Abstract
The adaptor protein Srcin1 is a novel Src-binding protein that regulates Src activation through C-terminal Src kinase (Csk). Srcin1 behaves as a tumour suppressor in breast cancer, but the role of Srcin1 in the development of colorectal cancer (CRC) remains unknown. In the present study, Srcin1 expression in normal tissue was examined by tissue microarray and assessed by immunohistochemistry in 10 patients. In addition, the biological impact of Srcin1 knockdown on CRC cells was investigated in vitro and in vivo. The results showed that Srcin1 was expressed in different types of normal human tissues, whereas its expression was increased in human CRC tissues. Srcin1 expression also correlated with tumour progression. The suppression of Srcin1 induced cell differentiation and G0/G1 cell cycle arrest. Furthermore, Srcin1 increased cell growth as well as the capacity of migration and invasion in CRC cells. Srcin1 induced the activation of the Wnt/β-catenin signalling pathway. Moreover, Srcin1 suppression sensitized cancer cells to 5-fluorouracil (5-FU)-induced apoptosis in vitro and in vivo. Together, these results demonstrate that Srcin1 contributes to CRC carcinogenesis, invasion and metastasis. These findings provide a rationale for a mechanistic approach to CRC treatment based on the development of Srcin1-targeted therapies.
Keywords: Srcin1, colorectal cancer, carcinogenesis, metastasis, invasion
Introduction
Colorectal carcinoma (CRC) is one of the most common types of solid malignancies and is a primary cause of cancer-related mortality worldwide (1). Colorectal carcinogenesis is a complex multistep process that involves the progressive disruption of proliferation (2), differentiation (3), apoptosis (4) and survival mechanisms (5) of intestinal epithelial cells. Although the survival rates are reasonably good for patients who are diagnosed with the disease in its very early stages (6), the majority of patients present with advanced disease. Therefore, chemotherapeutic agents have limited efficacy in patients who have relapsed and in those who present with metastatic disease (7). Therefore, new strategies are needed to improve patient survival and quality of life in this setting.
With substantial advances in our understanding of tumour biology, key signalling pathways that are involved in the mediation of CRC growth and progression have been identified. Dominant oncogenes and tumour suppressor genes that are involved in the pathogenesis of CRC have attracted substantial interest (8), and their central roles and fundamental contribution to the dysregulation of cancer cells have been elucidated (9). These genes offer new targets for biological therapies.
One such target is the v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (Src) (10), a tyrosine kinase that is frequently overexpressed in cancer. Expression of this gene leads to the increased migration and invasiveness of various types of cancer cells (11,12), including CRC cells (13). Src kinase signalling inhibitor 1 (Srcin1), which is also known as Srcin1 (p140 Cas-associated protein) (14), contains two coiled-coil domains, two proline-rich regions and two regions of highly charged amino acids. According to the characteristic domain structure, Srcin1 is thought to act as an adaptor protein (15).
Recently, it was determined that Srcin1 negatively controls tumour cell properties via the inhibition of in vivo tumour growth and metastasis of human breast cancers (16,17). This result suggests that Srcin1 functions as a tumour suppressor gene and may slow tumour progression (18). However, the manner in which Srcin1 acts at the molecular and cellular levels in CRC has yet to be elucidated.
Materials and methods
Reagents and antibodies
Sodium butyrate and 5-FU (5-fluorouracil) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sodium butyrate has various effects on cultured mammalian cells including inhibition of proliferation, induction of differentiation and induction or repression of gene expression (19). As such, it can be used in lab to bring about any of these effects. Specifically, butyrate treatment of cells results in histone hyper acetylation, and butyrate itself inhibits class I histone deacetylase (HDAC) activity (20), specifically HDAC1, HDAC2, HDAC3 and HDAC8. Butyrate is an essential vehicle for determining the role of histone acetylation in chromatin structure and function. Inhibition of HDAC activity is estimated to affect the expression of only 2% of mammalian genes (21).
Mouse anti-human Srcin1, cyclin D1, CDK6, cyclin B and mouse anti-human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which were used for western blotting, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Mouse anti-human Srcin1, which was used for western blotting and/or immunohistochemistry, was purchased from Novus Biologicals LLC (Littleton, CO, USA). Goat anti-rabbit immunoglobulins/HRP and rabbit anti-mouse immunoglobulins/HRP were purchased from Dako (Carpinteria, CA, USA).
Cell lines, vectors and transfection
Human colorectal carcinoma LS174T, SW620, SW1116, LoVo, W480, Caco-2, DLD1 and HT29 cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin in a humidified incubator at 37°C in an atmosphere of 5% CO2. Complementary DNA (cDNA) that corresponds to full-length Srcin in a pcDNA3.1 plasmid was obtained by RT-PCR amplification of cDNA from normal human testis. The clones were digested with NheI and KpnI (Invitrogen, Carlsbad, CA, USA), and the constructs were transiently transfected into LoVo cells in the presence of 8 µg/ml polybrene (Sigma-Aldrich) using Lipofectamine 2000 (Invitrogen). Construct expression was then confirmed by western blotting. For stable selection, 800 µg/ml G418 (Gibco, Carlsbad, CA, USA) was used.
Tissue microarray (TMA) and immunohistochemistry (IHC)
A tissue microarray (TMA) containing normal human tissue samples was purchased from Alenabio, Co., Ltd. (Xian, China). In accordance with the manufacturer's recommendations, all tissue samples were fixed in formalin, processed in a tissue processor (after no more than 24 h of fixation) and finally embedded in paraffin. The TMA FDA807-1 contains 80 cores (1.5 mm diameter) of the following 16 normal tissue types (5 cores of each tissue type): breast, brain, colon, oesophagus, kidney, liver, lung, ovary, pancreas, prostate, skin, small intestine, stomach, testis, uterine and rectum. In all, 8 patients with CRC metastatic lymph nodes and 10 patients with CRC tissues were excised in the Department of Surgery of Nanfang Hospital, Southern Medical University. The Ethics Committee of the Southern Medical University, China approved the experimental protocols. Histopathological analyses confirmed the malignant and adjacent normal mucosa tissues.
Western blot analysis
LoVo cell lysates were prepared by homogenization in RIPA buffer. The protein concentrations were determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA). The protein lysates were then resolved by SDS-PAGE. The blots were probed with the appropriate primary antibody followed by a horseradish peroxidase-conjugated secondary antibody. Antigen-antibody complexes were visualized by an enhanced chemiluminescence system (Amersham Biosciences, Ltd., Little Chalfont, UK).
WST-1 assay
The measurement was based on the ability of viable cells to cleave the sulfonated tetrazolium salt WST-1 (4-(3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio)-1, 3-benzene disulfonate) by mitochondrial dehydrogenases. LoVo cells (5,000 cells/well) were plated in a 96-well plate in regular growth medium. After 12, 24 and 48 h 10 µl of WST-1 reagent was added in each well followed by additional incubation for 1–2 h. The absorbance at 450 nm was measured using a microplate reader. We considered OD of vector (or treated) cells at time 12 h as 100%, and converted OD values by the following calculation: % growth = [(average OD treated cells/average OD untreated cells) × 100].
