Abstract
B7-homolog 4 (B7-H4), one of the costimulatory molecules of the B7 family, has been reported to be widely expressed in multiple types of cancer tissues, and to be important in tumor progression and poor prognosis. However, the role of B7-H4 in esophageal precancerous lesions has not been elucidated yet. In the present study, a model of esophageal squamous cell carcinogenesis was established in 208 C57BL/6 mice by 4-nitroquinoline-1-oxide (4NQO) drinking water of mice, and the changes in the expression of B7-H4 during the whole pathological process were investigated. Hematoxylin and eosin staining results demonstrated that the pathological stage was exacerbated with the increase in time of 4NQO-mediated carcinogenesis induction, and the pathological features were similar to those observed in humans. Immunohistochemistry results revealed that B7-H4 expression was upregulated and positively correlated with pathological stage (P<0.0001) as well as with infiltration of cluster of differentiation (CD) 11b+ macrophage cells (P=0.0002). In addition, B7-H4 messenger RNA expression increased in the esophagi of model mice compared with that of control mice, which was positively associated with the gene expression of interleukin (IL)-6 and signal transducer and activator of transcription 3 (STAT3), according to the results of reverse transcription-quantitative polymerase chain reaction analysis. Similarly, B7-H4 protein expression was upregulated in the esophageal tissues of model mice in comparison with that of control mice, and was positively associated with IL-6 expression and STAT3 phosphorylation. In conclusion, the present data suggested that B7-H4 expression increased during esophageal squamous cell carcinogenesis in mice in association with IL-6/STAT3 signaling pathway activation.
Keywords: B7-homolog 4, esophageal carcinogenesis, 4-nitroquinoline-1-oxide, precancerous condition, interleukin-6/signal transducer and activator of transcription 3 signaling pathway
Introduction
Esophageal cancer is the eighth most common type of cancer and the sixth leading cause of cancer-associated mortality worldwide (1). There are two histological types of esophageal cancer: Squamous cell carcinoma (SCC) and adenocarcinoma (ADE) (2). Although the incidence of ADE has increased rapidly in Western countries during the last 30 years, SCC is still the major type of esophageal cancer-related threat in Asia, Southern and Eastern Africa, and Northern France (3). In Africa and Asia, 90% of esophageal cancer cases are SCC, with a 5-year survival rate of only ~20%, since the majority of cases are diagnosed in the advanced stage, when the best treatment opportunity has been missed (4). Therefore, it is urgently required to investigate the mechanism of SCC formation and to explore novel biomarkers for the diagnosis and treatment of esophageal precancerous conditions.
4-Nitroquinoline-1-oxide (4NQO), a water-soluble quinolone derivative, can induce squamous cell carcinogenesis through oxidative stress and inhibition of DNA synthesis (5). The tumor formation induced by 4NQO is tissue-specific. Lung cancer, for example, can be induced by subcutaneous injection of 4NQO, while oral tumors can be induced by oral administration of 4NQO (6–9). However, the study of esophageal precancerous lesions induced by 4NQO remains undetermined.
B7-homolog 4 (B7-H4) is a member of the B7 family, which comprises costimulatory molecules (10). In 2003, B7-H4, a type-I transmembrane protein, was identified by three different laboratories simultaneously, and was assigned three different names: B7-H4 (11), B7s1 (12) and B7x (13). More recently, it was reported that B7-H4 messenger RNA was broadly expressed in human peripheral tissues, while B7-H4 protein expression was confined only to certain tumor tissues or cancer cells, including lung (14), ovarian (15), prostate (16), melanoma (17), stomach (18), breast (19) and kidney cancer (20). Besides, B7-H4 expression is induced in activated T cells, B cells and antigen presenting cells (APCs), including dendritic cells and macrophages (21). Although it has been demonstrated that B7-H4 expression in APCs induced by interleukin (IL)-6 inhibited the function and proliferation of T cells (22), the association between B7-H4 expression in tumor cells and the IL-6/signal transducer and activator of transcription 3 (STAT3) signaling pathway remains unknown. In addition, it was suggested that B7-H4 was overexpressed in SCC, which was positively correlated with tumor progression and a poor prognosis (23), and that B7-H4 facilitated SCC cell proliferation (24); however, the expression of B7-H4 in esophageal precancerous conditions has not been investigated to date. In the present study, therefore, a model of SCC formation was established in mice by 4NQO drinking water to explore changes in B7-H4 expression and its association with the IL-6/STAT3 signaling pathway.
Materials and methods
Animals and carcinogen treatment
A total of 208C57BL/6 mice (age, 5–6 weeks; weight, 16±2 g), including the same number of male and female mice, were obtained from Beijing Weitong Lihua Laboratory Animal Technology Ltd. Co., Beijing, China. The mice were raised in the specific pathogen-free animal laboratory of the Fourth Hospital of Hebei Medical University (Shijiazhuang, China), where the housing conditions were strictly controlled (temperature, 18–24°C; humidity, 40–60%; and light/dark cycle, 12/12 h). The carcinogen 4NQO (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) was dissolved and diluted in sterile distilled water to 50 µg/ml. All experiments followed the guidelines of the welfare committee of the Fourth Hospital of Hebei Medical University.
