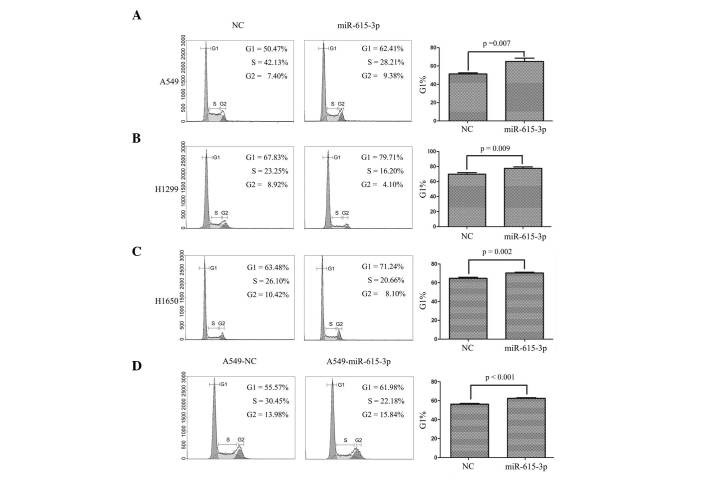
miR-615-3p arrests the cell cycle in the G1 phase in NSCLC cells
The finding that miR-615-3p inhibited the proliferation and oncogenic growth of NSCLC cells suggested that cell cycle progression may be inhibited by miR-615-3p. The A549, H1299 and H1650 cells were stained with PI and then examined by flow cytometry 48 h subsequent to transient transfection with miR-615-3p or NC. The results revealed that the cell cycle progression of the NSCLC cells was arrested in the G1 phase following transfection with miR-615-3p mimics. The percentage of cells in the G1 phase was significantly increased in the A549, H1299 and H1650 cells with miR-615-3p overexpression (all P<0.01; Fig. 4). The cell cycle assays conducted in the stable A549-miR-615-3p and A549-NC cell lines revealed similar results (all P<0.001; Fig. 4). These findings indicated that one of the mechanisms of miR-615-3p inhibiting the proliferation and growth of the NSCLC cells attributed to G1 phase arrest.
Figure 4.
miR-615-3p arrested the cell cycle at the G1 phase in NSCLC cells. The cell cycle distribution of (A) A549, (B) H1299 and (C) H1650 cells transiently transfected and (D) A549 cells stably transfected with miR-615-3p and NC was examined by flow cytometry. The representative images are presented. Statistical analysis showed that the percentage of cells in the G1 phase was significantly increased in the NSCLC cells with overexpression of miR-615-3p compared with the cells with overexpression of NC. NSCLC, non-small cell lung cancer; miR-615-3p, microRNA-615-3p; NC, negative control sequence.
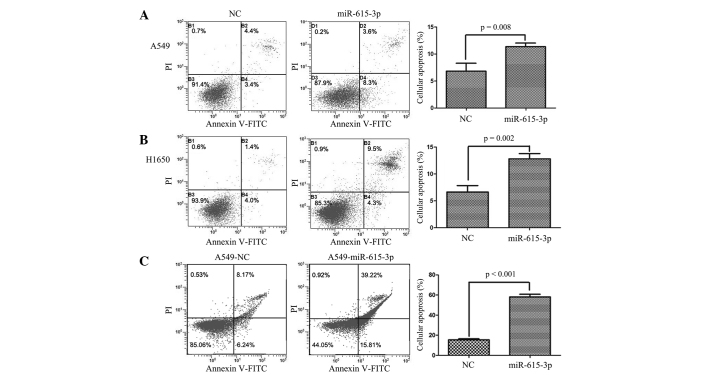
miR-615-3p induces apoptosis in NSCLC cells
Apoptosis has been recognized to be another cause of inhibition of cancer cell proliferation, and miR-615-3p was found to induce apoptosis in NSCLC cells in the present study (Fig. 5). To assess whether apoptosis was involved in the inhibition of lung cancer cell proliferation induced by miR-615-3p, an apoptosis assay was performed using flow cytometry in A549, H1299 and H1650 cells transfected transiently or stably with the miR-615-3p mimic. The results revealed that apoptosis in the A549 and H1650 cell lines overexpressing of miR-615-3p was notably increased compared with the cells transfected with the NC (all P<0.01; Fig. 5), which indicated that miR-615-3p may induce apoptosis in NSCLC cells. However, the apoptosis rate in the H1299 cells transfected with the miR-615-3p mimic was not significantly different compared with the H1299 cells transfected with NC.
