Abstract
At present, survivin is one of the most cancer-specific proteins that has been identified. The present study aimed to investigate the antitumor effects of novel survivin small interfering RNA (siRNA) nanoliposomes targeting survivin in human hepatocellular carcinoma MHCC-97H cells and xenograft mouse models. Survivin-targeted siRNA nanoliposomes were prepared and transfected into MHCC-97H cells and MHCC-97H-bearing nude mice. Survivin expression was analyzed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting. Cell viability was analyzed using an MTT assay and apoptosis was evaluated using Hoechst and Annexin V-fluorescein isothiocyanate/propidium iodide staining. Tumor growth in MHCC-97H-bearing mice was monitored following treatment and tumor samples were obtained for survivin expression analysis using RT-qPCR, western blotting and immunohistochemistry staining. Survivin expression levels were significantly downregulated by nanoliposome-mediated survivin siRNA delivery and this was associated with a significant inhibition of cell growth and an increase in the apoptosis of MHCC-97H cells. Downregulation of survivin expression using survivin siRNA nanoliposomes inhibited tumor growth in the MHCC-97H xenograft models without significant treatment-associated toxicity. Therefore, a cationic nanoliposome-based survivin siRNA delivery system was constructed and demonstrated to be efficient for survivin siRNA delivery in in vitro and in vivo studies. These results demonstrate that survivin downregulation was able to significantly attenuate cell proliferation and induce the apoptosis of MHCC-97H cells, as well as inhibit tumor cell growth in MHCC-97H xenograft models, indicating that survivin suppression using siRNA may contribute to the inhibition of tumor development by suppressing cell proliferation and promoting apoptosis.
Keywords: survivin, small interfering RNA, nanoliposome, hepatocellular carcinoma cells, xenograft
Introduction
Hepatocellular carcinoma (HCC) is one of the most common types of malignancy with high rates of recurrence and metastasis, and is ranked as the third leading cause of cancer-associated mortality worldwide (1). The global incidence of HCC has continuously increased in 2008 according to the statistics of GLOBOCAN 2008 (2), and Asian countries alone account for ~80% of cases worldwide (3). Surgical resection is an optional choice for resectable HCC (4). However, <20% of patients are eligible for surgical management involving hepatic resection or transplantation (5). The majority of patients with non-resectable HCC may benefit from multimodal treatment options (6). However, their efficacy is limited by recurrence (7) and at present, no effective treatment is available for the majority of patients with HCC, except liver transplantation for certain selected patients (8).
Molecular-targeted therapy has emerged as a novel potential treatment paradigm for HCC (9). Survivin, a member of the inhibitor of apoptosis family, is established as one of the most cancer-specific proteins identified (10). It is well documented to be highly and selectively overexpressed in almost all types of human malignancy, including HCC (11,12). Overexpression of survivin in tumors has been associated with chemotherapy and radiotherapy resistance, tumor recurrence and poor prognosis (13,14). Previous studies have focused on developing strategies for the use of survivin as a target for cancer therapy. However, intracellular modulation of survivin using small molecules is not feasible (9). In HCC preclinical models, the use of small interfering RNA (siRNA) and antisense oligonucleotide-based approaches to reduce survivin expression levels has demonstrated a certain level of efficacy (15). However, another study indicated that survivin knockdown only slightly affected the survival rate of HCC cells (16).
RNA interference is an effective post-transcriptional gene silencing technique with high specificity and efficiency, as well as low toxicity, and is widely utilized in cancer research. When used as a molecular-targeted cancer therapy, surviving-targeted siRNA is effective in inducing apoptosis and reversing chemoresistance in tumor cells (17,18). Cationic liposomes have previously been developed for siRNA delivery and exhibit beneficial effects on the induction of chemosensitivity and radiosensitivity in xenograft mouse models (19,20). However, the biological effects of nanoliposome-mediated survivin silencing using siRNA on human HCC have not been fully evaluated. In the present study, cationic nanoliposome-mediated survivin siRNAs were constructed and transfected into MHCC-97H cells and into a mouse xenograft model of HCC. Levels of cell proliferation and apoptosis were evaluated, as was survivin expression and the proliferation of tumors in nude mice.
Materials and methods
Cell lines and animals
The MHCC-97H HCC cell line was purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at 37°C in a humidified incubator containing 5% CO2.
Male athymic BALB/c nude mice, weighing 20–24 g (7–9 weeks old), were obtained from Shanghai Laboratory Animal Center (Shanghai, China). Mice were maintained in specific pathogen-free conditions with free access to food and water, under a constant temperature of 22±2°C and a 12 h light/dark cycle (7:00 a.m. to 7:00 p.m.). All animal experiments were approved by the Ethics Committee of the Capital Institute of Pediatrics (Beijing, China).
Preparation of survivin siRNA nanoliposomes
siRNA targeting survivin was synthesized by Biomics Biotechnologies Co., Ltd. (Nantong, China), with a sense sequence of 5′-GCAUCUCUACAUUCAAGAAdTdT-3′ and an antisense sequence of 5′-UUCUUGAAUGUAGAGAUGCdTdT-3′. An unrelated silencing sequence was synthesized as a negative control (NC), and the sequences of NC siRNA were as follows: Sense, 5′-UUCUCCGAACGUGUCACGUdTdT-3′; antisense, 5′-ACGUGACACGUUCGGAGAAdTdT-3′. siRNA molecules were encapsulated into nanoliposomes in ethanol-water solutions with a lipid composition of 1,2-distearoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Inc., Alablaster, AL, USA), cholesterol, dimethyldioctadecylammonium chloride and poly(ethylene glycol) ceramide C16 (all from Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) in 100% ethanol (Sigma-Aldrich; Merck Millipore) at a molar ratio of 25:45:25:2.5, respectively. The mean diameter of the nanoliposomes was determined using a nanoparticle size analyzer (Zetasizer Nano ZS90; Malvern Instruments Ltd., Malvern, UK) and the data was 70.7±29.077 nm for survivin siRNA and 64.9±26.128 nm for NC siRNA.
