Abstract
Pyruvate kinase isoenzyme M2 (PKM2) has previously been identified as a tumor biomarker and potential therapeutic target for the treatment of cancer. In the present study, FFJ-3, a structurally modified version of mollugin, an extract of the Traditional Chinese herbal medicine Rubia tinctorum (madder) was used in order to determine the anticancer activity of the compound and investigate the potential mechanisms underlying this effect in human cancer cells. The results of the present study revealed that FFJ-3 inhibited the survival of HepG2 human hepatoma cells, MCF-7 human breast cancer cells and A549 human lung adenocarcinoma cells using the MTT assay. In addition, FFJ-3 arrested cell cycle progression at G2/M and G1 in HepG2 and A549 cells, respectively. Further analyses demonstrated that FFJ-3 attenuated the expression of PKM2 protein via the inhibition of the phosphoinositide 3-kinase (PI3K)/Akt serine/threonine kinase (Akt) signaling pathway. Furthermore, treatment of all three cell types with FFJ-3 significantly increased apoptosis and decreased the mitochondrial membrane potential compared with the untreated control group. In addition, FFJ-3 treatment increased the ratio of B-cell lymphoma-2 (Bcl-2)/Bcl-2 associated X and activated the caspase-3 cascade. In conclusion, the inhibition of the PI3K/Akt signaling pathway and activation of the caspase-3 cascade by FFJ-3 were primarily responsible for the inhibition of cell proliferation and induction of apoptosis in MCF-7, HepG2 and A549 cells. The results of the present study suggest a potential therapeutic role for FFJ-3 in the treatment of human cancer.
Keywords: mollugin, cancer, pyruvate kinase isoenzyme M2, phosphoinositide 3-kinase, Akt serine/threonine kinase, caspase-3sp
Introduction
Cancer cells sustain rapid proliferation by altering normal metabolic signaling pathways. Metabolic changes in cancerous tissue and cells may be caused by the upregulation of pyruvate kinase isoenzyme M2 (PKM2) (1). PKM2, a tumor-associated dimeric form of the enzyme pyruvate kinase, is involved in aerobic glycolysis and regulates ATP production, consequently promoting the growth of cancer cells (2,3). PKM2 has previously been identified as a tumor biomarker and a potential therapeutic target for the treatment of cancer (4,5).
The phosphoinositide 3-kinase (PI3K)/Akt serine/threonine kinase (Akt) signaling pathway is a classical and important signaling pathway, which is frequently constitutively activated in numerous human cancer types. Furthermore, this signaling pathway is associated with characteristics of carcinogenesis, including changes in cell proliferation, survival and metabolism (6–9). In cancer cells, increased activation of Akt (phosphorylation) or the upregulation of Akt expression can promote cell growth and suppress apoptosis by regulating cell-cycle regulators, such as p21 and p27, in addition to pro-apoptotic proteins, such as of B-cell lymphoma-2 (Bcl-2)-associated agonist of cell death (Bax).
The underlying mechanism of PKM2 upregulation in cancer cells remains unclear. In the present study, the association between akt activity and PKM2 gene expression in human breast cancer MCF-7 cells was investigated.
FFJ-3 is a compound that is a structurally modified version of mollugin, an extract of the Traditional Chinese herbal medicine Rubia tinctorum (madder). Previous studies have demonstrated that madder exhibits anticancer activity (10,11). In the present study, the effects of FFJ-3 on PKM2 expression and the PI3K/Akt signaling pathway were investigated. Furthermore, the role and mechanism of the novel compound FFJ-3 in cancer cells, and a novel mechanism attributed to the role of the PI3K/Akt signaling pathway in the regulation of PKM2 that is associated with tumorigenesis are presented in the current study.