Figure 5.
miR-615-3p induced apoptosis in NSCLC cells. Apoptosis of (A) A549 and (B) H1650 cells with transient overexpression and (C) A549 cells with stable overexpression of miR-615-3p and NC was detected by flow cytometry. The representative images of apoptosis in NSCLC cells are shown. The histogram indicates the percentage of apoptotic cells in the cells transfected with miR-615-3p or NC. NSCLC, non-small cell lung cancer; miR-615-3p, microRNA-615-3p; NC, negative control sequence; FITC, fluorescein isothiocyanate; PI, propidium iodide.
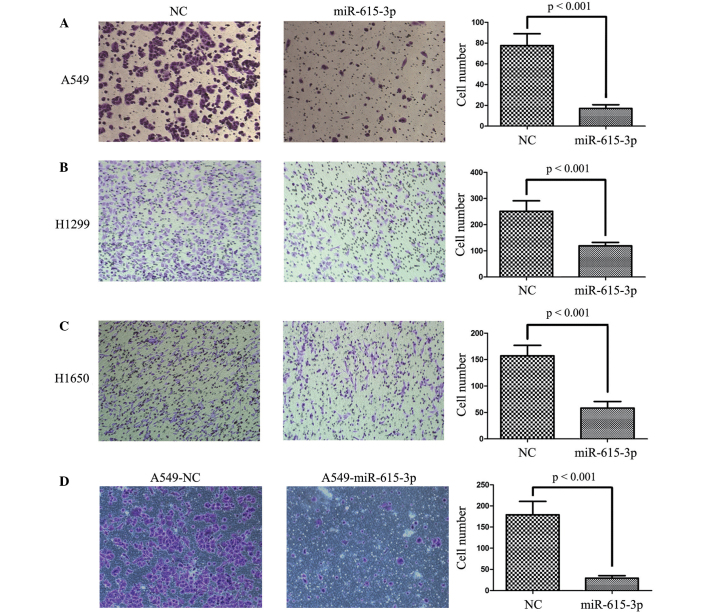
miR-615-3p suppresses migration and invasion in NSCLC cells
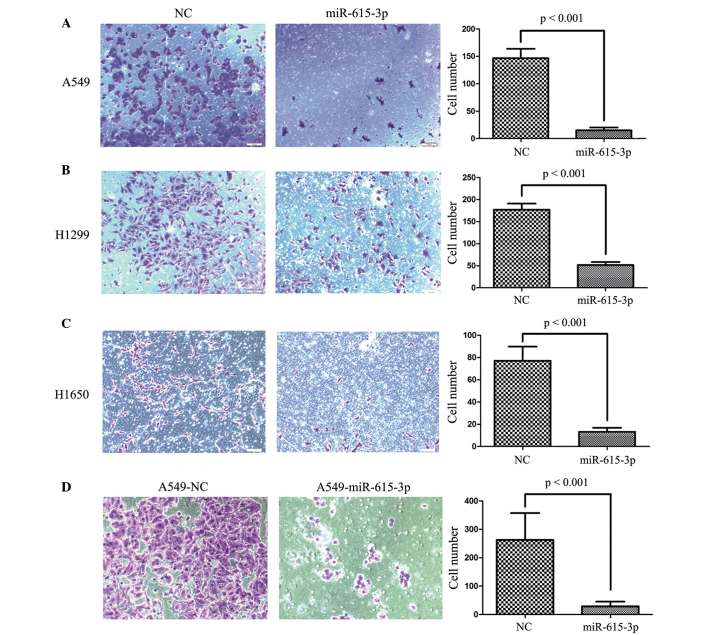
To explore the effect of miR-615-3p on the migration and invasion of NSCLC cells, Transwell migration and invasion assays were conducted using A549, H1299 and H1650 cells transiently or stably transfected with miR-615-3p. As expected, ectopic expression of miR-615-3p significantly suppressed the migration (all P<0.001; Fig. 6) and invasion (all P<0.001; Fig. 7) of the A549, H1299 and H1650 cells transiently transfected with miR-615-3b, and this was also observed in A549-miR-615-3p cells, a stable overexpression cell line. Overall, miR-615-3p may play a significant role in NSCLC development and progression.
Figure 6.
miR-615-3p suppressed NSCLC cell migration in vitro. The migrated NSCLC (A) A549, (B) H1299 and (C) H1650 cell lines with transient overexpression of miR-615-3p and NC, and (D) A549 cells with stable overexpression of miR-615-3p and NC were counted under a high-power field. The average number of migrated cells was counted in 5 randomly-selected fields of view. Representative images of migrated cells are shown in the left and center images and the average numbers of migrated cells are shown in the right panel. NSCLC, non-small cell lung cancer; miR-615-3p, microRNA-615-3p; NC, negative control sequence.