Gene transfection in MHCC-97H cells
For siRNA nanoliposome transfection, MHCC-97H cells (1.5×105 /ml, 500 µl) were seeded in a 24-well plate one day prior to transfection and divided into three groups following overnight cultivation: Normal group (without transfection), NC group (NC siRNA nanoliposome) and survivin siRNA group (survivin siRNA nanoliposome). Cells were then incubated with NC and survivin siRNA nanoliposomes in Opti-MEM (Life Technologies, Inc, Gaithersburg, MD) at 37°C for 4–6 h. Mediums were then changed and cells were then incubated for another 48 h. The mRNA and protein expression levels of survivin were subsequently determined.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using RISO™ RNA reagent (Biomics Biotechnologies Co., Ltd.) according to the manufacturer's protocol. cDNA synthesis and subsequent polymerization was performed using a SensiMix™ SYBR One-Step kit (Quantace; Bioline Reagents Ltd., London, UK). RT-qPCR was performed on the ABI PRISM real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using 2xOne-Step qPCR Mix (12.5 µl), 50x SYBR Green I (0.5 µl), primers (0.5 µl), and cDNA template (100 ng, 4 µl). The thermocycler conditions were as follows: Pre-incubation at 95°C for 10 min, followed by 45 amplification cycles at 95°C for 20 sec, 58°C for 30 sec, and 72°C for 30 sec. The housekeeping gene GAPDH was used as an internal control. Relative gene expression was calculated by the 2−ΔΔCq method (21). The primers used are summarized in Table I.
Table I.
Sequences of RT-qPCR primers.
| Gene | Sequence (5′-3′) | Length |
|---|---|---|
| Survivin | Forward: ACGACCCCATAGAGGAACAT | 175 bp |
| Reverse: TCCGCAGTTTCCTCAAATTC | ||
| GAPDH | Forward: GAAGGTGAAGGTCGGAGTC | 225 bp |
| Reverse: GAAGATGGTGATGGGATTTC |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction; bp, base pairs.
Western blotting
A total of 48 h following gene transfection, the MHCC-97H cells in each group were harvested for survivin expression analysis. The cells were washed twice with cold PBS and lysed on ice in SDS lysis buffer. Protein samples were separated by 8% SDS-PAGE and electrotransferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA) and blocked in 5% non-fat dry milk at room temperature for 2 h. The membranes were incubated overnight at 4°C with rabbit polyclonal anti-survivin antibody (dilution, 1:500; cat. no. ab469; Abcam, Cambridge, UK) and mouse anti-β-actin monoclonal antibody (dilution, 1:400; cat. no. BM0005; Wuhan Boster Biological Technology, Ltd., Wuhan, China), followed by goat anti-rabbit or -mouse horseradish peroxidase (HRP)-conjugated secondary immunoglobulin G antibodies (dilution, 1:500, cat. no. BA1054 and BA1051; Wuhan Boster Biological Technology, Ltd., Wuhan, China) at room temperature for 2 h. The detection was performed using an enhanced chemiluminescence system (Sino-American Biotechnology Co., Ltd., Luoyang, China). Image J software version 1.441 (National Institutes of Health, Bethesda, MD, USA) was used for band intensity quantification.
MTT assay
The proliferation of MHCC-97H cells following survivin siRNA nanoliposome transfection was detected using an MTT assay. Briefly, 1×103 cells were plated in 96-well plates and cultured overnight at 37°C. Following transfection, 10 µl MTT (Nanjing Shengxing Biotechnology Co. Ltd., Nanjing, China) was added to each well and the cells were incubated at 37°C for an additional 4 h. A total of 150 ml of dimethyl sulfoxide was added to each well to dissolve the formazan crystals. Following incubation at 37°C for 10 min, the absorbance of each sample was recorded at a wavelength of 570 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Hoechst and Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining
For Hoechst staining, 3×105 ml cells were plated in a 24-well plate, incubated with survivin siRNA nanoliposome at 37°C for 48 h and stained with Hoechst 33258 (Thermo Fisher Scientific, Inc.) for 10–20 min. Cells were analyzed using a fluorescence microscope (x100; Nikon E400; Nikon Corporation, Tokyo, Japan). Cells with condensed and fragmented nuclei were considered to be apoptotic.
The apoptosis of MHCC-97H cells following survivin siRNA nanoliposome transfection was further evaluated using flow cytometry (FCM) with an Annexin V-FITC/PI kit (Beyotime Institute of Biotechnology, Haimen, China). The adherent cells were collected using centrifugation and resuspended at a density of 1.5×105/ml. Cell suspensions of 2 ml were seeded into each well of 6-well culture plates and cultured at 37°C for 48 h, then stained with 5 µl Annexin V-FITC and 10 µl PI in the dark. The cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The results were displayed as quadrant dot plots with intact cells (Annexin V-/PI-), late apoptotic cells (Annexin V+/PI+), early apoptotic cells (Annexin V+/PI-) and necrotic cells (Annexin V-/PI+). The number of each type of cell was expressed as a percentage of the total number of stained cells.
MHCC-97H xenograft model and treatment
MHCC-97H cells (4–6×106/0.2 ml) were subcutaneously injected into male BALB/c nude mice. When the tumors reached ~1 cm in diameter, subcutaneous tumors were collected, cut into sections of 1×1×1 mm3 and transplanted into the subaxillary tissue of male BALB/c nude mice.
When the tumors reached a mean diameter of 3–5 mm, the mice were randomly assigned to four groups (n=6): the NC-IV group, intravenous injection of NC siRNA nanoliposome (3 mg/kg, twice a week for 5 weeks); the SU-IT group, intratumoral injection of survivin siRNA nanoliposome (50 µg/mouse, twice a week for 5 weeks); the SU-IV group, intravenous injection of siRNA nanoliposome (3 mg/kg, twice a week for 5 weeks); the DOX group, intraperitoneal injection of doxorubicin hydrochloride (2.5 mg/kg, weekly for 5 weeks; Shenzhen Wanle Pharmaceutical Co., Ltd., Wuhan, China).