Model protocol
After 1 week of adaptive feeding, the 208 mice were assigned to the control group (n=64) or to the 4NQO model group (n=144). The experiment lasted 28 weeks. The control mice were administered sterile distilled water during these 28 weeks, while the 4NQO model mice were fed with 4NQO (50 µg/ml) as drinking water from the 1st to the 15th week, and then with sterile distilled water from the 16th to the 28th week. The body weight of all mice was tested once every 2 weeks in order to assess their physical condition. In the period ranging from week 16 to 28, 9 mice from the control group and 20 mice from the 4NQO model group were sacrificed by removing the cervical vertebra every 2 weeks.
Hematoxylin and eosin (H&E) staining and immunohistochemistry tests
For every sacrifice, samples derived from 3 control mice and 7 4NQO model mice were subjected to H&E staining and immunohistochemistry tests. Following sacrifice, the thoracic cavity and abdominal cavity of the mice were exposed, and the esophageal tissues were separated and dissected longitudinally, rolled up from cardiac end to tongue end, and then fixed with pins. Subsequently, the esophagi were fixed in 4% formaldehyde, embedded in paraffin and sectioned into 4-µm-thick sections. Sections were stained with H&E. The H&E staining result was defined according to the highest pathological stage for each sample.
Immunohistochemistry tests were performed to detect the expression of B7-H4 as well as the infiltration of cluster of differentiation CD4+T, CD8+T and CD11b+ macrophage cells in esophageal tissues. The sections were dewaxed, rehydrated and exposed to 3% H2O2 solution for 20 min to block endogenous peroxidase. Then, antigens were hot-retrieved with high pressure (B7-H4, 4.5 min heating time and pH=6.0 citrate buffer; CD4 and CD8, 8 min heating time and pH=8.0 EDTA buffer; and CD11b, 10 min heating time, and pH=6.0 citrate buffer). Sections were incubated with primary antibodies against B7-H4 (1:500; GTX62090; GeneTex, Inc., Irvine, CA, USA), CD4 (1:50; orb4241; Biorbyt, Ltd., Cambridge, UK), CD8 (1:50; orb10325; Biorbyt, Ltd.) or CD11b (1:100; cb1211; Abcam, Shanghai, China) at 4°C overnight. Standard protocols for immunohistochemistry were followed as described previously (25). Scores of B7-H4 expression were derived from the multiplication of the area and intensity scores (26). The prevalence of CD4+T, CD8+T or CD11b+ macrophage cells was semi-quantitatively evaluated according to the number of each type of cells present per vision field (magnification, ×400) (27). Of note, the pathological grade of the observation field was in line with the H&E pathological result in each sample.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
For every sacrifice, tissue samples derived from 3 control mice and 7 4NQO model mice were subjected to RT-qPCR analysis to evaluate the gene expression of B7-H4 and associated molecules. Total RNA was extracted from the tissue samples using TRIzol Reagent (Sigma-Aldrich; Merck Millipore) according to the manufacturer's protocol. Total RNA (2 µg for each sample) was reverse transcribed to cDNA with a Reverse Transcription System (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol. qPCR was performed in a total volume of 20 µl, and the reaction mixture contained cDNA product (1 µg), specific primers (0.2 µmol/l), SYBR-Green qPCR Master Mix (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and nuclease-free water. The sequences of the primers were as follows: B7-H4 sense, 5′-CACTCCTATCTCTCTCCTGTTGCCT-3′ and antisense, 5′-CACATCTTGTTCCTGTCTGCTCTCT-3′; interleukin (IL)-6 sense, 5′-CCATCCAGTTGCCTTCTTGG-3′ and antisense, 5′-CTCCTCTCCGGACTTGTGAA-3′; IL-10 sense, 5′-CTGAAGACCCTCAGGATGCG-3′ and antisense, 5′-TGGCCTTGTAGACACCTTGG-3′; transforming growth factor (TGF)-β sense, 5′-GCTGAACCAAGGAGACGGAA-3′ and antisense, 5′-AGAAGTTGGCATGGTAGCCC-3′; interferon (IFN)-γ sense, 5′-AGCCTCAGGAAGCGGAAAAG-3′ and antisense, 5′-TGAGTTCAGTCAGCCGCTTG-3′; STAT3 sense, 5′-TTGAGTCGCTCACGTTTGAC-3′ and antisense, 5′-GGCACTTGTCTAACAACCAACC-3′; and GAPDH sense, 5′-CTTTGGCATTGTGGAAGGGCTC-3′ and antisense, 5′-GCAGGGATGATGTTCTGGGCAG-3′. The number of specific transcripts was normalized to the housekeeping gene GAPDH. The qPCR cycling conditions were as follows: 95°C for 10 min, followed by 42 cycles of 95°C for 15 sec, 55°C for 15 sec and 60°C for 1 min. Differences in the expression levels of the genes were determined by calculating the fold-change in expression according to the 2−ΔΔCq method (28).