Materials and methods
Antibodies, reagents and drugs
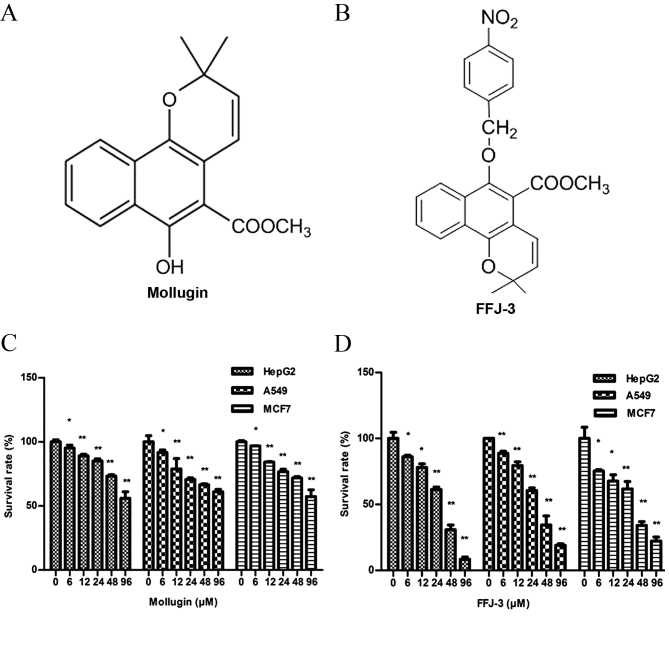
Rabbit anti-human monoclonal antibodies directed against Akt (cat. no. 4685), phosphorylated Akt (pSer473; cat. no. 4060), Bcl-2 (cat. no. 2870; dilution), Bax (cat. no. 5023), PKM2 (cat. no. 4053; dilution), caspase-3 (cat. no. 9665), cleaved caspase-3 (Asp175; cat. no. 9664), Poly (ADP-ribose) polymerase (PARP; cat. no. 9532) and cleaved PARP (Asp214; cat. no. 5625) (all dilution, 1:1,000) were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). Horseradish peroxidase-conjugated Affinipure goat anti-mouse and anti-rabbit immunoglobulin Gs (IgGs) (cat. nos. SA00001-1 and SA00001-2; dilution, 1:10,000) were obtained from Proteintech Group, Inc. (Chicago, IL, USA). The PI3K inhibitor LY294002 (cat. no. 9901) was purchased from Cell Signaling Technology, Inc. and the Akt activator insulin (cat. no. 40112ES25) was obtained from Yeasen Biotechnology Co., Ltd. (Shanghai, China). The PrimeScript™ RT reagent kit with gDNA Eraser (cat. no. RR047A) and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (cat. no. RR820A) were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). Propidium iodide (PI), RNase A, Hoechst 33342 and Rhodamine 123 (Rh123) were purchased from Sigma-Aldrich (Merck Millipore, Darmstadt, Germany). Mollugin (cat. no. 110884–201405; Fig. 1A) was purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). FFJ-3 (Fig. 1B) is a novel chemically modified compound based on the structure of Mollugin and was synthesized by introducing paranitrotoluene to the hydroxyl of Mollugin at the Institute of Traditional Chinese Medicine of Henan University (Kaifeng, China).
Figure 1.
Anti-proliferative effects of FFJ-3 and mollugin. Schematic diagrams of the chemical structure of (A) mollugin and (B) FFJ-3. Survival rate of HepG2, A549 and MCF-7 cells following treatment with (C) mollugin and (D) FFJ-3. Cells were exposed to the compounds for 48 h prior to the MTT assay. *P<0.05, **P<0.01 vs. the corresponding untreated control groups.
Cell lines and cell culture
Human HepG2 hepatocellular carcinoma, A549 lung cancer and MCF-7 breast cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were maintained in RPMI-1640 media (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin in a 37°C incubator with 5% CO2.
Cell viability assay
Cell proliferation and viability were detected using the MTT assay. All three types of cancer cells at a density of 1×104/ml were seeded into 96-well plates. After 12 h, the cells were treated with final concentrations of 6, 12, 24, 48 and 96 µM of FFJ-3 for 48 h at 37°C and the same concentrations of Mollugin were used as positive controls. Subsequently, media was replaced with fresh medium containing MTT (0.5 mg/ml; Sigma-Aldrich; Merck Millipore) and the plates were incubated at 37°C for 4 h, followed by further media replacement with 150 µl dimethyl sulfoxide. The absorbance values at a wavelength of 490 nm were measured using a microplate reader (Sunrise™, Tecan Group Ltd., Männedorf, Switzerland). Relative cell viability was expressed as a percentage relative to the untreated control cells.
Cell cycle analysis
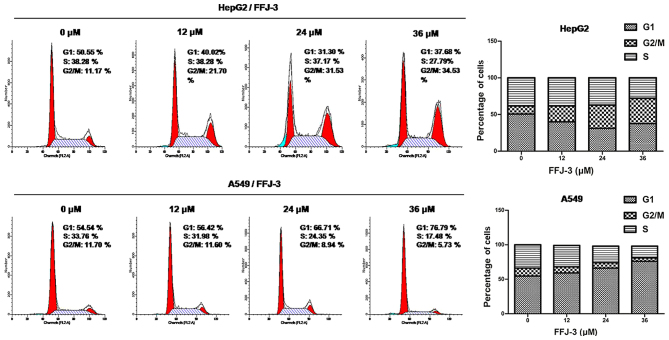
Cell cycle analysis was performed using flow cytometry. HepG2 and A549 cells were treated with FFJ-3 for 48 h, collected by centrifugation (1,000 × g for 5 min at room temperature) and washed twice with ice-cold PBS. Cells (1×106) were fixed with 75% ethanol at 4°C for ≥4 h. Subsequently, cells were washed twice with ice-cold PBS, followed by incubation with a DNA staining solution (PI, 50 µg/ml; RNase, 50 µg/ml; Triton X-100, 0.5%) at 37°C for 20 min. The DNA content of the cells was determined using flow cytometry (FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA). The percentage of cells in G0/G1, S and G2/M phases were determined using ModFit LT (version 4.1; Verity Software House, Topsham, ME, USA).