The tumor size and body weight of each mouse were measured every 3 days for a period of 31 days. The length and width of the tumor was measured using vernier calipers. Tumor volume was calculated to obtain the relative tumor volume (RTV). Tumor volume and the inhibition rate of survivin siRNA nanoliposomes on the tumor growth were calculated as described previously (22). The animals were sacrificed by cervical dislocation following the final treatment and tumors were excised and weighed. Tumor tissues were collected in each group for survivin expression analysis using RT-qPCR, western blot analysis and immunohistochemistry including hematoxylin and eosin (H&E) staining analysis.
Histological and immunohistochemistry analysis
Tumor tissues were fixed in 10% formalin phosphate buffer at room temperature for 24 h and consequently processed for paraffin embedding. The 5 µm sections of tissue were cut, dewaxed in xylene, and rehydrated with a series of graded alcohols (100, 95, 80 and 75%). The sections were then stained with hematoxylin eosin. For immunohistochemistry analysis, antigen retrieval was achieved by pressure cooking in citrate buffer (0.01 mol/l) for 10 min. The sections were incubated with rabbit anti-survivin monoclonal antibody (1:500, cat. no. ab76424, Abcam), followed by incubation with the secondary antibody, HRP-conjugated AffiniPure Goat Anti-Rabbit IgG (cat. no. 111-035-003; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) at a dilution of 1:1,000. The sections were visualized with 3,3-diaminobenzidine and counterstained with hematoxylin. The sections were photographed under a light microscope (x40; Nikon 50i; Nikon Corporation).
Statistical analysis
All experiments were performed in triplicate as a minimum and data are expressed as the mean ± standard deviation. One-way analysis of variance was used to compare the significance between groups. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA).
Results
siRNA nanoliposomes downregulate survivin expression in MHCC-97H cells
RT-qPCR analysis demonstrated that, compared with MHCC-97H cells treated with NC siRNA, the mRNA expression levels of survivin were effectively downregulated following survivin siRNA liposome transfection (P=0.006; Fig. 1A). The level of survivin protein was also significantly decreased (P=0.009; Fig. 1B) in comparison with the survivin protein levels in NC siRNA-treated MHCC-97H cells.
Figure 1.
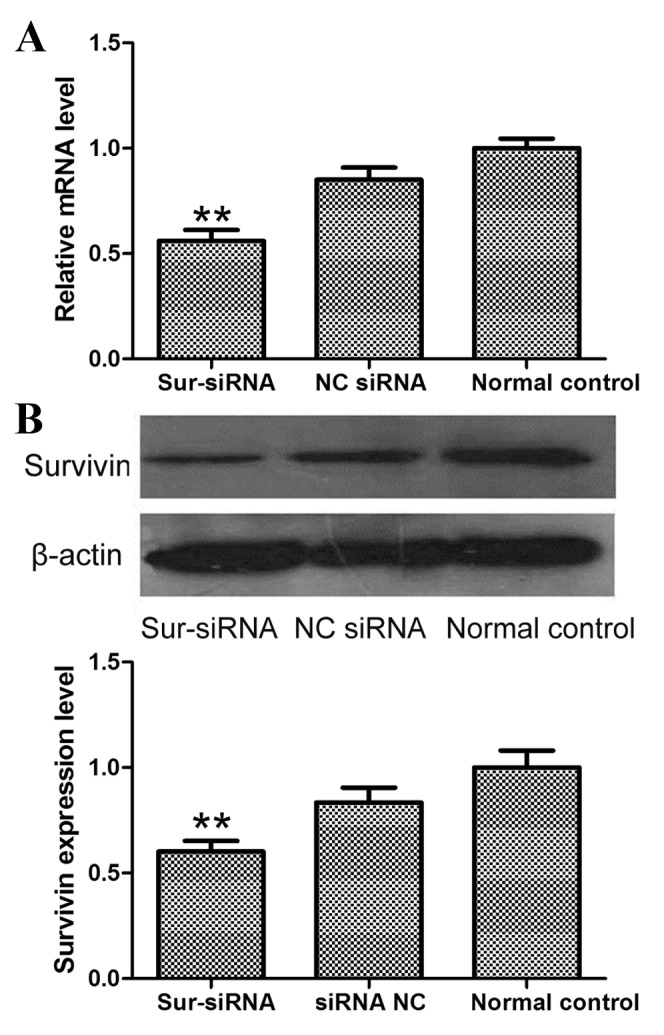
mRNA and protein expression levels of survivin following siRNA nanoliposome transfection of MHCC-97H cells. (A) mRNA levels were determined using RT-qPCR and (B) protein levels were evaluated using western blot analysis. **P<0.01, vs. the normal control group. siRNA, small interfering RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; sur-siRNA, survivin-targeted siRNA; NC, negative control.