Migration and Matrigel invasion assays
Cell migration was analysed by a conventional wound-healing assay, as previously described (22). Briefly, Srcin1-siRNA or scrambled-siRNA (Scr-siRNA) stably infected cells were grown in 6-well plates to confluence. Wounds were generated using the tip of a sterile micropipette, and the detached cells were removed by washing with PBS. The medium was then refreshed to remove cell debris, and the cells were cultured in the presence of 10 µg/ml mitomycin C to inhibit cell proliferation. Then, the cells were incubated and allowed to migrate for up to 48 h. Images were captured, and the migration index was calculated as follows: migration index = [(initial wound width − width of wound at time-point tested)/initial wound width] × 100%. A Matrigel invasion assay was performed using Matrigel invasion chambers (BD Biosciences) according to the manufacturer's instructions. Matrigel membranes were rehydrated for 2 h in RPMI-1640 medium, and all inserts were then placed in wells that contained 10% FBS in RPMI-1640. For each experiment, a total of 5×104 cells suspended in 0.5 ml of serum-free RPMI-1640 was added to the top of each of three chambers and incubated at 37°C in 5% CO2. After 24 h, the chambers were washed with phosphate-buffered saline (PBS), and the cells were removed from the top of the membranes with a cotton swab. Invading cells were fixed in 4% paraformaldehyde for 30 min and stained with 2% crystal violet. In each chamber, five representative fields were documented by photo-microscopy and the number of invading cells was counted. The average number of invading cells in each chamber in the triplicate samples is presented.
Flow cytometric analysis
To analyse the cell cycle, the cells were collected and fixed in ice-cold 70% ethanol in PBS and stored at −4°C until use. After resuspension, the cells were incubated with 100 µl of RNase I (1 g/ml) and 100 µl of propidium iodide (400 µg/ml) at 37°C and were then analysed by flow cytometry (FCM; BD Biosciences, San Jose, CA, USA). The cell cycle phase distribution was calculated from the resultant DNA histogram using MultiCycle AV software (Phoenix Flow Systems, San Diego, CA, USA). Apoptosis was detected with an Annexin V-FITC kit according to the manufacturer's instructions (Trevigen, Inc., Gaithersburg, MD, USA). Briefly, the cells that received various treatments were collected and stained with Annexin V-FITC and PI in the dark for 15 min at room temperature. After the addition of binding buffer, the cells were subjected to flow cytometry and analysed using WinMDI 2.9.
Immunofluorescence and the morphological detection of apoptosis
For the staining of F-actin, the cells were fixed in 3.7% formaldehyde and incubated with rhodamine-conjugated phallotoxin (5 U/ml; Molecular Probes, Eugene, OR, USA) in PBS at room temperature. The coverslips that contained the cells were washed, mounted and visualized.
A morphological evaluation of apoptotic cell death was performed as previously described, with some modifications. The cells were fixed for 5 min in 3% paraformaldehyde in PBS. After the cells were air-dried, they were stained for 10 min in Hoechst 33258 (10 µg/ml), mounted in 50% glycerol containing 20 mM citric acid and 50 mM orthophosphate, and stored at -20°C. Prior to analysis, nuclear morphology was evaluated using a Zeiss IM 35 fluorescence microscope. Illustration of apoptotic cells after staining with Hoechst 33258. The rate of apoptotic cells was caculated by the average value of apoptotic cells in 100 cells in three different fields.
Caspase-3, -8 and -9 activity assay
The activity of caspases-3, -8 and 9 was determined using the ApoAlert caspase colorimetric assay kit according to the manufacturer's instructions (One unit is the amount of enzyme that will cleave 1.0 nmol of the colorimetric substrate Ac-DEVDpNA per hour at 37°C under saturated substrate concentrations. Both the concentrations of pNA and the value of OD405 are proportional) (Clontech Laboratories Inc., Mountain View, CA, USA). Briefly, the assays were performed in 96-well microtiter plates; 10 µ of protein from cell lysates of each sample was incubated in 80 µl of the reaction buffer [1% NP-40, 20 mmol/l Tris-HCl (pH 7.5), 137 mmol/l Nad, and 10% glycerol] containing 10 µl of caspase-3, -8 or -9 substrate (Ac-DEVDpNA; 2 mmol/l). The cell lysates were incubated at 37°C for 4 h. The samples were then measured by an enzyme-linked immunosorbent assay (ELISA) reader at an absorbance of 405 nm. All experiments were performed at least 3 times.
Transient transfection and Dual-luciferase reporter assay
The reporter constructs pTOPFLASH and pFOPFLASH, in which the luciferase gene was driven by wild-type or dominant-negative mutant consensus 3x T-cell factor/lymphoid enhancer factor binding element (TBE), respectively, were kindly provided by Professor J. Schneikert of the University Erlangen-Nürnberg, Germany. Transient transfection was performed with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The firefly and Renilla luciferase activities were measured using the Dual-luciferase reporter assay kit (Promega, Madison, WI, USA) with a luminometer (Lumat LB 9507; Berthold Technologies GmbH, Bad Wildbad, Germany).
Construction and transfection of lentiviral vectors with Srcin1 short hairpin RNA
To investigate the effect of small interfering RNA (siRNA)-induced downregulation of Srcin1 expression on tumour growth in vivo, a Srcin1 RNAi lentiviral vector (pGCSIL-Srcin1 shRNA) was constructed (Shanghai GeneChem, Co., Ltd, Shanghai, China). Double-stranded oligonucleotides encoding human Srcin1-vshRNA CCGGAAGCTGTGTCTGTTGAGGCTGTCAAGAGCAGCCTCAACAGACACAGCTTTTTTTG) were inserted into the short hairpin RNA (shRNA) expression vector pGCSIL. A scrambled sequence that shared no homology with the mammalian genome was used as a control. The successful cloning of these sequences into the pGCSIL-GFP lentivector was confirmed by Sanger sequencing. The expression of shRNA was driven by a U6 promoter. This vector also encoded green fluorescent protein (GFP). The recombinant lentiviral vector was produced by co-transfection of HEK293FT cells with the lentiviral expression vector and the packing plasmid mix using Lipofectamine 2000 according to the manufacturer's instructions. LoVo cells were then transduced at a multiplicity of infection (MOI) of 5.
In vivo tumourigenesis
Female BALB/c nude mice (Laboratory Animal Unit, Southern Medical University, Guangzhou, China) were housed in a specific pathogen-free (SPF) environment. In all, 5×106 LoVo cells were subcutaneously injected into twenty 4-week-old male nude mice (five mice per group). Seven days after subcutaneous tumour cell injection, mice were anesthetized and 1.5×1011 v.g. of lenti-Src shRNA, lenti-Srcin1 shRNA lenti-Src shRNA + 5-FU, lenti-Srcin1 shRNA + 5-FU was injected (23). The size of the tumours was measured every 7 days using a microcaliper. The volumes of the tumours were calculated as follows: V = (4/3) R12R2, where R1 is radius 1, R2 is radius 2, and R1<R2 (the smallest and the largest tumours were deleted). The mice were sacrificed by CO2 euthanasia 30 days after the injection. All tumours were removed and dissociated. The Committee on the Use of Live Animals in Teaching and Research of Southern Medical University approved the protocol.
Statistical analyses
The results are representative of at least three independent experiments that were performed in triplicate and are expressed as the mean ± SD. A statistical analysis of the data was performed using Student's t-test. A value of P<0.05 was considered significant.