Western blot analysis
Following seeding (1×106/well) in 6-well plates, the cancer cells were treated with 0, 12, 24 and 36 µM of FFJ-3 for 48 h, 0, 10 and 50 µM of LY294002 for 24 h or 48 h and 0, 50 and 100 ng/ml of insulin for 24 h or 48 h at 37°C. Total protein was extracted using radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 8.0; 150 mM sodium chloride; 1.0% NP-40; 0.5% sodium deoxycholate; 0.1% SDS) with 10 µg/ml of the protease inhibitor phenylmethylsulfonyl fluoride. The lysates were collected through centrifugation at 4°C, 12,000 × g for 10 min. Protein concentration of the cell lysates was determined using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Haimen, China). Equal amounts of protein (40 µg) were separated by SDS-PAGE. PKM2, Akt, pAkt, PARP and cleaved PARP were separated using an 8% gel. Bax, Bcl-2, Caspase-3 and cleaved caspase-3 were separated using a 12% gel. Gels were subsequently transferred electrophoretically to a polyvinylidene fluoride membrane (Merck Millipore) at 70 mA for 2 h. Subsequently, the membrane was blocked using 5% skimmed milk and the membrane was incubated with specific primary antibodies (described above) at 4°C overnight. Following washing in PBS with Tween-20, the membranes were incubated with anti-mouse or anti-rabbit IgG secondary antibody (described above) at room temperature for 2 h. Bands were visualized using an EasyBlot Enhanced Chemiluminescence kit (Sangon Biotech Co., Ltd., Shanghai, China) and detected using a FluorChem Q Multifluor system with AlphaView Q software (version 3.0) (both ProteinSimple, San Jose, CA, USA). β-actin was used as a loading control.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 2 µg RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit. qPCR was performed using SYBR Premix Ex Taq II and the PikoReal™ Real-Time PCR system (Thermo Fisher Scientific, Inc.) was used to measure mRNA expression. A total of 0.1 µg cDNA template was used to perform the qPCR reaction. The following thermocycling conditions were used: 95°C for 5 min; 40 cycles of 95°C for 10 sec, 59°C for 30 sec and 72°C for 30 sec; 60°C for 30 sec. The reactions for each sample-primer set were performed in triplicate. The sequences of the qPCR primers were as follows: PKM2 forward, 5′-AGAACTTGTGCGAGCCTCAA-3′ and reverse, 5′-GAGCAGACCTGCCAGACTC-3′ (product length, 128 bp); GAPDH forward, 5′-CTCTGCTCCTCCTGTTCGAC-3′ and reverse, 5′-ACCAAATCCGTTGACTCCGA-3′ (product length, 109 bp). Relative quantification analysis was performed using the comparative Cq (2−ΔΔCq) method (12). All data were normalized to the internal control (GAPDH).
Cell apoptosis assay
MCF-7, HepG2 and A549 cells (1×104/well) were plated into 96-well plates. Following 12 h culturing, the cells were treated with 0, 12, 24 and 36 µM of FFJ-3 at 37°C for 48 h, followed by washing with ice-cold PBS. Cells were subsequently incubated with 100 µl of Hoechst 33342 (5 mg/l) for 30 min, followed by incubation with PI (5 mg/l) for 30 min at 37°C. The cells were washed with ice-cold PBS twice. Cell apoptosis analysis was immediately performed using an ArrayScan™ VTI 600 High-Content live cell imaging system (Thermo Fisher Scientific, Inc.), in which the second channel's mean fluorescence intensity (MFI) represented late apoptosis.
itochondrial membrane potential assay
Changes in mitochondrial membrane potential (MMP) were analyzed through measuring changes in the fluorescent intensity of Rh123 staining of the cells. MCF-7, HepG2 and A549 cells exposed to 0, 12, 24 and 36 µM of FFJ-3 for 48 h at 37°C were collected by centrifugation (1,000 × g, 5 min, room temperature) and washed with ice-cold PBS, followed by incubation with 5 µg/ml Rh123 at 37°C for 20 min. The Rh123 content of the cells was determined using an EnSpire® Multimode plate reader (PerkinElmer, Inc., Waltham, MA, USA).