Western blotting for protein analysis
For every sacrifice, samples derived from 3 control mice and 6 4NQO mice were used to evaluate the expression of B7-H4 and associated proteins. The proteins were extracted from homogenates of esophageal tissues and quantified using the Bicinchoninic Acid kit (Sigma-Aldrich; Merck Millipore), as described previously (29). Western blot analysis was performed as described previously (30). All proteins were subjected to 10% SDS-PAGE and electrophoretically transferred onto a nitrocellulose membrane. The transfer times were 40 min, 1, 1.5 and 1.5 h for β-actin, B7-H4, STAT3 and phosphorylated (p)-STAT3, respectively. The membrane was blocked with 5% bovine serum albumin (Sigma-Aldrich; Merck Millipore) at 37°C for 1 h and incubated with primary antibodies, including rabbit anti-mouse β-actin (1:5,000; 600-401-886s; Rockland Immunochemicals, Inc., Philadelphia, PA, USA), anti-B7-H4 (1:2,000; GeneTex, Inc.), anti-STAT3 (1:2,000; 79D7; Cell Signaling Technology, Inc., Danvers, MA, USA) and anti-p-STAT3 (1:500; 3771; Cell Signaling Technology, Inc.) monoclonal antibodies overnight at 4°C, followed by incubation with a fluorescent rabbit antibody (1:5,000; sc2020; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 1 h at 37°C. Finally, the gray value was detected with an Odyssey fluorescence scanner (LI-COR Biotechnology GmbH, Bad Homburg, Germany).
ELISA for protein analysis
ELISA for IL-6 evaluation was detected according to the protocol of the Mouse IL-6 ELISA kit (Dakewe Biotech Co., Ltd., Beijing, China). A total of 50 µl protein solution was required for each sample. Protein samples and IL-6 standard solutions were added to enzyme-labeled wells. Subsequently, 100 µl biotinylated antibody was added to each well and the plates were incubated for 90 min at 37°C. After washing five times, 100 µl streptavidin-horseradish peroxidase was added to each well and incubated for 30 min at 37°C. Then, 100 µl 3,3′,5,5′-tetramethylbenzidine was added to each well and incubated for 20 min at 37°C. Finally, the reaction was stopped by 100 µl/well stop solution. The absorbance value was detected at 620 nm.
Statistical analysis
Results are presented as the mean ± standard deviation. All the experimental data were analyzed with GraphPad Prism 5.0 statistical software package (GraphPad Software, Inc., La Jolla, CA, USA). Spearman correlation analysis was performed to compare the correlation between different parameters. Student's t-test and Kruskal-Wallis analysis were performed to compare the difference between ≥2 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
SCC is induced by 4NQO in mice
Firstly, with the prolongation of the time of inducing cancer, the physical conditions of 4NQO model mice became poor, and their body weight increased more slowly compared with that of control mice (Fig. 1A). In addition, 1 control mouse and 4 4NQO model mice died without additional intervention and were excluded from the study.
Figure 1.
Changes in body weight and esophageal pathological conditions during squamous cell carcinoma formation induced by 4NQO in C57BL/6 mice. (A) The body weight of female and male 4NQO model mice increased more slowly than that of control mice (*P<0.05, **P<0.01, Student's t-test). (B) The esophageal pathological conditions of 4NQO model mice were exacerbated during the carcinogenesis process. (C) Hematoxylin and eosin staining of esophageal precancerous lesions (magnification, ×400). LGIN, low-grade intraepithelial neoplasia; HGIN, high-grade intraepithelial neoplasia; 4NQO, 4-nitroquinoline-1-oxide.
Secondly, the esophageal tissues of all mice were dissected. Among these tissues, the esophagi of control mice exhibited smooth surface, normal thickness and flexibility, without any abnormal convex or concave appearance. However, the esophagi of 4NQO model mice exhibited exacerbation of the pathological conditions with the prolongation of the time (Fig. 1B). There were white patches scattered in the epidermis of esophagi from weeks 16 to 18. Subsequently, the esophageal tissues commenced to appear thicker and rough, with abnormal touchable protuberances from weeks 20 to 22. Furthermore, the thickness of the esophagi increased further and the surface became uneven, with an increased number of obvious masses of different size from weeks 24 to 28. In the present study, H&E staining revealed that, during the whole process of carcinogenesis, the pathological stage of the esophageal epithelium underwent normal, low-grade intraepithelial neoplasia (LGIN) and high-grade intraepithelial neoplasia (HGIN), as shown in Fig. 1C. Normal esophageal epithelial cells displayed regular distribution, natural nuclear size and polarity. In LGIN, however, the mucosal epithelium became thicker, and less than half of the cells lost polarity, their nuclei became enlarged and were hyperchromatic. Furthermore, in HGIN, the epithelial layer became even thicker, and >1/2 of cells lost polarity and presented a more enlarged nucleus. Additionally, carcinoma in situ, which occurs when the cells in the whole epithelium lose polarity and become immature (31), was also observed in HGIN. All the above pathological characteristics were similar to those observed in humans (31).