Results
Srcin1 expression is significantly increased in human normal tissues, CRC and metastatic lymph node tissues
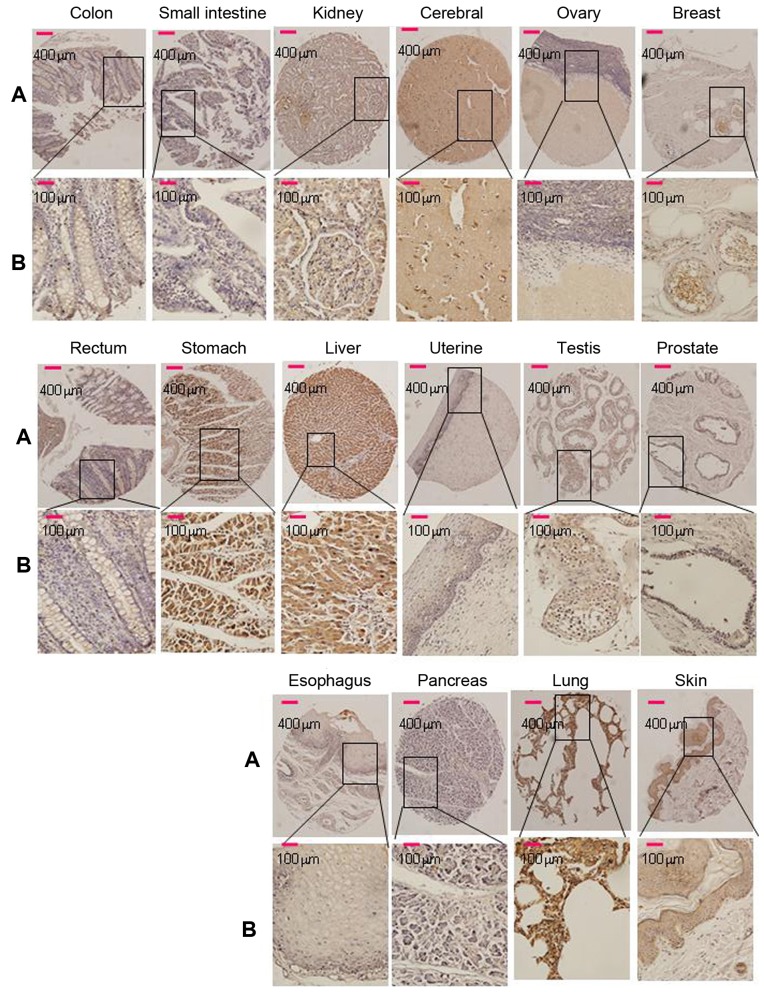
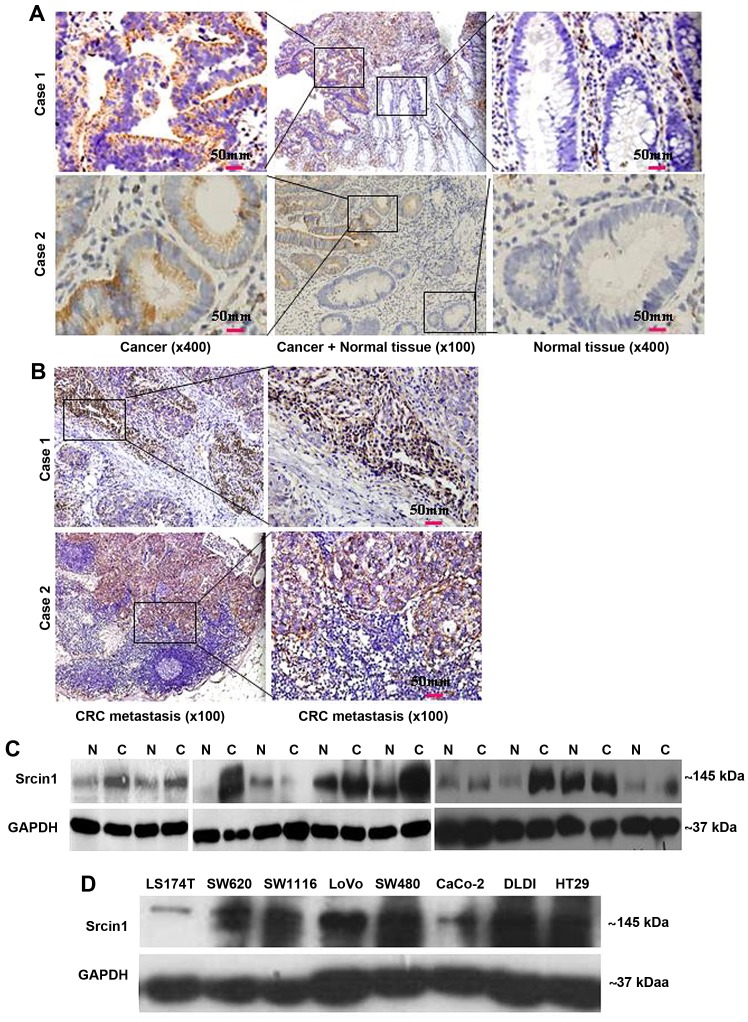
Of the 16 normal tissue types (5 cores of each tissue type) that were tested for Srcin1 expression in this TMA, cytoplasmic immunoreactivity was observed in 100% (all 5 cores) of the breast, cerebrum, liver and skin samples. Strong cytoplasmic immunoreactivity was observed in 80% of the kidney, testis and stomach samples; cytoplasmic immunoreactivity was observed in 60% of the lung and 20% of the pancreas samples (Fig. 1). Cells of the esophagus, ovary, prostate and uterus were negative. Among them, 80% (4/5) of cores that represented normal colon and rectal tissues were negative (Fig. 1). The paraffin tissue sections of normal colorectal mucosa showed either negative or weak staining for Srcin1 protein. However, the expression of Srcin1 protein, as exemplified in two patients by IHC, was predominantly localized within the cytoplasm of colorectal cancer cells (21/25, 84% of the cases) (Fig. 2A). In addition, all of CRC metastatic lymph nodes were positive for Srcin1 protein (8/8, 100% of the cases) (Fig. 2B).
Figure 1.
Srcin1 expression in 16 normal human tissues was detected by immunohistochemistry. Breast, brain, colon, oesophagus, kidney, liver, lung, ovary, pancreas, prostate, skin, small intestine, stomach, testis, uterine and rectum, of 16 normal human tissues stained with anti-Srcin1 antibody by means of TMA. Original magnification, A, ×100 and B, ×400.
Figure 2.
CRC cells express higher levels of Srcin1 than normal cells. (A) Expression of Srcin1 in normal and malignant human colorectal tissues were detected by IHC. (B) Expression of Srcin1 in metastatic lymph nodes were detected by IHC. (C) Srcin1 expression in colon cancer (C) tissues and matched normal colon (N) tissues as detected by western blot analysis. GAPDH was used as the internal control for western blot analysis. (D) Expression of Srcin1 in colorectal cancer cell lines. All of these experiments were repeated 3 times with identical findings.
We then measured Srcin1 expression in 10 pairs of matched normal colon (N) and cancerous colon (T) tissues by western blotting. Of the 10 cancerous tissue specimens, 7 expressed higher levels of Srcin1 than normal tissues (7/10, 70% of the cases) (Fig. 2C). Next, Srcin1 protein was observed in the colorectal cells lines LS174T, SW1116, SW620, LoVo, SW480, CaCo, DLD1 and HT29 by western blotting (Fig. 2D).