Statistical analysis
Each experiment was repeated three times. Results are presented as the mean ± standard deviation. Statistical analyses were conducted using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA). Data was analyzed using analysis of variance (one-way) for each two-group comparison. P<0.05 was considered to indicate a statistically significant difference.
Results
FFJ-3 inhibits the proliferation of human cancer cells
As the Traditional Chinese medicine madder extracts and its derivatives have been demonstrated to exhibit anticancer activity, the structure of mollugin (an active ingredient of madder) was modified (Fig. 1A) to produce the novel compound FFJ-3 (Fig. 1B). In the present study, the anticancer effects of FFJ-3 were examined in human cancer cells. Treatment of HepG2, A549 and MCF-7 cells with mollugin (6–96 µM; Fig. 1C) or FFJ-3 (6–96 µM; Fig. 1D) resulted in a significant dose-dependent decrease in cell survival compared with the untreated control groups (all P<0.05). The half maximal inhibitory concentration (IC50) of FFJ-3 in HepG2, A549 and MCF-7 cells was 25.56, 36.57 and 27.16 µM, respectively. The anticancer effect, as indicated by the IC50, of FFJ-3 was stronger compared with that of Mollugin (HepG2, 124 µM; A549, 168 µM; MCF-7, 134 µM; Fig. 1D).
Furthermore, the effects of FFJ-3 on cell proliferation were examined. There was a marked increase in the percentage of HepG2 cells in the G2/M phase (4n DNA) following treatment with different concentrations of FFJ-3 (12 µM, 21.70%; 24 µM, 31.53%; 36 µM, 34.53%) compared with the untreated control group (11.70%) (Fig. 2). In A549 cells, the percentage of cells in the G1 phase was increased following treatment with 12, 24 and 36 µM of FFJ-3 compared with the untreated control group (Fig. 2). These results suggest that FFJ-3 is a potential candidate for the inhibition of human cancer cell survival and proliferation.
Figure 2.
Cell cycle analysis. Cells were treated with FFJ-3 (0, 12, 24 and 36 µM) for 48 h. Graphs illustrate the percentage of cells distributed across each cell cycle phase. Following treatment with FFJ-3, a significant increase in the percentage of HepG2 cells in the G2/M (4N DNA) phase was observed. In A549 cells, the G1 phase cell population increased following treatment with FFJ-3. FL, fluorescence; A, area; number, number of cells.
FFJ-3 inhibits PKM2 expression and Akt activity
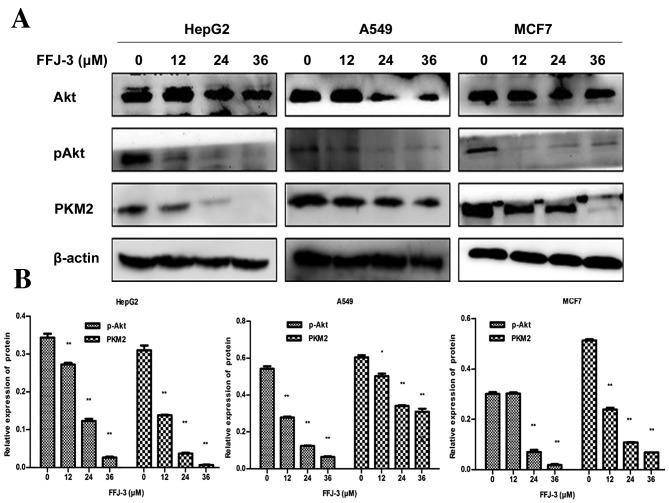
PKM2 has been identified as a potential therapeutic target for the treatment of cancer (13,14). Downregulation of PKM2 or interaction with PKM2 can suppress tumorigenicity and tumor progression (15,16). In the present study, the results of the western blot analysis revealed that FFJ-3 significantly decreased the expression of PKM2 in HepG2, A549 and MCF-7 cells compared with the untreated control group in a dose-dependent manner (all P<0.05; Fig. 3). PI3K/Akt is a classical and important signaling pathway in numerous human cancer types, and is associated with cell proliferation, survival and metabolism. Thus, the effects of FFJ-3 on the expression and activity of Akt were investigated. The results of the western blot analysis demonstrated that 24 and 36 µM FFJ-3 significantly decreased the amount of phosphorylated (p-)Akt in a dose-dependent manner in all three cell lines compared with the untreated control group (all P<0.01; Fig. 3).
Figure 3.
Inhibitory effects of FFJ-3 on PKM2 and p-Akt protein expression. (A) p-Akt and PKM2 protein expression were examined in HepG2, A549 and MCF-7 cells following 12–36 µM FFJ-3 treatment for 48 h using western blot analysis. (B) The results demonstrated that FFJ-3 significantly inhibited the expression of PKM2 and p-Akt in a dose-dependent manner. *P<0.05, **P<0.01 vs. the untreated control groups. PKM2, pyruvate kinase isoenzyme M2; p-Akt, phosphorylated Akt serine/threonine kinase.