Figure 7.
miR-615-3p suppressed NSCLC cell invasion in vitro. The NSCLC (A) A549, (B) H1299 and (C) H1650 cell lines with transient overexpression of miR-615-3p and NC, and (D) A549 cells with stable overexpression of miR-615-3p and NC were seeded to the upper well of Transwell chambers coated with Matrigel. The cells that invaded into the Matrigel and moved to the bottom of the chamber were fixed and counted under a high-power field. The average number of cells was calculated from 5 randomly-selected fields of view. Representative images of the invaded cells are presented in the left and center images and the average numbers of invaded cells are shown in the right panel. NSCLC, non-small cell lung cancer; miR-615-3p, microRNA-615-3p; NC, negative control sequence.
Discussion
Using a custom miRNA microarray, miRNA expression was profiled in 397 NSCLC and 151 matched normal lung tissues, and it was found that miR-615-3p was downregulated in NSCLC (Pu et al, unpublished data). The reduced miR-615-3p expression was then validated in the present study using 40 matched pairs of NSCLC and normal lung tissues, and the findings were consistent with the results reported in a previous study investigating malignant mesothelioma, which identified that miR-615-3p was downregulated in NSCLC tissues (14). Another study reported that miR-615-3p was upregulated and significantly associated with overall survival time in mantle-cell lymphoma (15). In the study by Goswami et al (15), miR-615-3p overexpression was observed to be upregulated in non-Hodgkin lymphoma (NHL) formalin-fixed, parafffin-embedded samples, and was significantly associated with the overall survival time of NHL patients; these findings are different from the present results. This discrepancy may be generated due to two reasons: First, in the study by Goswami et al, miR expression was detected with the TaqMan Low-Density Array, which was different from our Custom MicroRNA Microarray. Second, it is a normal phenomenon that one gene serves opposite roles in different tumors. However, at present, no studies on the biological function of miR-615-3p in cancer have been reported. To the best of our knowledge, the present study reported for the first time that miR-615-3p was downregulated in NSCLC tissues and acted as a tumor suppressor in NSCLC cells. Experiments performed in vitro exhibited that miR-615-3p markedly inhibited cell growth, arrested cell cycle progression, induced apoptosis and repressed cell migration and invasion in NSCLC cells. The present results suggest that miR-615-3p is important in the tumorigenesis and progression of NSCLC. On the basis of the results from the present study, additional investigation of the target gene of miR-615-3p in NSCLC cells is required.
In summary, to the best of our knowledge, the present study demonstrated for the first time that miR-615-3p acts as a tumor suppressor in NSCLC. In addition, the present findings revealed the biological function of miR-615-3p in NSCLC and provided an insight into a potential molecular mechanism of NSCLC carcinogenesis.
Acknowledgements
This study was supported by the State Key Laboratory of Oncology in South China (grant no. 2013010811).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.NSCLC Meta-Analysis Collaborative Group, corp-author. Preoperative chemotherapy for non-small-cell lung cancer: A systematic review and meta-analysis of individual participant data. Lancet. 2014;383:1561–1571. doi: 10.1016/S0140-6736(13)62159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 7.Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, Wan J, Zhou F, Shao K, Sun Y, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res. 2011;17:6802–6811. doi: 10.1158/1078-0432.CCR-11-0419. [DOI] [PubMed] [Google Scholar]
- 8.Vosa U, Vooder T, Kolde R, Fischer K, Välk K, Tõnisson N, Roosipuu R, Vilo J, Metspalu A, Annilo T. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50:812–822. doi: 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- 9.Wang R, Wang ZX, Yang JS, Pan X, De W, Chen LB. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14) Oncogene. 2011;30:2644–2658. doi: 10.1038/onc.2010.642. [DOI] [PubMed] [Google Scholar]
- 10.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Gao W, Yu Y, Cao H, Shen H, Li X, Pan S, Shu Y. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64:399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2 and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma-A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–623. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 15.Goswami RS, Atenafu EG, Xuan Y, Waldron L, Reis PP, Sun T, Datti A, Xu W, Kuruvilla J, Good DJ, et al. MicroRNA signature obtained from the comparison of aggressive with indolent non-Hodgkin lymphomas: Potential prognostic value in mantle-cell lymphoma. J Clin Oncol. 2013;31:2903–2911. doi: 10.1200/JCO.2012.45.3050. [DOI] [PubMed] [Google Scholar]