Thus, a high level of Srcin1 protein expression correlates with a transformed phenotype, which indicates that Srcin1 may play a role in CRC tumourigenesis.
Suppression with siRNA induces cell differentiation in CRC cells
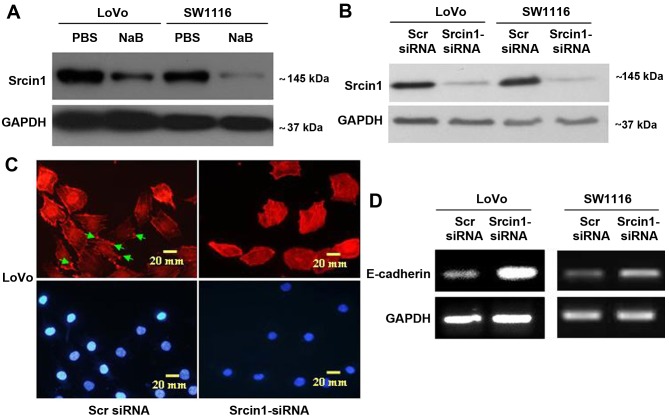
To examine the effect of Srcin1 on cell differentiation (Fig. 3A), we synthesized Srcin1-siRNA, the expression of which was confirmed by western blotting (Fig. 3B) in both CRC cell lines (LoVo and SW1116). We first stained for F-actin in Srcin-suppressed cells using phalloidin-conjugated rhodamine and immunofluorescence microscopy. Diffuse and generally uniform staining was observed throughout the cytoplasm in Srcin1-siRNA-transfected LoVo and SW1116 cells. However, after Scr siRNA transfection, F-actin was present throughout the cytoplasm and at the rim zone of the protrusion (arrow), an area that features actin polymerization (Fig. 3C).
Figure 3.
Suppression with siRNA induces cell differentiation in CRC cells. (A) Srcin1 expression in LoVo and SW1116 cells treated without or with sodium butyrate (NaB) for 48 h, as detected by western blot analysis. (B) LoVo and SW1116 cells were transfected with Srcin1-siRNA and Scr-siRNA (scramble siRNA) for 48 h following by western blot analysis. (C) LoVo cells transfected with Srcin1-siRNA or Src-siRNA were stained with rhodamine-phallotoxin for 48 h, with F-actin filaments being visualized under fluorescent microscopy. (D) Expression of E-cadherin in LoVo and SW1116 cells transfected with Srcin1-siRNA and Src-siRNA for 48 h, as detected by RT-PCR. These pictures are representatives of 3 independent experiments with identical results.
Furthermore, we showed by reverse-transcription polymerase chain reaction (RT-PCR) that the knockdown of Srcin1 increased the expression of differentiation marker E-cadherin in both LoVo and SW1116 cells (Fig. 3D). This result suggests that the knockdown of Srcin1 might induce the differentiation of CRC cells.
The impact of Srcin1 on cell cycle arrest in human CRC cells
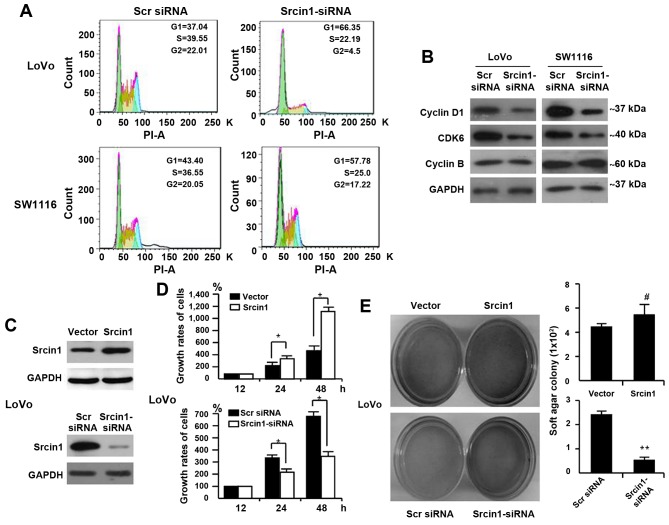
We performed flow cytometric analyses to detect whether Srcin1 could change the cell cycle distribution. As shown in Fig. 4A, Srcin1 knockdown resulted in an increase in cells in the G0/G1-phase and a concomitant decrease in cells in the S-phase and G2/M-phase of the cell cycle.
Figure 4.
Srcin1 expression modulates cell cycle and cell proliferation in human CRC cells. (A) Cell cycle analysis was examined by FACScan using LoVo and SW1116 cells. RNAi-mediated repression of Srcin1 modulated G0/G1 checkpoint. (B) Expression of cyclins and cyclin-dependent kinase (CDKs) proteins after treatment with Srcin1-shRNA. All these experiments were repeated two times with identical findings. (C) LoVo/vector and LoVo/Srcin1 or Srcin1-siRNA and Src-siRNA were detected by western blot analysis with GAPDH as the internal control. (D) Cells seeded in 96-well tissue culture plates in triplicate were collected 12, 24 and 48 h by WST-1 assay. The growth rates of the cells are expressed as means ± SEM (n=3; *P<0.05). (E) Cells were plated in a tissue culture dish with complete culture medium containing 0.35% agar on top and 0.5% agar at the bottom. After 14 days, cell colonies were visualized after staining with 0.005% crystal violet. Colonies containing >50 cells were considered viable. The results are expressed as means ± SD. #P>0.05 and **P<0.05.
Furthermore, cell cycle-related protein expression was assessed by western blotting. Consistent with the accumulation of cells in G0/G1 phase, the expression of cyclin D1 and CDK6 was significantly decreased in Srcin1-siRNA-transfected LoVo and SW1116 cells, whereas the expression of cyclin B remained unchanged compared with scrambled-siRNA-transfected cells (Fig. 4B).
Together, these results indicate that Srcin1 silencing in CRC is associated with a block in cell cycle progression.
Effect of Srcin1 on the proliferation of CRC cells
To demonstrate the effect of Srcin1 on cell growth, we first detected Srcin1 expression in the CRC cell line LoVo using a WST-1 assay. We then established stable transfectants of LoVo cells that expressed the vector pcDNA3.1, pcDNA3.1-Srcin1 and Scr siRNA and Srcin1-siRNA were confirmed by immunoblotting (Fig. 4C). We found that the growth rates of the vector-transfected cells were 100%, 260.80±9.33 and 477.84±35.09% at 12, 24 and 48 h, respectively, whereas those of Srcin1-expressing cells were 100%, 371.48±13.87 and 1168.31±176.82%, respectively (Fig. 4D). A significant difference between Srcin1- and the vector-transfected cells was found at two time-points (24 and 48 h).
Moreover, we assessed the effect of Srcin1 repression on CRC cell proliferation. We showed that the growth rates of Scr-siRNA-transfected cells were 100%, 357.47±39.76 and 673.97±72.43% at 12, 24 and 48 h, respectively, whereas Srcin1-siRNA-transfected cells were 100%, 210.98±15.68 and 332.70.97±24.79% at 12, 24 and 48 h, respectively (Fig. 4D). A significant difference between the Srcin1-siRNA- and Scr-siRNA-transfected cells was found at two time-points (24 and 48 h). Srcin1 overexpression significantly inhibited cancer cell growth (P<0.05).