Survivin siRNA nanoliposomes inhibit the proliferation of MHCC-97H cells
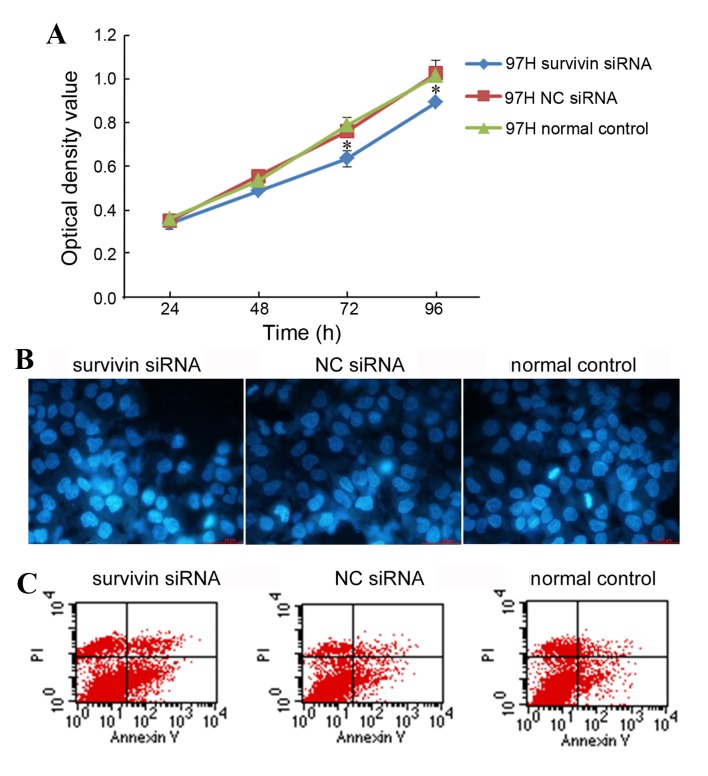
The viability of MHCC-97H cells following survivin siRNA nanoliposome transfection was evaluated using an MTT assay. The MTT results demonstrated that the downregulation of survivin expression using siRNA nanoliposome transfection significantly reduced the proliferation of MHCC-97H cells, as compared with that in the normal group (Fig. 2A; P<0.05). No significant differences were identified in cell proliferation between the NC siRNA nanoliposome-transfected cells and the normal group (Fig. 2A).
Figure 2.
Viability and apoptosis of MHCC-97H cells following survivin siRNA nanoliposome transfection. (A) Cell viability as determined using the MTT assay. (B) Apoptosis of MHCC-97H cells induced by survivin siRNA nanoliposome transfection. Nuclear morphology of cells stained with Hoechst-33258 was analyzed using fluorescence light microscopy (magnification, ×40). (C) Annexin V-FITC/PI staining as analyzed using flow cytometry. *P<0.05 vs. the normal control group. siRNA, small interefering RNA; PI, propidium iodide; FITC, fluorescein isothiocyanate; NC, negative control.
Survivin siRNA nanoliposomes enhance the apoptosis of MHCC-97H cells
MHCC-97H cells were transfected with survivin siRNA nanoliposomes and apoptosis was evaluated using Hoechst and Annexin V-FITC/PI staining. Hoechst staining revealed an increased nuclear condensation and morphological changes in MHCC-97H cells following survivin siRNA nanoliposome transfection, when compared with that of the NC siRNA nanoliposome-transfected or normal cells (Fig. 2B). The Annexin V-FITC/PI double staining analysis by flow cytometry indicated that the number of Annexin V-FITC/PI double-positive cells increased significantly following transfection with survivin siRNA nanoliposomes, whilst no significant difference was detected between NC siRNA nanoliposome-transfected cells and normal cells (P=0.003 and P=0.803, respectively; Fig. 2C).
Survivin siRNA nanoliposome inhibit the growth of MHCC-97H xenograft
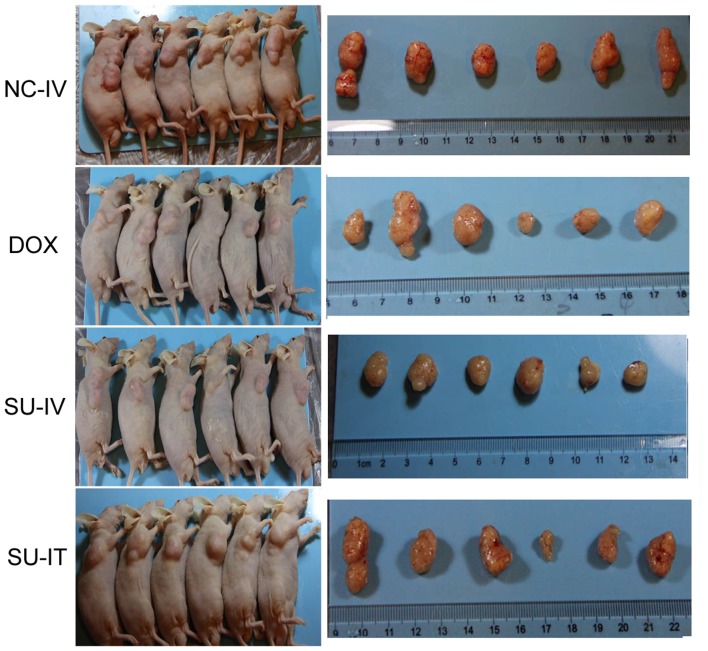
The antitumor effect of survivin siRNA nanoliposomes was further investigated in the MHCC-97H xenograft model. As presented in Table II, survivin siRNA nanoliposome treatment significantly inhibited tumor growth (P<0.001). Compared with the NC siRNA group, the survivin siRNA nanoliposome group exhibited inhibited tumor growth, similar to the effect observed in the DOX group. The images of the dissected tumor tissues and inhibition rate of tumor growth at day 31 are presented in Fig. 3. No significant differences were identified in the body weight among the groups (P=0.3144), whereas delivery of survivin siRNA nanoliposome intratumorally or intravenously significantly reduced the tumor weight to 32.2 and 36.67%, compared with NC-IV tumor weight (P=0.035 and P=0.021, respectively). The tumor inhibition rate of the survivin siRNA-treatment groups was similar to that of the DOX group (31.11%).
Table II.