The association between esophageal pathological stage and time of carcinogenesis induction was analyzed (Table I). In the present study, no mice exhibited abnormal pathological conditions in the control group. 4NQO model mice, on the contrary, exhibited all types of precancerous conditions (LGIN and HGIN). Furthermore, with increasing time, the pathological degree of their esophagi markedly increased (P<0.0001). There was no difference in the pathological stage between male and female mice, according to the results of Kruskal-Wallis analysis.
Table I.
Pathological changes in esophageal tissues during the whole carcinogenesis process in mice.
| Controla | 4NQOb | |||||
|---|---|---|---|---|---|---|
| Time (weeks) | Normal | LGIN | HGIN | Normal | LGIN | HGIN |
| 16 | 3 (2,1) | 0 (0,0) | 0 (0,0) | 1 (0,1) | 5 (3,2) | 1 (0,1) |
| 18 | 3 (1,2) | 0 (0,0) | 0 (0,0) | 0 (0,0) | 6 (3,3) | 1 (1,0) |
| 20 | 3 (2,1) | 0 (0,0) | 0 (0,0) | 0 (0,0) | 5 (2,3) | 2 (2,0) |
| 22 | 3 (1,2) | 0 (0,0) | 0 (0,0) | 0 (0,0) | 4 (2,2) | 3 (1,2) |
| 24 | 3 (2,1) | 0 (0,0) | 0 (0,0) | 0 (0,0) | 4 (2,2) | 3 (2,1) |
| 26 | 3 (1,2) | 0 (0,0) | 0 (0,0) | 0 (0,0) | 2 (1,1) | 5 (3,2) |
| 28 | 3 (2,1) | 0 (0,0) | 0 (0,0) | 0 (0,0) | 1 (1,0) | 6 (2,4) |
The values in parentheses refer to the number of female mice and male mice, respectively.
No abnormal pathological condition was identified in the esophagi of control mice.
The pathological grades of the esophagi in the 4NQO group were positively correlated with the time of induction (P<0.0001, Spearman correlation analysis). There was no significant difference regarding the pathological stage between male and female mice (P=0.3333, Kruskal-Wallis analysis). LGIN, low-grade intraepithelial neoplasia; HGIN, high-grade intraepithelial neoplasia; 4NQO, 4-nitroquinoline-1-oxide.
Overall, these findings indicated that 4NQO was a carcinogen able to induce SCC formation in mice, and the precancerous conditions exacerbated with increased time of induction. Therefore, the expression and significance of B7-H4 in the mouse model of SCC induced by 4NQO were evaluated.
B7-H4 expression is upregulated during SCC formation, according to the results of immunohistochemistry analysis
B7-H4 was expressed in the cytoplasm and membrane of the epithelial cells in the mucosal layer (Fig. 2). According to the final scores (low or high expression), there were only 2 of 22 normal samples (9.1%) with B7-H4 high expression. As to the precancerous conditions, the B7-H4 high-expression rate was 40.7% (11/27) in LGIN specimens and 81.0% (17/21) in HGIN cases (Table II). Spearman correlation analysis demonstrated that the expression of B7-H4 was positively correlated with the pathological stage (P<0.0001), but not with the gender of the mice (P=0.2959).
Figure 2.
B7-H4 expression, and CD4+T, CD8+T and CD11b+ macrophage cell infiltration in esophagi of mice, as estimated by immunohistochemistry staining. B7-H4 was expressed in the membrane and cytoplasm of epithelial cells in the mucosa layer. Infiltration of CD4+T, CD8+T and CD11b+ macrophage cells was predominantly located in the submucosa of esophageal tissues. Magnification, ×400. The arrows indicate the cells that are positive. B7-H4, B7-homolog 4; CD, cluster of differentiation.
Table II.