PI3K/Akt signaling regulates PKM2 gene expression in MCF-7 cells
To confirm whether the reduced level of PKM2 was due to a reduction in Akt activity, the association between the PI3K/Akt signaling pathway and PKM2 was investigated. MCF-7 cells were treated with LY294002, a specific chemical inhibitor of PI3K, for 24 and 48 h. It was demonstrated that LY294002 significantly downregulated PKM2 protein expression in a dose-dependent manner compared with the control group (P<0.01), concomitantly with inhibition of PI3K activity, as indicated by a significant reduction in p-Akt protein compared with the untreated control group (P<0.05) (Fig. 4A and B).
Figure 4.
Regulatory role of the PI3K/Akt signaling pathway on PKM2 expression in MCF-7 cells. (A) The effect of PI3K/Akt signaling pathway on PKM2 protein expression. LY294002, a specific chemical inhibitor of PI3K, downregulated PKM2 protein expression in a dose-dependent manner, and insulin, a potent activator of the PI3K/Akt signaling pathway, increased PKM2 protein expression. (B) The expression of Akt, p-Akt and PKM2 relative to β-actin. *P<0.05, **P<0.01 vs. the untreated control group. (C) The effect of the PI3K/Akt signaling pathway on PKM2 mRNA expression. Quantitative polymerase chain reaction analysis results demonstrated that the inhibition of PI3K by LY294002 did not decrease PKM2 mRNA expression. Furthermore, activation of Akt by insulin did not increase PKM2 mRNA expression. PKM2, pyruvate kinase isoenzyme M2; p-Akt, phosphorylated Akt serine/threonine kinase; PI3K, phosphoinositide 3-kinase.
MCF-7 cells were exposed to 50–100 ng/ml insulin, an activator of the PI3K/Akt signaling pathway, for 24 and 48 h. The results demonstrated that 100 ng/ml insulin caused a significant increase in PKM2 protein expression and Akt phosphorylation compared with the untreated control group (all P<0.05; Fig. 4A and B). Furthermore, to determine whether the PI3K/Akt signaling pathway regulated PKM2 expression at the transcriptional level, the effect of LY294002 and insulin on PKM2 mRNA expression was investigated. It was revealed that the inhibition of PI3K by 10–50 µM LY294002 did not lead to decreased PKM2 mRNA expression and that the activation of Akt by 50–100 ng/ml insulin did not result in increased PKM2 mRNA expression in MCF-7 cells (Fig. 4C). These findings indicate that PKM2 gene expression is regulated by the PI3K/Akt signaling pathway at the translational level in MCF-7 cells.
FFJ-3 induces cancer cell apoptosis and reduces MMP
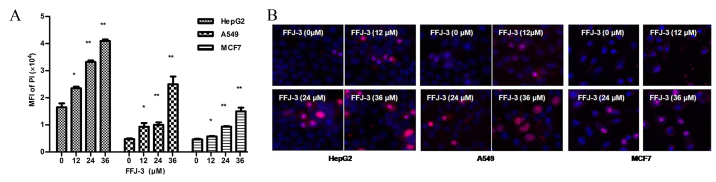
The effect of FFJ-3 on cancer cell apoptosis was investigated in the current study. Treatment of MCF-7, HepG2 and A549 cells with 12–36 µM FFJ-3 resulted in a significant increase in cell apoptosis in a dose-independent manner compared with the untreated control group, as indicated by the fluorescence intensity of PI staining (P<0.05; Fig. 5). To investigate the mechanism underlying FFJ-3-induced cancer cell apoptosis, the change in MMP was determined using Rh123 staining. The results demonstrated a significant increase in Rh123 fluorescence intensity in all three cell lines following treatment with 12–36 µM FFJ-3 compared with the untreated control group (P<0.05; Fig. 6). These results suggest that FFJ-3 reduces MMP, consequently inducing cell apoptosis through the mitochondrial apoptosis signaling pathway.
Figure 5.
Induction of cancer cells apoptosis by FFJ-3 in HepG2, A549 and MCF-7 cells. (A) FFJ-3 induced a significant increase in the MFI of PI. (B) Imaging of PI and Hoechst 33342 double-staining. *P<0.05, **P<0.0 vs. the untreated control groups. PI, propidium iodide; MFI, mean fluorescence intensity.
Figure 6.