In vivo antitumor effects of survivin siRNA nanoliposome on Balb/c nude mice bearing MHCC-97H tumor cells.
| NC-IV | DOX | SU-IV | SU-IT | |||||
|---|---|---|---|---|---|---|---|---|
| Day | RTV | BW/g | RTV | BW/g | RTV | BW/g | RTV | BW/g |
| Day 0 | / | 20.50±1.26 | / | 21.00±0.82 | / | 21.33±0.62 | / | 21.33±0.62 |
| Day 3 | 1.85±0.39 | 22.00±0.41 | 1.28±0.29 | 22.17±0.80 | 1.33±0.59 | 22.17±0.37 | 1.31±0.20 | 22.50±0.82 |
| Day 6 | 2.88±0.72 | 22.75±0.56 | 2.08±1.03 | 23.08±0.89 | 1.98±1.02 | 23.17±0.37 | 2.12±0.69 | 23.00±0.65 |
| Day 10 | 5.07±1.33 | 23.42±0.84 | 3.67±2.03 | 23.58±1.20 | 2.99±1.63a | 24.08±0.53 | 2.69±0.93b | 23.92±1.27 |
| Day 13 | 10.38±2.97 | 23.42±1.06 | 6.40±3.60 | 24.00±1.35 | 5.39±3.47a | 24.08±0.53 | 5.74±1.70b | 24.08±1.20 |
| Day 17 | 14.71±2.76 | 23.50±1.00 | 8.85±4.58a | 23.58±1.20 | 8.77±6.24 | 24.50±0.58 | 7.90±2.25b | 24.25±1.18 |
| Day 20 | 29.86±8.76 | 24.17±0.69 | 16.28±8.98a | 23.50±1.12 | 13.72±9.82a | 24.75±0.25 | 14.47±4.62b | 24.92±1.20 |
| Day 24 | 39.54±11.55 | 25.25±0.75 | 23.34±12.45a | 24.17±0.85 | 19.62±12.47a | 26.08±0.34 | 19.64±3.61b | 26.00±0.91 |
| Day 27 | 61.77±17.38 | 25.58±0.89 | 32.09±21.34a | 3.58±0.73 | 24.62±15.27b | 26.50±0.65 | 31.68±10.34b | 26.08±0.79 |
| Day 31 | 76.94±23.28 | 25.17±1.03 | 40.86±25.85a | 24.00±1.29 | 34.53±21.35b | 26.25±0.48 | 39.75±11.75b | 26.17±0.85 |
P<0.05
P<0.01 vs. the NC-IV group. RTV, relative tumor volume; BW/g, body weight/gram; NC-IV, negative control small interfering RNA administered intravenously; DOX, intraperiotoneal injection of doxorubicin hydrochloride; SU-IV, intravenous injection of survivin small interfering RNA nanoliposome; SU-IT, intratumoral injection of survivin small interfering RNA nanoliposome.
Figure 3.
Survivin siRNA nanoliposomes inhibited tumor growth in a xenograft mouse model. Representative images of dissected tumor tissues are presented. siRNA, small interfering RNA; NC-IV, negative control small interfering RNA administered intravenously; DOX, intraperiotoneal injection of doxorubicin hydrochloride; SU-IV, intravenous injection of survivin small interfering RNA nanoliposome; SU-IT, intratumoral injection of survivin small interfering RNA nanoliposome.
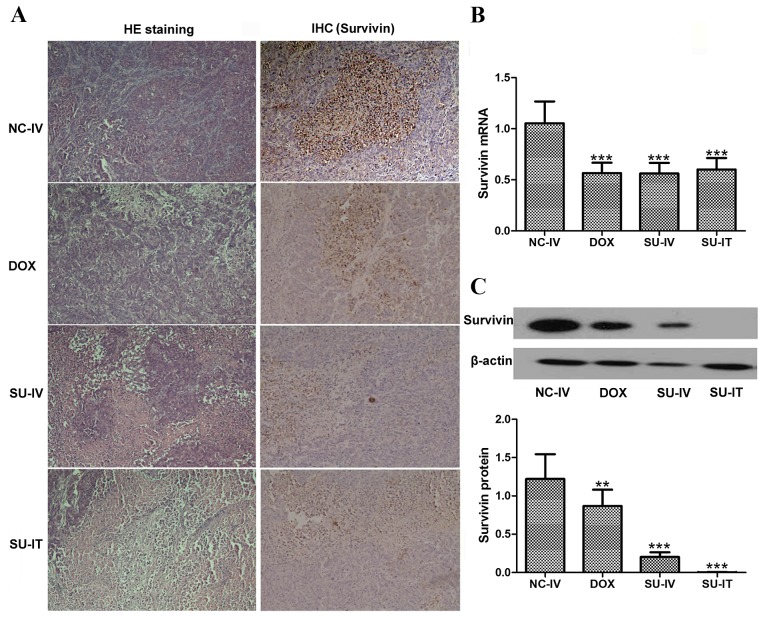
The tumor samples were harvested for survivin expression analysis. As presented in Fig. 4, the gross distribution of immunoreactive survivin in the tumors was analyzed using immunohistochemistry and revealed a general decrease of survivin staining in tumors from mice treated with survivin siRNA nanoliposome, whereas the tumors of mice treated with NC siRNA nanoliposomes exhibited significantly increased survivin staining. Survivin mRNA and protein was highly expressed in the transplanted tumors treated with NC siRNA nanoliposomes, and survivin siRNA nanoliposomes significantly downregulated the tumor-induced upregulation of survivin mRNA and protein expression (P<0.001 and P=0.001, respectively).
Figure 4.
Tumor samples were harvested for (A) histological analysis (H&E and survivin staining), (B) RT-qPCR and (C) western blot analysis of survivin expression levels following survivin siRNA nanoliposome treatment. The mRNA expression levels of survivin are measured relative to GAPDH (magnification ×40). ***P<0.001, vs. the NC-IV group. H&E, hematoxylin and eosin; IHC, immuohistochemistry; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; NC-IV, negative control small interfering RNA administered intravenously; DOX, intraperiotoneal injection of doxorubicin hydrochloride; SU-IV, intravenous injection of survivin small interfering RNA nanoliposome; SU-IT, intratumoral injection of survivin small interfering RNA nanoliposome.