Association between B7-H4 expression and pathological stage or lymphocytes infiltration in the epithelia of esophageal tissues during the carcinogenesis process.
| B7-H4 expressiona | ||||
|---|---|---|---|---|
| Relative parameters | Number of mice | Low | High | P-value |
| Pathological stage | <0.0001 | |||
| Normal | 22 | 20 | 2 | |
| LGIN | 27 | 16 | 11 | |
| HGIN | 21 | 4 | 17 | |
| Gender | 0.2959 | |||
| Female | 36 | 20 | 16 | |
| Male | 34 | 10 | 14 | |
| Lymphocytes infiltration | 0.8507 | |||
| CD4+T cells | ||||
| Low | 59 | 34 | 25 | |
| High | 11 | 6 | 5 | |
| CD8+T cells | 0.3166 | |||
| Low | 51 | 31 | 20 | |
| High | 19 | 9 | 10 | |
| CD11b+macrophages | 0.0002 | |||
| Low | 49 | 35 | 14 | |
| High | 21 | 5 | 16 | |
B7-H4 expression was positively correlated with pathological stage (P<0.0001) and CD11b macrophages infiltration (P=0.0002), according to Spearman correlation analysis. LGIN, low-grade intraepithelial neoplasia; HGIN, high-grade intraepithelial neoplasia; B7-H4, B7-homologue 4; CD, cluster of differentiation.
CD4, CD8 and CD11b are membrane molecules mainly expressed in CD4+T, CD8+T and macrophage cells (32). As represented in Fig. 2, the infiltrations of these three types of lymphocytes were predominantly located in the submucosa of esophageal epithelia instead of the mucosa layer. It was speculated that the reason for this observation maybe that the extent of the lesion was not severe enough to be infiltrated by lymphocytes into the mucosa layer. In addition, it was observed that, during the precancerous condition, the effect of lymphocytes on the lesion was indirect, which may be due to the secretion of cytokines. The association between B7-H4 expression in the mucosa and lymphocytes infiltration in the pertinent submucosa was also analyzed. As indicated in Table II, the results of Spearman correlation analysis revealed that B7-H4 expression was positively correlated with CD11b+ macrophages infiltration (P=0.0002), but not with CD4+T (P=0.8507) or CD8+T cells infiltration (P=0.3166).
Collectively, these data indicated that B7-H4 protein expression increased significantly and was positively correlated with pathological variables and macrophages infiltration, but was not associated with gender or with CD4+T or CD8+T cell infiltration during the carcinogenesis process induced by 4NQO. Thus, the present data suggested that B7-H4 probably is important in promoting the progression of esophageal precancerous lesions, which may be attributed to its association with certain type of cytokines secreted by macrophages.
B7-H4 gene expression is increased and is associated with IL-6 and STAT3 gene expression during SCC formation, according to the results of RT-qPCR analysis
To evaluate the association between B7-H4 expression and esophageal precancerous lesions or cytokines secreted by macrophage cells, B7-H4, IL-6, IL-10, TGF-β, IFN-γ and STAT3 gene expression was assessed by RT-qPCR. Firstly, there was no obvious change in the expression of these genes in the control mice from the 16th to the 28th week (Fig. 3). Therefore, the control value of one gene comes from the data of all esophagi of the control mice during the whole carcinogenesis. Secondly, it was shown that the expression level of B7-H4, IL-6, IL-10, TGF-β and STAT3 genes in the esophagi of 4NQO model mice increased significantly compared with that of control mice, while IFN-γ gene expression did not change obviously (Fig. 4). Additionally, there were significantly positive correlations between B7-H4 gene expression and IL-6 (r=0.5952, P<0.0001), IL-10 (r=0.6561, P<0.0001), TGF-β (r=0.6586, P<0.0001) and STAT3 (r=0.5799, P<0.0001) gene expression. Therefore, the present data supported the hypothesis that B7-H4 participated in the formation of esophageal precancerous lesions via interaction with the IL-6, IL-10, TGF-β or STAT3 signal transduction pathways.
Figure 3.
Gene expression of B7-H4 and associated molecules in control mice. The expression of genes in the esophagi of control mice did not change obviously from weeks 16 to 28 (B7-H4: P=0.0650; IL-6: P=0.2842; IL-10: P=0.2816; TGF-β, P=0.0743; IFN-γ: P=0.3399; STAT3: P=0.3225; Kruskal-Wallis analysis). mRNA, messenger RNA; Cq, quantification cycle; B7-H4, B7-homolog 4; CD, cluster of differentiation; IL, interleukin; TGF, transforming growth factor; IFN, interferon; STAT3, signal transducer and activator of transcription 3.
Figure 4.
Gene expression of B7-H4, IL-6, IL-10, TGF-β and STAT3 in the esophagi of 4NQO model mice increased during the carcinogenesis process (*P<0.05, **P<0.01, vs. the control group; Kruskal-Wallis analysis). mRNA, messenger RNA; Cq, quantification cycle; B7-H4, B7-homolog 4; CD, cluster of differentiation; IL, interleukin; TGF, transforming growth factor; IFN, interferon; STAT3, signal transducer and activator of transcription 3; 4NQO, 4-nitroquinoline-1-oxide.