In addition, we examined the anchorage-independent growth of the stable transfectants (empty vector and Srcin1) in soft agar after 12 days. As expected, LoVo empty vector cells formed colonies in soft agar, and the forced expression of Srcin1 in LoVo cells markedly enhanced colony formation in soft agar (Fig. 4E). In contrast, we tested the effect of RNAi-mediated repression of Srcin1 in LoVo cells. The application of Srcin1-siRNA significantly inhibited the ability of these cells to form colonies (Fig. 4E), which indicates that Srcin1 downregulation suppressed the proliferation of colon cancer cells.
The role of Srcin1 in CRC cell metastasis and invasion
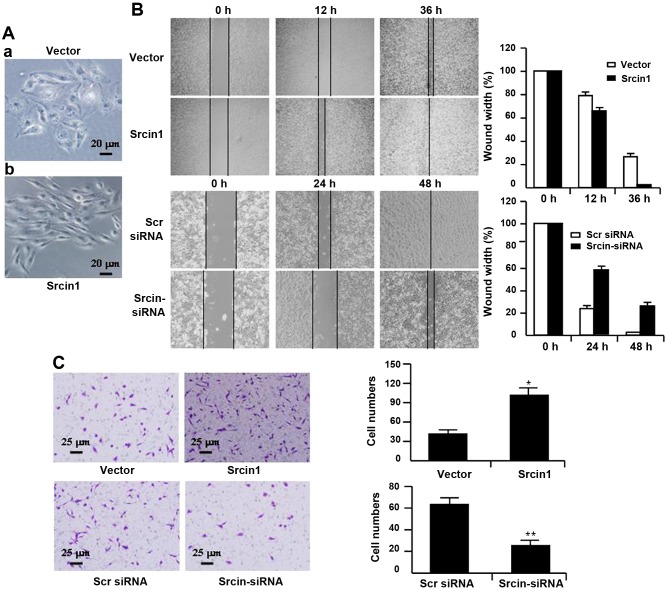
To characterize Srcin1 transfectants, we first examined the morphologic features of these cells. The parental cells (i.e., cells that were stably transfected with vector) displayed a flat morphology with a short cytoplasmic process (Fig. 5A–a). However, the cells that were stably transfected with Srcin1 demonstrated diffuse Srcin1 expression in the cytoplasm and had an elongated or shuttle-shape morphology with a loss of cell-cell contacts. In addition, increased cell scattering was observed according to phase-contrast microscopy, which may be associated with an EMT-like conversion (Fig. 5A–b) (24).
Figure 5.
Srcin1 expression modulates cell migration and invasive of LoVo cells. (A) LoVo cells stably expressing Srcin1 show spindle-like, fibroblastic morphology under phase-contrast microscope; (magnification, ×20). (B) Images of the wound closure of pooled monolayer of stable transfectants or knockdown of Srcin1 of LoVo cells; magnification, ×10. (C) Invasive potential of pooled stable transfectants or RNAi-mediated repression of Srcin1 of CRC cells; *P<0.05; **P<0.01. The experiments were repeated three times with identical findings.
Therefore, we studied the effect of Srcin1 on the metastatic and invasive potential of CRC-derived LoVo cells. Cell migration was determined using a wound-healing assay. As shown in Fig. 5B, Srcin1 upregulation significantly increased LoVo cell migration, whereas Srcin1 downregulation significantly suppressed migration. The migration index of Srcin1 transfectants was increased by 12 and 24% at 12 and 36 h, respectively. In contrast, the migration index of Srcin1 knockdown cells was decreased by 30 and 28% at 24 and 48 h, respectively (Fig. 5B).
To examine cell invasion in vitro, we used Transwell inserts coated with Matrigel matrix. The invasiveness of LoVo cells that were transfected with Srcin1 increased by 110.6% compared with the control cells. In contrast, after Srcin1 was knocked down, the invasiveness of LoVo cells decreased by 269.5% compared with the Scr siRNA-transfected cells (Fig. 5C).
These data indicate that the role of Srcin1 in invasion and metastasis are associated with an increase in the metastatic potential of cancer cells.
Srcin1 induces the activation of the Wnt/β-catenin signalling pathway
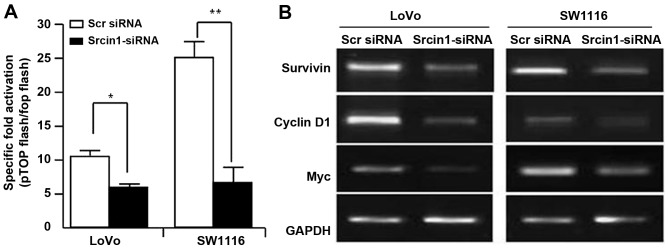
The Wnt/β-catenin signalling pathway governs cell proliferation, differentiation, migration and invasion in CRC (25,26). We assessed the effect of Srcin1 on the trans-activation of β-catenin. pTOPFLASH and pFOPFLASH, contained the wild-type and dominant-negative TBE binding element, respectively. We knocked down the expression of Srcin1 in LoVo and SW1116 cells using siRNA, and the luciferase reporter assay was then used. As shown in Fig. 5A, Srcin1-siRNA decreased the expression of Srcin1 compared with Scr siRNA, and additionally, the TOP-Flash plasmid, but not the FOP-Flash plasmid, was downregulated by Srcin1-siRNA, but not by Scr siRNA (Fig. 6A).
Figure 6.
Knockdown of Srcin1 decreases β-catenin transcriptional activity. (A) Transcription activities of pTOPFLASH and pFOPFLASH cotransfected with Scr siRNA or Srcin1-siRNA in LoVo and SW1116 cells. The results expressed relative luciferase unit ratios between pTOPFLASH and pFOPFLASH; *P<0.05; **P<0.01. (B) RNA expression of survivin, cyclin D1 and Myc were detected in cells with transfection of Scr siRNA or Srcin1-siRNA for 24 h by RT-PCR.
To further investigate whether Srcin1-siRNA regulated the transcriptional activity of Wnt/β-catenin signalling, we examined the expression of survivin, c-Myc and cyclin D1. Indeed, the knockdown of Srcin1 by shRNA also inhibited the expression of survivin, c-Myc and cyclin D1 (Fig. 6B). These results indicate that Srcin1 may directly or indirectly regulate Wnt/β-catenin signalling in CRC.
Srcin1-specific silencing induces apoptosis and sensitizes cancer cells to the chemotherapeutic 5-FU
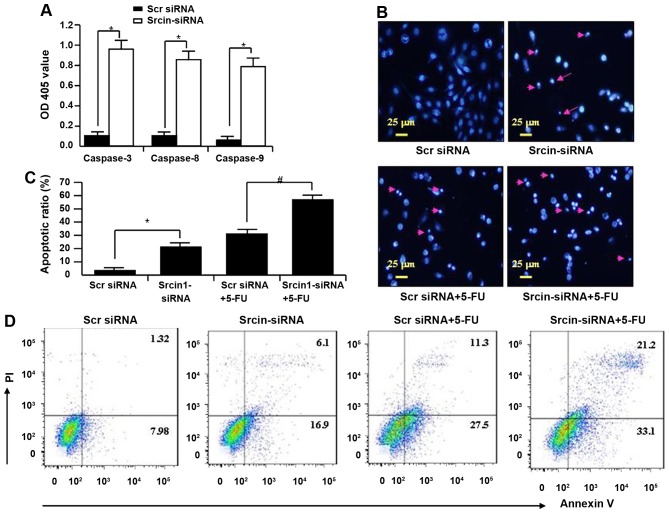
To investigate the mechanism of knockdown of Srcin1-induced growth suppression, apoptosis was assayed by ELISA. The activities of caspase-3, -8 and -9 were also analysed. The activities of caspase-3, -8 and -9 were significantly increased in Srcin1-siRNA-transfected cells compared with Src siRNA-transfected cells (Fig. 7A).