Discussion
Targeted molecular therapy using siRNA has been investigated in a number of studies as a method of treating tumors due to its specific and potent silencing of targeted genes (23–25). Targeted therapy has been reported to induce apoptosis, block cell proliferation and even suppress tumor formation and growth (26–28). Survivin is an established apoptosis-inhibiting factor that is selectively expressed in numerous types of cancer cells but not in normal tissues (29), and is therefore a potential target gene for therapeutic intervention. Using siRNA-mediated knockdown of survivin inhibited cancer cell proliferation, enhanced apoptotic susceptibility and decreased tumorigenicity in human xenografts (22,30). Despite the potential therapeutic advantages, effective delivery of siRNA to tumor cells remains a major barrier for RNA-based cancer therapy, and the success of gene therapy is highly dependent on the delivery vector (19). An effective delivery system that is able to protect the siRNA from enzymatic degradation and promote specific cellular uptake is required (31). Use of a cationic liposome/micelle-based system is a possible solution to this problem and has been applied for siRNA delivery (32). Survivin downregulation using a siRNA/cationic liposome complex resulted in significant cell growth inhibition, increased apoptotic rate and radiosensitivity in human HCC (20).
In the current study, an effective siRNA sequence targeting survivin was selected and cationic liposome-based nanoliposomes were constructed to deliver survivin-targeted siRNA into human MHCC-97H HCC cells. The phenotypic changes following cationic liposome-mediated transfection were analyzed. The results demonstrated that the transfection of HCC cells with survivin siRNA nanoliposomes downregulated the mRNA and protein expression levels of the survivin gene, as demonstrated using RT-qPCR and western blot analysis. This result indicated that this nanoliposome construct may facilitate survivin siRNA transfection into transplanted hepatic tumor cells in a mouse model to block survivin expression. Delivery of survivin siRNA nanoliposome into MHCC-97H xenografts significantly inhibited the growth of tumor cells. The findings of the current in vitro and in vivo studies are concordant, suggesting that the stable knockdown of survivin using siRNA nanoliposome abrogated its function in cell proliferation and apoptosis. To evaluate treatment-associated toxicity, body weight was used as an index for the general health status of the mice, and delivery of the survivin siRNA nanoliposome by intratumoral or intravenous injection caused no significant reduction in body weight of the mice in comparison with that of the NC group and DOX group. This suggests that there is no treatment-associated toxicity as a result of survivin siRNA nanoliposome administration in a mouse model.
Apoptosis is the main mode of cell death that is induced by numerous classes of anticancer agent (33,34). Survivin was originally proposed to perform an antiapoptotic function (35,36). Knockdown of survivin using genetic deletion, anti-sense oligonucleotides, dominant negative inhibitors or siRNAs is able to induce apoptosis in tumor cells (25,26,37,38). A recent study by Han et al (39) also indicated that the transfection of survivin antisense oligonucleotide using polyamidoamine dendrimer liposomes inhibited hepatic cell proliferation by inducing apoptosis, concordant with previous studies, which have suggested that survivin has an important role in cellular proliferation (40,41). A previous study detected survivin overexpression in 70% of patients with HCC, which was correlated with poor prognosis (42). The potential underlying molecular mechanism was suggested to involve the proliferation-promoting effect of survivin in HCC cells (42). Downregulation of survivin in the present study was associated with significant cell growth inhibition, as detected using an MTT assay, and increased apoptotic rate, as revealed using Hoechst and Annexin V-FITC/PI staining. This is concordant with the results of previous studies, which indicated that survivin expression levels correlate with reduced tumor growth and increased apoptosis in several types of tumor cells (43–45). However, Zhang et al (30) indicated that tumor growth-inhibition due to survivin downregulation was associated with decreased cell proliferation; however survivin downregulation did not affect the levels of apoptosis. The authors hypothesized that, as there are numerous anti-apoptotic mechanisms involved in the prevention of apoptosis in HCC cells, one of these alternate underlying mechanisms may be the reason for this discrepancy (30). These conflicting results may also be partially explained by the varied strategies used for gene knockout and the differing delivery system and vectors used. siRNA, a double-stranded RNA, was suggested to be more resistant to nuclease degradation, providing a longer duration of therapeutic effects and, therefore, being more efficient and durable in cell culture compared with the antisense oligonucleotide strategy (46). Future studies are required to further elucidate the potential underlying mechanisms of siRNA delivery.
Efficient introduction of siRNA into target cells is necessary to facilitate the application of siRNA in cancer gene therapy. Several independent studies have investigated the systemic delivery strategies for the introduction of survivin siRNA into mice (30,47,48). The local injection method was considered to be feasible for the clinical application of siRNA and was generally performed in these previous studies. Zhang et al (30) injected survivin short hairpin RNA into the local tumor region and monitored tumor growth and survival rate. In the present study, intratumoral and intravenous injections were performed to introduce survivin siRNA nanoliposome into the MHCC-97H xenograft models. The results indicate that the RTV and tumor inhibition rate, as well as the treatment-associated toxicity, were not significantly different between the SU-IT and SU-IV mice. This suggests that survivin-targeted siRNA delivered using nanoliposomes is safe for intratumoral local or intravenous systemic administration.
In conclusion, a cationic nanoliposome-based survivin siRNA delivery system was constructed and demonstrated to be efficient for survivin siRNA delivery in in vitro and in vivo assays. These results demonstrate that survivin downregulation may significantly attenuate cell proliferation and induce the apoptosis of MHCC-97H HCC cells, as well as inhibiting tumor cell growth in MHCC-97H xenograft models, indicating that survivin suppression with siRNA may contribute to tumor inhibition through the inhibition of proliferation and promotion of apoptosis.
Acknowledgements
The present study was supported by the National High Technology Research and Development Program of China (863 program; grant no. 2012AA020804).