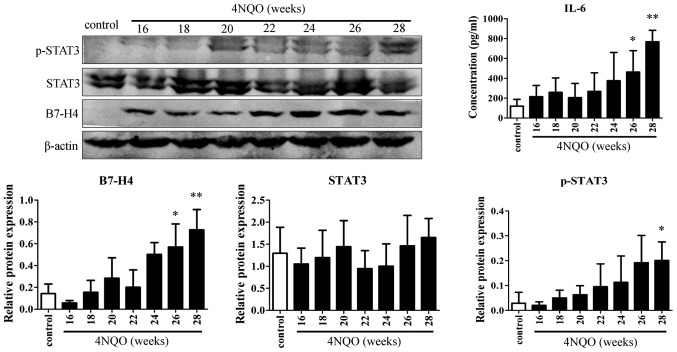
B7-H4 protein expression is increased, and is associated with IL-6 and p-STAT3 expression during SCC formation
The present study focused on the association between B7-H4 expression and the IL-6/STAT3 signaling pathway. The protein levels of B7-H4, STAT3, p-STAT3 and IL-6 were determined by western blotting and ELISA. No obvious changes in the expression levels of these proteins were observed in the control mice from weeks 16 to 28 (Fig. 5). Thus, the control value of one protein came from the data of all esophagi of control mice during the whole carcinogenesis in the following statistical analysis. As presented in Fig. 6, in comparison with control mice, B7-H4 and IL-6 protein expression was upregulated in the model group during the esophageal carcinogenesis process, and increased significantly in weeks 26 and 28 (B7-H4: P=0.0159 and 0.0025, respectively; IL-6: P=0.0265 and 0.0057, respectively). Additionally, p-STAT3 levels also increased significantly in the 28th week (P=0.0127), despite the fact that the change in STAT3 expression was not significant (P=0.1033). During the whole process of carcinogenesis, B7-H4 expression was positively correlated with IL-6 and p-STAT3 expression (r=0.6145, P<0.0001; and r=0.6180, P<0.0001, respectively). Overall, these results implicated that the high expression of B7-H4 and its function enhancing esophageal precancerous lesions is associated, at least in part, with IL-6/STAT3 activation.
Figure 5.
Expression of B7-H4 and associated proteins in control mice. Protein expression in the esophagi of control mice did not change obviously from weeks 16 to 28 (all P>0.05, Kruskal-Wallis analysis). B7-H4, B7-homolog 4; IL, interleukin; STAT3, signal transducer and activator of transcription 3; p-, phosphorylated.
Figure 6.
B7-H4, p-STAT3 and IL-6 protein expression in the esophagi of 4NQO model mice increased during the carcinogenesis process (*P<0.05, **P<0.01 vs. the control group; Kruskal-Wallis analysis). B7-H4, B7-homolog 4; IL, interleukin; STAT3, signal transducer and activator of transcription 3; p-, phosphorylated; 4NQO, 4-nitroquinoline-1-oxide.
Discussion
In the present study, a model of esophageal carcinogenesis was successfully established in mice by 4NQO drinking water. Although Tang et al (33) and Tseng et al (34) have reported that 4NQO could induce SCC in mice, the present study is the first to profile in detail the precancerous conditions in esophageal carcinogenesis. In future studies, the present model could be used to investigate the mechanism of SCC, to explore additional biomarkers of esophageal precancerous conditions and to evaluate the efficacy of genes targeted or chemotherapy drugs, which may provide a significant basis for early prevention and treatment of SCC.
In the present study, the results of immunohistochemistry demonstrated that B7-H4 expression was significantly correlated with pathological stage. In addition, in contrast to CD4+T and CD8+T cells, B7-H4 expression was markedly associated with macrophage cell infiltration. This finding was not strictly in accordance with a recent report from Mugler et al, who noticed that B7-H4 expression was inversely correlated with CD8+T cell infiltration (19). This discrepancy may be due to the extent of the pathological lesions. In the precancerous stage, the majority of lymphocyte cells are predominantly located in the submucosa instead of the mucosa layer, which impedes the interaction between B7-H4 and lymphocytes. Additionally, other types of T cells, including regulatory and memory T cells, were not evaluated in the present study. Overall, the present results suggested that the function of B7-H4 in inhibiting T cell proliferation or inducing T cell apoptosis to mediate tumor escape was limited during the formation of esophageal precancerous lesions.
The results of RT-qPCR demonstrated that B7-H4 gene expression increased markedly with the increase in time during carcinogenesis. Additionally, B7-H4 gene expression was positively correlated with IL-6, IL-10, TGF-β and STAT3 gene expression, but not with IFN-γ gene expression. It is well known that IL-6 (35), IL-10 (36) and STAT3 (37) are important in exerting antitumor inhibitory effects, while IFN-γ is a tumor-killing factor (38). Therefore, the data of the present study suggested that B7-H4 gene expression was positively correlated with antitumor inhibitory cytokines and factors, including IL-6, IL-10 and STAT3, in the tumor microenvironment or in cancer cells. Additionally, the results of ELISA and western blot analysis for esophageal tissues revealed that B7-H4 protein expression was positively correlated with IL-6 and p-STAT3 expression. Collectively, the present findings indicated that B7-H4 expression increased during SCC formation in association with IL-6/STAT3 activation.