Figure 7.
Suppression of Srcin1-siRNA increased cell susceptibility to apoptotic stimuli. (A) The activity of caspase-3, -8 and -9 in LoVo after Srcin1-siRNA for 72 h. OD405 nm of transfected cells and parental cells was obtained and calculated individually in each group (*P<0.05, comparing to Scr-siRNA). (B) LoVo cells were treated with Scr-siRNA, Srcin1-siRNA, Scr-siRNA + 5-FU or Srcin1-siRNA + 5-FU. Nuclei were stained with Hoechst 33258 and visualized under a fluorescent microscope (arrow indicates cells with nuclear fragmentation and condensed chromatin). Scale bars, 25 µm. (C) Illustration of apoptotic cells after staining with Hoechst 33258. The values are expressed as the means ± SEM from 3 separate experiments. *P<0.05; #P<0.05 between the 2 transfectants. (D) Cells with Scr siRNA or Srcin1-siRNA were treated with 5-FU for 48 h, double stained with Annexin V-FITC and PI, followed by flow cytometric analysis to determine apoptosis. These figures are representatives of two independent experiments with same findings.
Apoptotic induction was further confirmed by DAPI staining at the single-cell level. Bright blue-fluorescent condensed nuclei and chromatin fragmentation were considered indicative of apoptosis by fluorescence microscopy (Fig. 7B). Similarly, the ratios of cells with condensed nuclei were higher in Srcin1-siRNA-transfected LoVo cells compared with the Src siRNA-transfected LoVo cells (P<0.05 compared with Src siRNA; Fig. 7B).
After an evaluation of the role of Srcin1-siRNA in chemotherapy-induced apoptosis, the ratios of cells with condensed nuclei were higher in Srcin1 + 5-FU-treated LoVo cells compared with the Src siRNA + 5-FU-treated controls (P<0.05; Fig. 7C).
To validate the results of DAPI staining, Src siRNA-transfected cells were treated with 5-FU for 48 h; the cells were then stained with Annexin V and PI and analysed by flow cytometry. As shown in Fig. 7D, the apoptotic index of Srcin1-siRNA + 5-FU-treated cells was significantly increased relative to Src siRNA-transfected cells.
These findings suggest that Srcin1-siRNA enhances the susceptibility of cancer cells to apoptotic triggers induced by 5-FU.
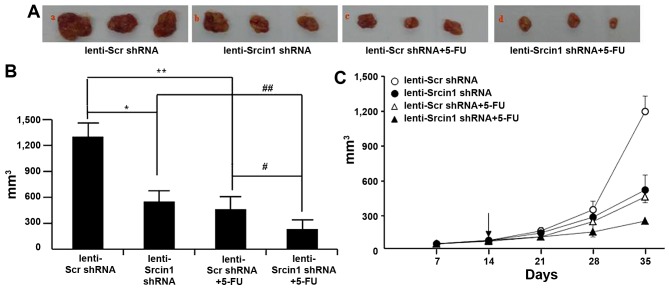
Suppression of Srcin1 sensitizes cancer cells to 5-FU-induced apoptosis in vivo
Finally, we investigated the potential utility of lenti-Srcin1 shRNA as a therapy for established tumours. Lentivirus-transduced LoVo cells were subcutaneously injected into the right dorsal flank of nude mice, some of which were treated with 5-FU. When the tumour nodules became visible (~3–5 mm in diameter), lenti-Src shRNA or lenti-Srcin1 shRNA was injected directly into the tumour; 5-FU was intraperitoneally injected as a co-treatment. The tumour sizes were continuously monitored on a weekly basis. As shown in Fig. 8A and B, the tumour volumes of the lenti-Srcin1 shRNA-injected mice were markedly smaller than those of the lenti-Src shRNA-injected mice. Similar results were obtained in the 5-FU-treated (lenti-Scr shRNA + 5-FU or lenti-Srcin1 shRNA + 5-FU) mice compared with untreated (lenti-Scr shRNA or lenti-Srcin1 shRNA) mice (Fig. 8A and B). Importantly, mice that were injected with both lenti-Src shRNA and 5-FU presented with the smallest tumour nodules (Fig. 8C).
Figure 8.
Suppression of Srcin1 with shRNA sensitized cancer cells to 5-FU-induced apoptosis in vivo. (A) LoVo cells (5×106) were injected subcutaneously in the right flanks of the nude mice. When the tumor nodules became visible, 5-FU (50 µg/kg, once per two days, 4 times) or NS were administered by intraperitoneal injection. Images shown were taken on day 35. (B) Tumor growth was monitored in 3 dimensions and expressed as tumor volume in cubic millimeters. Data are the pooled average ± standard error of mean of the tumor volumes for each of 3 animals per group. *P<0.05, lenti-Scr shRNA vs. lenti-Srcin1 shRNA, #P<0.05, lenti-Scr shRNA + 5-FU vs. lenti-Srcin1 shRNA+ 5-FU, **P<0.05, lenti-Scr shRNA vs. lenti-Scr shRNA + 5-FU, ##P<0.05, lenti-Srcin1 shRNA vs. lenti-Srcin1 shRNA+ 5-FU. (C) Tumor size was measured weekly after tumor cell inoculation in each group. The arrow indicates the time when intratumoral and intravenous injections were performed.
These results demonstrate that targeting with lenti-Srcin1 shRNA has an inhibitory effect on tumorigenesis in vivo. Furthermore, inhibition of Srcin1 siRNA has a synergistic effect with 5-FU treatment in the treatment of colorectal cancer.
Discussion
In the present study, we describe a novel role of Srcin1 in the regulation of the pathogenesis and progression of CRC. The evidence that supports our conclusions is as follows: i) by western blotting and IHC, CRC cells expressed higher levels of Srcin1 than normal tissues; CRC metastatic lymph nodes were positive for Srcin1 protein by IHC; ii) RNAi-mediated silencing of Srcin1 in LoVo cells suppressed the proliferation, metastasis and invasiveness of these cells; and iii) lentivirus-Srcin1-shRNA increased cell susceptibility to apoptotic stimuli by 5-FU in vitro and in vivo. Together, these findings provide strong evidence for the oncogenic activity of Srcin1 in CRC.
Despite the high expression of Srcin1 in normal human breast tissues, as reported in previous studies (15,27), Srcin1 expression in other tissue types is unknown. This study showed that Srcin1 is expressed in human somatic tissues according to IHC of a TMA. The present study revealed the unequivocal presence of Srcin1 in 7 of 16 tissues examined. In particular, 80% (4/5) of normal colon and rectal tissues were negative. However, Scrin1 may be a novel negative regulator of tumour growth because it strongly impaired breast cancer cell growth (17). Thus, Srcin1 is particularly intriguing because it can function as either a repressor or an activator of target proteins in a cell type-dependent fashion. Further study could be of interest.