References
- 1.Dong Y, Zou J, Su S, Huang H, Deng Y, Wang B, Li W. MicroRNA-218 and microRNA-520a inhibit cell proliferation by downregulating E2F2 in hepatocellular carcinoma. Mol Med Report. 2015;12:1016–1022. doi: 10.3892/mmr.2015.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008 GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Machi J, Bueno RS, Wong LL. Long-term follow-up outcome of patients undergoing radiofrequency ablation for unresectable hepatocellular carcinoma. World J Surg. 2005;29:1364–1373. doi: 10.1007/s00268-005-7829-6. [DOI] [PubMed] [Google Scholar]
- 5.Said A, Wells J. Management of hepatocellular carcinoma. Minerva Med. 2009;100:51–68. [PubMed] [Google Scholar]
- 6.Park HC, Seong J, Tanaka M, Zeng ZC, Lim HY, Guan S, Bae SH, Tak WY. Multidisciplinary management of nonresectable hepatocellular carcinoma. Oncology. 2011;81(Suppl 1):134–140. doi: 10.1159/000333276. [DOI] [PubMed] [Google Scholar]
- 7.Bouza C, López-Cuadrado T, Alcázar R, Saz-Parkinson Z, Amate JM. Meta-analysis of percutaneous radiofrequency ablation versus ethanol injection in hepatocellular carcinoma. BMC Gastroenterol. 2009;9:31. doi: 10.1186/1471-230X-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serizawa N, Tian J, Fukada H, Baghy K, Scott F, Chen X, Kiss Z, Olson K, Hsu D, Liu FT, et al. Galectin 3 regulates HCC cell invasion by RhoA and MLCK activation. Lab. Invest. 2015;95:1145–1156. doi: 10.1038/labinvest.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn RS. Development of molecularly targeted therapies in hepatocellular carcinoma: Where do we go now? Clin Cancer Res. 2010;16:390–397. doi: 10.1158/1078-0432.CCR-09-2084. [DOI] [PubMed] [Google Scholar]
- 10.Ryan BM, O'Donovan N, Duffy MJ. Survivin: A new target for anti-cancer therapy. Cancer Treat Rev. 2009;35:553–562. doi: 10.1016/j.ctrv.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 12.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 13.Mobahat M, Narendran A, Riabowol K. Survivin as a preferential target for cancer therapy. Int J Mol Sci. 2014;15:2494–2516. doi: 10.3390/ijms15022494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodel F, Sprenger T, Kaina B, Liersch T, Rödel C, Fulda S, Hehlgans S. Survivin as a prognostic/predictive marker and molecular target in cancer therapy. Curr Med Chem. 2012;19:3679–3688. doi: 10.2174/092986712801661040. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Lin R, Dai J, Jin D, Wang SQ. Suppression of tumor growth using antisense oligonucleotide against survivin in an orthotopic transplant model of human hepatocellular carcinoma in nude mice. Oligonucleotides. 2006;16:365–374. doi: 10.1089/oli.2006.16.365. [DOI] [PubMed] [Google Scholar]
- 16.Tai CJ, Chin-Sheng H, Kuo LJ, Wei PL, Lu HH, Chen HA, Liu TZ, Liu JJ, Liu DZ, Ho YS, et al. Survivin-mediated cancer cell migration through GRP78 and epithelial-mesenchymal transition (EMT) marker expression in Mahlavu cells. Ann Surg Oncol. 2012;19:336–343. doi: 10.1245/s10434-011-1692-5. [DOI] [PubMed] [Google Scholar]
- 17.Song X, Wang JB, Yin DL, Yang HY, Liu LX, Jiang HC. Down-regulation of lung resistance related protein by RNA interference targeting survivin induces the reversal of chemoresistances in hepatocellular carcinoma. Chin Med J(Engl) 2009;122:2636–2642. [PubMed] [Google Scholar]
- 18.Cheng SQ, Wang WL, Yan W, Li QL, Wang L, Wang WY. Knockdown of survivin gene expression by RNAi induces apoptosis in human hepatocellular carcinoma cell line SMMC-7721. World J Gastroenterol. 2005;11:756–759. doi: 10.3748/wjg.v11.i5.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian H, Liu S, Zhang J, Zhang S, Cheng L, Li C, Zhang X, Dail L, Fan P, Dai L, et al. Enhancement of cisplatin sensitivity in lung cancer xenografts by liposome-mediated delivery of the plasmid expressing small hairpin RNA targeting Survivin. J Biomed Nanotechnol. 2012;8:633–641. doi: 10.1166/jbn.2012.1419. [DOI] [PubMed] [Google Scholar]
- 20.Yang W, Sun T, Cao J, Liu F. Survivin downregulation by siRNA/cationic liposome complex radiosensitises human hepatoma cells in vitroin vivo. Int J Radiat Biol. 2010;86:445–457. doi: 10.3109/09553001003668006. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Juanjuan Li, Mei Y, Xiaodong S, Shunqiao F, Ruihong T, Xia Z, Rong L, Ziqin L, Tianyou W. Evaluation of anti-cancer activity of survivin siRNA delivered by folate receptor-targeted polyethylene-glycol liposomes in K562-bearing xenograft mice. BBiomed. Eng. Appl. Basis Commun. 2014;26 [Google Scholar]
- 23.Shen Q, Cicinnati VR, Zhang X, Iacob S, Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G, Beckebaum S. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer. 2010;9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Q, Hu B, An HM, Shen KP, Xu L, Deng S, Wei MM. Synergistic anticancer effects of curcumin and resveratrol in Hepa1-6 hepatocellular carcinoma cells. Oncol Rep. 2013;29:1851–1858. doi: 10.3892/or.2013.2310. [DOI] [PubMed] [Google Scholar]