Despite the fact that our study indicated that B7-H4 maybe a potential biomarker of esophageal precancerous lesions in association with IL-6/STAT3 signaling pathway activation, additional studies are required to investigate in depth the mechanism of B7-H4 in the process of esophageal carcinogenesis and to provide further evidence for the diagnosis and treatment of esophageal precancerous conditions. Thus, further studies are required: i) To detect the expression of B7-H4 in esophageal squamous cancer cells and to evaluate the effect of B7-H4 expression on cell proliferation and its association with the activation of the IL-6/STAT3 signaling pathway via B7-H4 RNA interference treatment; ii) to establish an esophageal carcinogenesis model induced by 4NQO in B7-H4-positive and B7-H4-negative mice in order to further analyze the effect of B7-H4 on esophageal carcinogenesis; and iii) to analyze the expression of B7-H4 in esophageal precancerous tissues and blood of patients in order to elucidate the correlation between B7-H4 expression and pathological stage.
In conclusion, the present study is the first to analyze the expression of B7-H4 in esophageal precancerous conditions in mice, and to provide a novel association between B7-H4 and the IL-6/STAT3 signaling pathway, which is classically recognized as an important stimulator in carcinogenesis and cancer progression. However, further studies are required in order to clarify the association between B7-H4 expression and esophageal carcinogenesis.
Glossary
Abbreviations
- B7-H4
B7-homolog 4
- 4NQO
4-nitroquinoline-1-oxide
- ESCC
esophageal squamous cell carcinoma
- STAT3
signal transducer and activator of transcription 3
- p-STAT3
phosphorylated signal transducer and activator of transcription 3
- LGIN
low-grade intraepithelial neoplasia
- HGIN
high-grade intraepithelial neoplasia
- IL-6
interleukin-6
- IL-10
interleukin-10
- TGF-β
transforming growth factor-β
- IFN-γ
interferon-γ
References
- 1.Herszényi L, Tulassay Z. Epidemiology of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci. 2010;14:249–258. [PubMed] [Google Scholar]
- 2.Henry MA, Lerco MM, Ribeiro PW, Rodrigues MA. Epidemiological features of esophageal cancer: Squamous cell carcinoma versus adenocarcinoma. Acta Cir Bras. 2014;29:389–393. doi: 10.1590/S0102-86502014000600007. [DOI] [PubMed] [Google Scholar]
- 3.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncolol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Siewert JR, Ott K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol. 2007;17:38–44. doi: 10.1016/j.semradonc.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Kanojia D, Vaidya MM. 4-Nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol. 2006;42:655–667. doi: 10.1016/j.oraloncology.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Imaida K, Sato H, Okamiya H, Takahashi M, Hayashi Y. Enhancing effect of high fat diet on 4-nitroquinoline 1-oxide-induced pulmonary tumorigenesis in ICR male mice. Jpn J Cancer Res. 1989;80:499–502. doi: 10.1111/j.1349-7006.1989.tb01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallenius K, Lekholm U. Oral cancer in rats induced by the water-soluble carcinogen 4-Nitrochinoline N-oxide. Odontol Revy. 1973;24:39–48. [PubMed] [Google Scholar]
- 8.Vered M, Yarom N, Dayan D. 4NQO oral carcinogenesis: Animal models, molecular markers and future expectations. Oral Oncol. 2005;41:337–339. doi: 10.1016/j.oraloncology.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. 2004;10:301–313. doi: 10.1158/1078-0432.CCR-0999-3. [DOI] [PubMed] [Google Scholar]
- 10.Ceeraz S, Nowak EC, Noelle RJ. B7 family checkpoint regulators in immune regulation and disease. Trends Immunol. 2013;34:556–563. doi: 10.1016/j.it.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sica GL, Choi IH, Zhu G, Tamada K, Wang SD, Tamura H, Chapoval AI, Flies DB, Bajorath J, Chen L. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 2003;8:849–861. doi: 10.1016/S1074-7613(03)00152-3. [DOI] [PubMed] [Google Scholar]
- 12.Prasad DV, Richards S, Mai XM, Dong C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity. 2003;18:863–873. doi: 10.1016/S1074-7613(03)00147-X. [DOI] [PubMed] [Google Scholar]