It has been reported that Srcin1 is essential for the regulation of cell proliferation and motility (16,18). Little is known, however, regarding the role of Srcin1 in CRC. Studies have reported that Srcin1 expression in normal human breast tissues inversely correlates with its expression in breast cancer tissues (18). We showed that Srcin1 was expressed at higher a level in CRC cells than in cells from normal tissues. We determined that Srcin1 is a mediator of NaB-induced pro-differentiation of CRC cells. Our finding that Srcin1 suppression induced the maturation of F-actin filaments in cancer cells implicates Srcin1 in the dedifferentiation of cancer cells. Moreover, the suppression of Srcin1 increased the expression of a differentiation marker for colorectal epithelial cells (E-cadherin). Taken together, our data here show that the suppression of Srcin1 increased differentiation and tumorigenesis of CRC.
The cell cycle is regulated by a series of checkpoints that monitor the genomic integrity and ensure that DNA replication proceeds in a coordinated manner (28). Aberrations in cell cycle progression occur in the majority of human malignancies (29). Different combinations of cyclin and CDK subunits operate at checkpoint controls during the cell cycle to integrate mitogenic and antiproliferative signals (30). Cyclin D1 has a critical role in the control of the G1/S transition (31). The present study indicates that downregulation of Srcin1 causes G0/G1 phase cell cycle arrest via a reduction in cyclin D1 levels, which appears to be the underlying mechanism in CRC growth inhibition. Therefore, this might also contribute to G0/G1 phase arrest.
The canonical Wnt/β-catenin signalling pathway plays an essential role in the regulation of developmental and adult tissue homeostasis (25,26). This pathway is centred on β-catenin, which binds to TCF/LEF transcription factors and leads to the transcription of target genes (32). In this study, we proposed that Srcin1 could regulate Wnt/β-catenin. The data that were obtained from both experiments where Srcin1 was overexpressed or knocked down by siRNA supported our hypothesis. Moreover, Srcin1 expression affected the location and function of the TCF/β-catenin complex, which results in the induction of genes that are downstream of β-catenin. These studies and our results suggest that Srcin1 may play a critical role in cell to-cell adhesion through its regulation of the TCF/β-catenin complex. We are currently evaluating the regulation of Wnt/β-catenin signaling by Srcin1 in vivo.
Recently, the catalytic activity of Src kinases was found to regulate proliferation, migration and invasiveness of MDA-MB-231 breast cancer cells, as well as decrease their susceptibility to the chemotherapy agent oxaliplatin in CRC (33,34). However, whether Srcin1 contributes to chemoresistance remains to be elucidated. In the present study, we demonstrated that Srcin1 knockdown in LoVo cells remarkably suppressed cell proliferation and enhanced cell apoptosis in response to 5-FU treatment in vitro. In vivo, Srcin1 knockdown inhibited xenograft tumorigenesis and resulted in the near eradication of established CRC xenografts when combined with 5-FU. Collectively, these findings imply that Srcin1 inhibition by RNAi along with 5-FU-based chemotherapy may be exploited as a potential synergistic therapy for patients with CRC. Srcin1 interacts with Src kinase and Srcin1 and Src may differentially regulate CRC cell differentiation, proliferation, migration, invasion, and survival, we are evaluating the effects of Srcin1 overexpression and knockdown on Src activity.
Taken together, the results of the present study demonstrate that Srcin1 is crucial for cancer growth and metastasis. Srcin1 knockdown induces differentiation, inhibits cell growth, causes cell cycle arrest in G0/G1 phase, promotes spontaneous apoptosis, and enhances the chemosensitivity of CRC LoVo cells to 5-FU. Our data suggest that Srcin1 may serve as a promising therapeutic target for CRC.
Acknowledgments
The present study was supported by grants from the National Natural Science Funds of China (nos. 81172057, 81272761 and 81470036), the President Foundation of Nanfang Hospital, Southern Medical University (2012B009 and 2013Z007), the High-Level Topic-Matching Funds of Nanfang Hospital (201347 and G201227), the Projects of Science and Technology of Guangdong (2012B050600020), the Guangdong Provincial Key Laboratory of Gastroenterology, Department of Gastroenterology, Nanfang Hospital, Southern Medical University and the Guangzhou Pilot Project of Clinical and Translational Research Center (early gastrointestinal cancer, no. 7415696196402).
Abbreviations
- ATCC
American Type Culture Collection
- qRT-PCR
real-time RT-PCR
- CRC
colorectal cancer
- IHC
immunohistochemistry
- TMA
tissue microarray
- AJCC
American Joint Committee on Cancer
- siRNA
small interfering RNA
- RLU
relative luciferase unit
- MOI
multiplicity of infection
- Srcin1
SRC kinase signalling inhibitor 1
References
- 1.Gill S, Sinicrope FA. Colorectal cancer prevention: Is an ounce of prevention worth a pound of cure? Semin Oncol. 2005;32:24–34. doi: 10.1053/j.seminoncol.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 2.Chinery R, Beauchamp RD, Shyr Y, Kirkland SC, Coffey RJ, Morrow JD. Antioxidants reduce cyclooxygenase-2 expression, prostaglandin production, and proliferation in colorectal cancer cells. Cancer Res. 1998;58:2323–2327. [PubMed] [Google Scholar]
- 3.Wang J, Yang Y, Xia HH, Gu Q, Lin MC, Jiang B, Peng Y, Li G, An X, Zhang Y, et al. Suppression of FHL2 expression induces cell differentiation and inhibits gastric and colon carcinogenesis. Gastroenterology. 2007;132:1066–1076. doi: 10.1053/j.gastro.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Gu Q, Wang JD, Xia HH, Lin MC, He H, Zou B, Tu SP, Yang Y, Liu XG, Lam SK, et al. Activation of the caspase-8/Bid and Bax pathways in aspirin-induced apoptosis in gastric cancer. Carcinogenesis. 2005;26:541–546. doi: 10.1093/carcin/bgh345. [DOI] [PubMed] [Google Scholar]
- 5.He H, Xia HH, Wang JD, Gu Q, Lin MC, Zou B, Lam SK, Chan AO, Yuen MF, Kung HF, et al. Inhibition of human telomerase reverse transcriptase by nonsteroidal antiinflammatory drugs in colon carcinoma. Cancer. 2006;106:1243–1249. doi: 10.1002/cncr.21694. [DOI] [PubMed] [Google Scholar]
- 6.Capozzi E, Della Puppa L, Fornasarig M, Pedroni M, Boiocchi M, Viel A. Evaluation of the replication error phenotype in relation to molecular and clinicopathological features in hereditary and early onset colorectal cancer. Eur J Cancer. 1999;35:289–295. doi: 10.1016/S0959-8049(98)00317-7. [DOI] [PubMed] [Google Scholar]