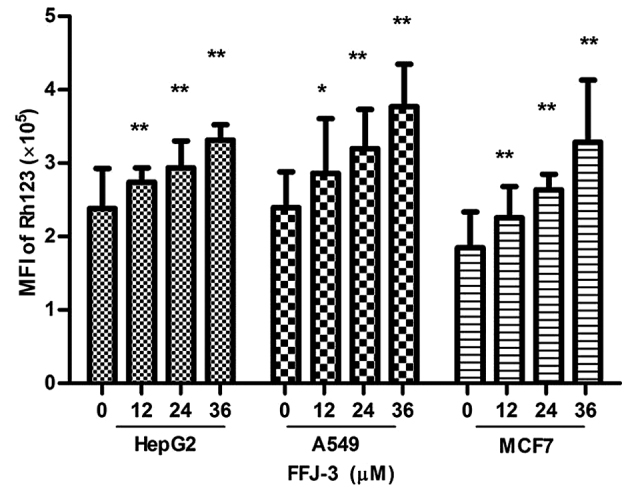
FFJ-3 reduces the mitochondrial membrane potential of cancer cells. Changes in mitochondrial membrane potential were detected with Rh123 staining. A significant increase in Rh123 fluorescence intensity in HepG2, A549 and MCF-7 cells following treatment with 12–36 µM FFJ-3 was demonstrated. *P<0.05, **P<0.01 vs. the untreated control group. Rh123, Rhodamine 123.
FFJ-3 activates the caspase-3 cascade and increases the ratio of Bax/Bcl-2
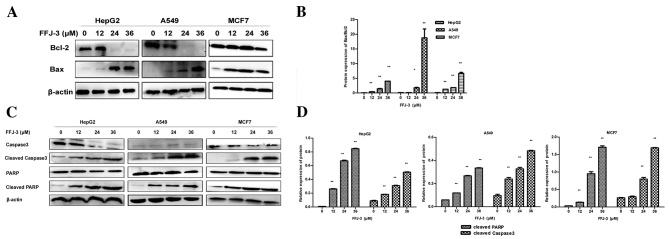
To further investigate the mechanism underlying FFJ-3-induced cancer cell apoptosis, the effects of 12–36 µM FFJ-3 on the expression of mitochondrial apoptotic signaling pathway proteins was investigated. An increase in Bax expression and decrease in Bcl-2 expression was identified in MCF-7, HepG2 and A549 cells treated with FFJ-3 (Fig. 7A). The Bax/Bcl-2 expression ratio was significantly increased following treatment with 24–36 µM FFJ-3 (P<0.05; Fig. 7B). Furthermore, the caspase-3 cascade was examined. The results of the western blot analysis revealed that 24–36 µM FFJ-3 activated caspase-3, as indicated by the significant increase in cleaved caspase-3 protein (P<0.01; Fig. 7C and D). Consequently, this induced the cleavage of PARP, as indicated by the simultaneous significant increase in cleaved PARP protein (P<0.05; Fig. 7C and D). These results support the hypothesis that FFJ-3-induced apoptosis occurs through the mitochondrial apoptosis signaling pathway in MCF-7, HepG2 and A549 cells.
Figure 7.
FFJ-3 increases the ratio of Bax/Bcl-2 expression and activates the caspase-3 cascade in cancer cells. Changes in Bcl-2, Bax, caspase-3, cleaved caspase-3, PARP and cleaved PARP expression were examined in HepG2, A549 and MCF-7 cells following FFJ-3 treatment for 48 h. (A) Western blot analysis of Bax and Bcl-2 protein expression following treatment with FFJ-3. (B) FFJ-3 significantly increased the ratio of Bax/Bcl-2 protein expression. (C) Western blot analysis of caspase-3, cleaved caspase-3, PARP and cleaved PARP protein expression following treatment with FFJ-3. (D) FFJ-3 activates the caspase-3 cascade in cancer cells. *P<0.05, **P<0.01 compared with the untreated control group. Bcl-2, B-cell lymphoma 2; Bax, Bcl-2 associated X apoptosis regulator; PARP, poly (ADP-ribose) polymerase; MFI, mean fluorescence intensity.
Discussion
Cancer has become a primary cause of human mortality. The development of therapeutics directed against specific targets or signaling pathways in cancer is essential. Madder is a type of Traditional Chinese herbal medicine, and previous studies have demonstrated that madder possesses anticancer activity. FFJ-3 is a structural modification of mollugin, an extract of Madder. In the present study, the effect of FFJ-3 on PKM2 and the PI3K/Akt signaling pathway in cancer cell lines was investigated.