- 25.Rychahou PG, Jackson LN, Silva SR, Rajaraman S, Evers BM. Targeted molecular therapy of the PI3K pathway: Therapeutic significance of PI3K subunit targeting in colorectal carcinoma. Ann Surg. 2006;243:833–844. doi: 10.1097/01.sla.0000220040.66012.a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho-Rok J, Yoo J, Jang YJ, Kim S, Chu IS, Yeom YI, Choi JY, Im DS. Adenovirus-mediated transfer of siRNA against PTTG1 inhibits liver cancer cell growth in vitroin vivo. Hepatology. 2006;43:1042–1052. doi: 10.1002/hep.21137. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Fu X, Chen Y, Hong Y, Tan Y, Cao H, Wu M, Wang H. Use of adenovirus-delivered siRNA to target oncoprotein p28 GANK in hepatocellular carcinoma. Gastroenterology. 2005;128:2029–2041. doi: 10.1053/j.gastro.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Li K, Lin SY, Brunicardi FC, Seu P. Use of RNA interference to target cyclin E-overexpressing hepatocellular carcinoma. Cancer Res. 2003;63:3593–3597. [PubMed] [Google Scholar]
- 29.Zhang Y, Zhang Y, Chen J, Zhang B, Pan Y, Ren L, Zhao J, Luo Y, Zhai D, Wang S, Wang J. Polybutylcyanoacrylate nanoparticles as novel vectors in cancer gene therapy. Nanomedicine. 2007;3:144–153. doi: 10.1016/j.nano.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Zhang R, Ma L, Zheng M, Ren J, Wang T, Meng Y, Zhao J, Jia L, Yao L, Han H, et al. Survivin knockdown by short hairpin RNA abrogates the growth of human hepatocellular carcinoma xenografts in nude mice. Cancer Gene Ther. 2010;17:275–288. doi: 10.1038/cgt.2009.68. [DOI] [PubMed] [Google Scholar]
- 31.Ahn HJ, Kwon IC, Choi KW. Recombinant protein for intracellular delivery of siRNA and composition comprising the same. Google Patents. 2015 [Google Scholar]
- 32.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 33.Aloya R, Shirvan A, Grimberg H, Reshef A, Levin G, Kidron D, Cohen A, Ziv I. Molecular imaging of cell death in vivo by a novel small molecule probe. Apoptosis. 2006;11:2089–2101. doi: 10.1007/s10495-006-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chodon D, Ramamurty N, Sakthisekaran D. Preliminary studies on induction of apoptosis by genistein on HepG2 cell line. Toxicol In Vitro. 2007;21:887–891. doi: 10.1016/j.tiv.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Ikeda M, Okamoto I, Tamura K, Satoh T, Yonesaka K, Fukuoka M, Nakagawa K. Down-regulation of survivin by ultraviolet C radiation is dependent on p53 and results in G(2)-M arrest in A549 cells. Cancer Lett. 2007;248:292–298. doi: 10.1016/j.canlet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Vivek R, Kannan S, Achiraman S, Thirumurugan R, Ganesh DS, Krishnan M. Survivin deficiency leads to imparalization of cytokinesis in cancer cells. Asian Pac J Cancer Prev. 2011;12:1675–1679. [PubMed] [Google Scholar]
- 37.Xu Z, Zhang Z, Chen Y, Chen L, Lin L, Li Y. The characteristics and performance of a multifunctional nanoassembly system for the co-delivery of docetaxel and iSur-pDNA in a mouse hepatocellular carcinoma model. Biomaterials. 2010;31:916–922. doi: 10.1016/j.biomaterials.2009.09.103. [DOI] [PubMed] [Google Scholar]
- 38.Chen XQ, Yang S, Li ZY, Lu HS, Kang M-Q, Lin TY. Effects and mechanism of downregulation of survivin expression by RNA interference on proliferation and apoptosis of lung cancer cells. Mol Med Rep. 2012;5:917–922. doi: 10.3892/mmr.2012.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han S, Cai Z, Peng L, Li Z, Zhou HB, Li XQ, Fang SZ, Huang ZH, Cui DX. Polyamidoamine dendrimer liposome-mediated survivin antisense oligonucleotide inhibits hepatic cancer cell proliferation by inducing apoptosis. Tumor Biol. 2014;35:5013–5019. doi: 10.1007/s13277-014-1661-2. [DOI] [PubMed] [Google Scholar]
- 40.Wakana Y, Kasuya K, Katayanagi S, Tsuchida A, Aoki T, Koyanagi Y, Ishii H, Ebihara Y. Effect of survivin on cell proliferation and apoptosis in gastric cancer. Oncol Rep. 2002;9:1213–1218. [PubMed] [Google Scholar]
- 41.Morinaga S, Nakamura Y, Ishiwa N, Yoshikawa T, Noguchi Y, Yamamoto Y, Rino Y, Imada T, Takanashi Y, Akaike M, et al. Expression of survivin mRNA associates with apoptosis, proliferation and histologically aggressive features in hepatocellular carcinoma. Oncol Rep. 2004;12:1189–1194. [PubMed] [Google Scholar]
- 42.Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M, et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31:1080–1085. doi: 10.1053/he.2000.6496. [DOI] [PubMed] [Google Scholar]
- 43.Uchida H, Tanaka T, Sasaki K, Kato K, Dehari H, Ito Y, Kobune M, Miyagishi M, Taira K, Tahara H, Hamada H. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitroin vivo. Mol Ther. 2004;10:162–171. doi: 10.1016/j.ymthe.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto T, Tanigawa N. The role of survivin as a new target of diagnosis and treatment in human cancer. Med Electron Microsc. 2001;34:207–212. doi: 10.1007/s007950100017. [DOI] [PubMed] [Google Scholar]
- 45.Li QX, Zhao J, Liu JY, Jia LT, Huang HY, Xu YM, Zhang Y, Zhang R, Wang CJ, Yao LB, et al. Survivin stable knockdown by siRNA inhibits tumor cell growth and angiogenesis in breast and cervical cancers. Cancer Biol Ther. 2006;5:860–866. doi: 10.4161/cbt.5.7.2893. [DOI] [PubMed] [Google Scholar]
- 46.Bertrand JR, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun. 2002;296:1000–1004. doi: 10.1016/S0006-291X(02)02013-2. [DOI] [PubMed] [Google Scholar]
- 47.Lobovkina T, Jacobson GB, Gonzalez-Gonzalez E, Hickerson RP, Leake D, Kaspar RL, Contag CH, Zare RN. In vivo sustained release of siRNA from solid lipid nanoparticles. ACS Nano. 2011;5:9977–9983. doi: 10.1021/nn203745n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendruschk S, Wiedemuth R, Aigner A, Töpfer K, Cartellieri M, Martin D, Kirsch M, Ikonomidou C, Schackert G, Temme A. RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo. Neuro Oncol. 2011;13:1074–1089. doi: 10.1093/neuonc/nor098. [DOI] [PMC free article] [PubMed] [Google Scholar]