- 7.Sinicrope FA, Sugarman SM. Role of adjuvant therapy in surgically resected colorectal carcinoma. Gastroenterology. 1995;109:984–993. doi: 10.1016/0016-5085(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 8.Kim IJ, Ku JL, Kang HC, Park JH, Yoon KA, Shin Y, Park HW, Jang SG, Lim SK, Han SY, et al. Mutational analysis of OGG1, MYH, MTH1 in FAP, HNPCC and sporadic colorectal cancer patients: R154H OGG1 polymorphism is associated with sporadic colorectal cancer patients. Hum Genet. 2004;115:498–503. doi: 10.1007/s00439-004-1186-7. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Liang L, Huang L, Ma X, Li D, Cai S. High expression of protein phosphatase 4 is associated with the aggressive malignant behavior of colorectal carcinoma. Mol Cancer. 2015;14:95. doi: 10.1186/s12943-015-0356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maroney AC, Qureshi SA, Foster DA, Brugge JS. Cloning and characterization of a thermolabile v-src gene for use in reversible transformation of mammalian cells. Oncogene. 1992;7:1207–1214. [PubMed] [Google Scholar]
- 11.Liu W, Yue F, Zheng M, Merlot A, Bae DH, Huang M, Lane D, Jansson P, Lui GY, Richardson V, et al. The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget. 2015;6:8851–8874. doi: 10.18632/oncotarget.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger KL, Learman BS, Boucherle AK, Sirintrapun SJ, Isom S, Díaz B, Courtneidge SA, Seals DF. Src-dependent Tks5 phosphorylation regulates invadopodia-associated invasion in prostate cancer cells. Prostate. 2014;74:134–148. doi: 10.1002/pros.22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker AM, Cox TR, Bird D, Lang G, Murray GI, Sun XF, Southall SM, Wilson JR, Erler JT. The role of lysyl oxidase in SRC-dependent proliferation and metastasis of colorectal cancer. J Natl Cancer Inst. 2011;103:407–424. doi: 10.1093/jnci/djq569. [DOI] [PubMed] [Google Scholar]
- 14.Di Stefano P, Cabodi S, Boeri Erba E, Margaria V, Bergatto E, Giuffrida MG, Silengo L, Tarone G, Turco E, Defilippi P. P130Cas-associated protein (p140Cap) as a new tyrosine-phosphorylated protein involved in cell spreading. Mol Biol Cell. 2004;15:787–800. doi: 10.1091/mbc.E03-09-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damiano L, Le Dévédec SE, Di Stefano P, Repetto D, Lalai R, Truong H, Xiong JL, Danen EH, Yan K, Verbeek FJ, et al. p140Cap suppresses the invasive properties of highly metastatic MTLn3-EGFR cells via impaired cortactin phosphorylation. Oncogene. 2012;31:624–633. doi: 10.1038/onc.2011.257. [DOI] [PubMed] [Google Scholar]
- 16.Damiano L, Di Stefano P, Camacho Leal MP, Barba M, Mainiero F, Cabodi S, Tordella L, Sapino A, Castellano I, Canel M, et al. p140Cap dual regulation of E-cadherin/EGFR cross-talk and Ras signalling in tumour cell scatter and proliferation. Oncogene. 2010;29:3677–3690. doi: 10.1038/onc.2010.128. [DOI] [PubMed] [Google Scholar]
- 17.Sharma N, Repetto D, Aramu S, Grasso S, Russo I, Fiorentino A, Mello-Grand M, Cabodi S, Singh V, Chiorino G, et al. Identification of two regions in the p140Cap adaptor protein that retain the ability to suppress tumor cell properties. Am J Cancer Res. 2013;3:290–301. [PMC free article] [PubMed] [Google Scholar]
- 18.Di Stefano P, Damiano L, Cabodi S, Aramu S, Tordella L, Praduroux A, Piva R, Cavallo F, Forni G, Silengo L, et al. p140Cap protein suppresses tumour cell properties, regulating Csk and Src kinase activity. EMBO J. 2007;26:2843–2855. doi: 10.1038/sj.emboj.7601724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruh J. Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol Cell Biochem. 1982;42:65–82. doi: 10.1007/BF00222695. [DOI] [PubMed] [Google Scholar]
- 20.Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 21.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 22.Kopetz S, Morris VK, Parikh N, Overman MJ, Jiang ZQ, Maru D, Elvin P, Gallick G. Src activity is modulated by oxaliplatin and correlates with outcomes after hepatectomy for metastatic colorectal cancer. BMC Cancer. 2014;14:660. doi: 10.1186/1471-2407-14-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Guo Z, Zhang D, Zhang W, Yan Q, Shi X, Zhang M, Zhao Y, Zhang Y, Jiang B, et al. A novel colon cancer gene therapy using rAAV-mediated expression of human shRNA-FHL2. Int J Oncol. 2013;43:1618–1626. doi: 10.3892/ijo.2013.2090. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Jiang B, Guo Z, Sardet C, Zou B, Lam CS, Li J, He M, Lan HY, Pang R, et al. Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis. 2010;31:1220–1229. doi: 10.1093/carcin/bgq094. [DOI] [PubMed] [Google Scholar]
- 25.Yan Q, Zhang W, Wu Y, Wu M, Zhang M, Shi X, Zhao J, Nan Q, Chen Y, Wang L, et al. KLF8 promotes tumorigenesis, invasion and metastasis of colorectal cancer cells by transcriptional activation of FHL2. Oncotarget. 2015;6:25402–25417. doi: 10.18632/oncotarget.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneikert J, Grohmann A, Behrens J. Truncated APC regulates the transcriptional activity of beta-catenin in a cell cycle dependent manner. Hum Mol Genet. 2007;16:199–209. doi: 10.1093/hmg/ddl464. [DOI] [PubMed] [Google Scholar]
- 27.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy S, Clynes M, Doolan P, Mehta JP, Rani S, Crown J, O'Driscoll L. SNIP/p140Cap mRNA expression is an unfavourable prognostic factor in breast cancer and is not expressed in normal breast tissue. Br J Cancer. 2008;98:1641–1645. doi: 10.1038/sj.bjc.6604365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eves EM, Rosner MR. MAP kinase regulation of the mitotic spindle checkpoint. Methods Mol Biol. 2010;661:497–505. doi: 10.1007/978-1-60761-795-2_31. [DOI] [PubMed] [Google Scholar]
- 30.Stewénius Y, Gorunova L, Jonson T, Larsson N, Höglund M, Mandahl N, Mertens F, Mitelman F, Gisselsson D. Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci USA. 2005;102:5541–5546. doi: 10.1073/pnas.0408454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Arsdale T, Boshoff C, Arndt KT, Abraham RT. Molecular pathways: Targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin Cancer Res. 2015;21:2905–2910. doi: 10.1158/1078-0432.CCR-14-0816. [DOI] [PubMed] [Google Scholar]
- 32.Kurokawa K, Akaike Y, Masuda K, Kuwano Y, Nishida K, Yamagishi N, Kajita K, Tanahashi T, Rokutan K. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene. 2014;33:1407–1417. doi: 10.1038/onc.2013.86. [DOI] [PubMed] [Google Scholar]
- 33.Chan DW, Mak CS, Leung TH, Chan KK, Ngan HY. Downregulation of Sox7 is associated with aberrant activation of Wnt/b-catenin signaling in endometrial cancer. Oncotarget. 2012;3:1546–1556. doi: 10.18632/oncotarget.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sánchez-Bailón MP, Calcabrini A, Gómez-Domínguez D, Morte B, Martín-Forero E, Gómez-López G, Molinari A, Wagner KU, Martín-Pérez J. Src kinases catalytic activity regulates proliferation, migration and invasiveness of MDA-MB-231 breast cancer cells. Cell Signal. 2012;24:1276–1286. doi: 10.1016/j.cellsig.2012.02.011. [DOI] [PubMed] [Google Scholar]