Pyruvate kinase is an essential enzyme in the glycolytic pathway, which modulates ATP generation. Pyruvate kinase exists as different isoforms, including L, R, M1 and M2, which have tissue-specific expression in various organisms. A number of studies have demonstrated that tumor cells exclusively express the PKM2 embryonic isoform of pyruvate kinase, which serves an important role in metabolic reprogramming during cancer progression (17,18). Furthermore, elevated expression of PKM2 has been identified in patients with solid tumors, such as colorectal carcinoma (19), thus PKM2 is considered a tumor biomarker. PKM2 signaling has been revealed to suppress tumorigenicity and tumor progression (20–22). Therefore, the present study investigated the effect of FFJ-3 on PKM2 expression in MCF-7, HepG2 and A549 cells. FFJ-3 was demonstrated to significantly reduce PKM2 protein expression in a dose-dependent manner.
PI3K/Akt is a classical and important signaling pathway that is frequently activated in numerous human cancer types, and is associated with features of carcinogenesis, including cell proliferation, survival and metabolism (23–25). In the current study, the role of FFJ-3 in the PI3K/Akt signaling pathway was determined. The results of the present study demonstrated that FFJ-3 significantly decreased the expression and activity of Akt in all three cancer cell lines in a dose-dependent manner.
Whether the FFJ-3-induced decrease in PKM2 expression was due to a reduction in Akt activity remained unclear. Thus, the association between the PI3K/Akt signaling pathway and PKM2 expression was investigated. MCF-7 cells were treated with LY294002, a specific chemical inhibitor of PI3K, and insulin, a potent activator of the PI3K/Akt signaling pathway. LY294002 was demonstrated to significantly downregulate PKM2 protein expression, concomitantly with inhibition of PI3K activity, as indicated by a reduction in p-Akt expression. Furthermore, a significant increase in PKM2 protein expression and Akt phosphorylation was identified in MCF-7 cells treated with 100 ng/ml insulin. In addition, experiments were performed to evaluate the effect of LY294002 and insulin on PKM2 mRNA expression in MCF-7 cells. It was revealed that inhibition of PI3K by LY294002 did not lead to decreased PKM2 mRNA expression, and activation of Akt by insulin did not result in increased PKM2 mRNA expression. These findings indicate that the PI3K/Akt signaling pathway regulates PKM2 expression at the translational level.
The association between FFJ-3 and cancer cell apoptosis was investigated in the present study. The results indicated that FFJ-3 significantly induced apoptosis and reduced the MMP of MCF-7, HepG2 and A549 cells in a dose-independent manner. Furthermore, it was demonstrated that FFJ-3 activates caspase-3, consequently inducing the cleavage of PARP. These findings suggest that FFJ-3 induces cancer cell apoptosis through the mitochondrial signaling pathway.
In conclusion, the results of the present study suggest that FFJ-3 inhibits PKM2 protein expression via the PI3K/Akt signaling pathway and activates the caspase-3 cascade in human cancer cells. FFJ-3 may be a promising therapeutic candidate for the treatment of patients with cancer.
Acknowledgments
The present study was supported by the Key Science and Technology Fund of Henan Province (grant no. 142300410128).
References
- 1.Iqbal MA, Gupta V, Gopinath P, Mazurek S, Bamezai RN. Pyruvate kinase M2 and cancer: An updated assessment. FEBS Lett. 2014;588:2685–2692. doi: 10.1016/j.febslet.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Wu N, Ma L, Liu M, Liu G, Zhang Y, Lin X. Oleanolic acid suppresses aerobic glycolysis in cancer cells by switching pyruvate kinase type M isoforms. PLoS One. 2014;9:e91606. doi: 10.1371/journal.pone.0091606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang JX, Zhou Y, Wang CH, Yuan WW, Zhang, Zd Zhang XF. Tumor M2-pyruvate kinase in stool as a biomarker for diagnosis of colorectal cancer: A meta-analysis. J Can Res Ther. 2014;10(Suppl):225–228. doi: 10.4103/0973-1482.145886. [DOI] [PubMed] [Google Scholar]
- 5.Wang F, Yang Y. RETRACTED: Inhibition of PKM2 sensitizes triple-negative breast cancer cells to doxorubicin. Biochem Biophys Res Commun. 2014;454:465–470. doi: 10.1016/j.bbrc.2014.02.133. [DOI] [PubMed] [Google Scholar]
- 6.Cheaib B, Auguste A, Leary A. The PI3K/Akt/mTOR pathway in ovarian cancer: Therapeutic opportunities and challenges. Chin J Cance. 2015;34:4–16. doi: 10.5732/cjc.014.10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manzo-Merino J, Contreras-Paredes A, Vázquez-Ulloa E, Rocha-Zavaleta L, Fuentes-Gonzalez AM, Lizano M. The Role of Signaling Pathways in ervical cancer and molecular therapeutic targets. Arch Med Res. 2014;45:525–539. doi: 10.1016/j.arcmed.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta. 2015;1855:104–121. doi: 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Markowska A, Pawałowska M, Lubin J, Markowska J. Signalling pathways in endometrial cancer. Contemp Oncol (Pozn) 2014;18:143–148. doi: 10.5114/wo.2014.43154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shilpa PN, Sivaramakrishnan V, Devaraj S Niranjali. Induction of apoptosis by methanolic extract of Rubia cordifolia Linn in HEp-2 cell line is mediated by reactive oxygen species. Asian Pac J Cancer Prev. 2012;13:2753–2758. doi: 10.7314/APJCP.2012.13.6.2753. [DOI] [PubMed] [Google Scholar]
- 11.Shilpa PN, Venkatabalasubramanian S, Devaraj SN. Ameliorative effect of methanol extract of Rubia cordifolia in N-nitrosodiethylamine-induced hepatocellular carcinoma. Pharm Biol. 2012;50:376–383. doi: 10.3109/13880209.2011.608073. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Kefas B, Comeau L, Erdle N, Montgomery E, Amos S, Purow B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010;12:1102–1112. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong G, Mao Q, Xia W, Xu Y, Wang J, Xu L, Jiang F. PKM2 and cancer: The function of PKM2 beyond glycolysis. Oncol Lett. 2016;11:1980–1986. doi: 10.3892/ol.2016.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Wang Z, Guo W, Zhang Z, Zhao F, Zhao Y, Jia D, Ding J, Wang H, Yao M, He X. TRIM35 interacts with pyruvate kinase isoform M2 to suppress the Warburg effect and tumorigenicity in hepatocellular carcinoma. Oncogene. 2015;34:3946–3956. doi: 10.1038/onc.2014.325. [DOI] [PubMed] [Google Scholar]
- 16.Chu B, Wang J, Wang Y, Yang G. Knockdown of PKM2 induces apoptosis and autophagy in human A549 alveolar adenocarcinoma cells. Mol Med Rep. 2015;12:4358–4363. doi: 10.3892/mmr.2015.3943. [DOI] [PubMed] [Google Scholar]
- 17.Papadaki C, Sfakianaki M, Lagoudaki E, Giagkas G, Ioannidis G, Trypaki M, Tsakalaki E, Voutsina A, Koutsopoulos A, Mavroudis D, et al. PKM2 as a biomarker for chemosensitivity to front-line platinum-based chemotherapy in patients with metastatic non-small-cell lung cancer. Br J Cancer. 2014;111:1757–1764. doi: 10.1038/bjc.2014.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, Wang Z, Guo W, Zhang Z, Zhao F, Zhao Y, Jia D, Ding J, Wang H, Yao M, He X. TRIM35 Interacts with pyruvate kinase isoform M2 to suppress the Warburg effect and tumorigenicity in hepatocellular carcinoma. Oncogene. 2015;34:3946–3956. doi: 10.1038/onc.2014.325. [DOI] [PubMed] [Google Scholar]
- 19.Cui R, Shi XY. Expression of pyruvate kinase M2 in human colorectal cancer and its prognostic value. Int J Clin Exp Pathol. 2015;8:11393–11399. [PMC free article] [PubMed] [Google Scholar]
- 20.Hitosugi T, Kang S, Heiden MG Vander, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: Multiple faces for conferring benefits on cancer cells. Clin Cancer Res. 2012;18:5554–5561. doi: 10.1158/1078-0432.CCR-12-0859. [DOI] [PubMed] [Google Scholar]
- 22.Yang W, Lu Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. 2013;339:153–158. doi: 10.1016/j.canlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamal A, Nayak V Lakshma, Nagesh N, Vishnuvardhan MV, Reddy NV Subba. Benzo[b]furan derivatives induces apoptosis by targeting the PI3K/Akt/mTOR signaling pathway in human breast cancer cells. Bioorg Chem. 2016;66:124–131. doi: 10.1016/j.bioorg.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Berrak Ö, Akkoç Y, Arısan ED, Çoker-Gürkan A, Obakan-Yerlikaya P, Palavan-Ünsal N. The inhibition of PI3K and NFκB promoted curcumin-induced cell cycle arrest at G2/M via altering polyamine metabolism in Bcl-2 overexpressing MCF-7 breast cancer cells. Biomed Pharmacother. 2016;77:150–160. doi: 10.1016/j.biopha.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Frasson C, rRampazzo E, aAccordi B, cBeggio G, ePistollato F, iBasso G, aPersano L. Inhibition of PI3K signalling selectively affects medulloblastoma cancer stem cells. Biomed Res Int. 2015;2015:973912. doi: 10.1155/2015/973912. [DOI] [PMC free article] [PubMed] [Google Scholar]