- 13.Zang X, Loke P, Kim J, Murphy K, Waitz R, Allison JP. B7x: A widely expressed B7 family member that inhibits T cell activation. Proc Natl Acad Sci USA. 2003;100:10388–10392. doi: 10.1073/pnas.1434299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Wang Y, Zhao J, Gu M, Giscombe R, Lefvert AK, Wang X. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer. 2006;53:143–151. doi: 10.1016/j.lungcan.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Fauci JM, Straughn JM, Jr, Ferrone S, Buchsbaum DJ. A review of B7-H3 and B7-H4 immune molecules and their role in ovarian cancer. Gynecol Oncol. 2012;127:420–425. doi: 10.1016/j.ygyno.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Zang X, Thompson RH, Al-Ahmadie HA, Serio AM, Reuter VE, Eastham JA, Scardino PT, Sharma P, Allison JP. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc Natl Acad Sci USA. 2007;104:19458–19463. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quandt D, Fiedler E, Boettcher D, Marsch WCh, Seliger B. B7-h4 expression in human melanoma: Its association with patients' survival and antitumor immune response. Clin Cancer Res. 2011;17:3100–3111. doi: 10.1158/1078-0432.CCR-10-2268. [DOI] [PubMed] [Google Scholar]
- 18.Arigami T, Uenosono Y, Ishigami S, Hagihara T, Haraguchi N, Natsugoe S. Clinical significance of the B7-H4 coregulatory molecule as a novel prognostic marker in gastric cancer. World J Surg. 2011;35:2051–2057. doi: 10.1007/s00268-011-1186-4. [DOI] [PubMed] [Google Scholar]
- 19.Mugler KC, Singh M, Tringler B, Torkko KC, Liu W, Papkoff J, Shroyer KR. B7-h4 expression in a range of breast pathology: Correlation with tumor T-cell infiltration. Appl Immunohistochem Mol Morphol. 2007;15:363–370. doi: 10.1097/01.pai.0000213159.79557.71. [DOI] [PubMed] [Google Scholar]
- 20.Jung SG, Choi KU, Lee SD, Lee ZZ, Chung MK. The relationship between B7-H4 expression and clinic pathological characteristics in clinical stage T1 conventional renal cell carcinoma. Korean J Urol. 2011;52:90–95. doi: 10.4111/kju.2011.52.2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kryczek I, Wei S, Zou L, Zhu G, Mottram P, Xu H, Chen L, Zou W. Cutting edge: Induction of B7-H4 on APCs through IL-10: Novel suppressive mode for regulatory T cells. J Immunol. 2006;177:40–44. doi: 10.4049/jimmunol.177.1.40. [DOI] [PubMed] [Google Scholar]
- 22.Fauci JM, Straughn JM, Jr, Ferrone S, Buchsbaum DJ. A review of B7-H3 and B7-H4 immune molecules and their role in ovarian cancer. Gynecol Oncol1. 2012;27:420–425. doi: 10.1016/j.ygyno.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Chen LJ, Sun J, Wu HY, Zhou SM, Tan Y, Tan M, Shan BE, Lu BF, Zhang XG. B7-H4 expression associates with cancer progression and predicts patient's survival in human esophageal squamous cell carcinoma. Cancer Immunol Immunother. 2011;60:1047–1055. doi: 10.1007/s00262-011-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Wang L, Wang W, Zhao L, Shan B. B7-H4 facilitates proliferation of esophageal squamous cell carcinoma cells through promoting interleukin-6/signal transducer and activator of transcription 3 pathway activation. Cancer Sci. 2016;107:944–954. doi: 10.1111/cas.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua D, Sun J, Mao Y, Chen LJ, Wu YY, Zhang XG. B7-H1 expression is associated with expansion of regulatory T cells in colorectal carcinoma. World J Gastroenterol. 2012;18:971–978. doi: 10.3748/wjg.v18.i9.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanderstraeten A, Luyten C, Verbist G, Tuyaerts S, Amant F. Mapping the immunosuppressive environment in uterine tumors: Implications for immunotherapy. Cancer Immunol Immunother. 2014;63:545–557. doi: 10.1007/s00262-014-1537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Zhang Q, Chen W, Shan B, Ding Y, Zhang G, Cao N, Liu L, Zhang Y. B7-H3 is overexpressed in patients suffering osteosarcoma and associated with tumor aggressiveness and metastasis. PLoS One. 2013;8:e70689. doi: 10.1371/journal.pone.0070689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Gao H, Yao Q, Lan X, Li S, Wu J, Zeng G, Xue Y. Elevated HABP1 protein expression correlates with progression and poor survival in patients with gastric cancer. Onco Target Ther. 2016;9:6711–6718. doi: 10.2147/OTT.S114756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori K, Le Goff B, Charrier C, Battaglia S, Heymann D, Rédini F. DU145 human prostate cancer cells express functional receptor activator of NFkappaB: New insights in the prostate cancer bone metastasis process. Bone. 2007;40:981–990. doi: 10.1016/j.bone.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu M, Zaninotto G, Nagata K, Graham DY, Lauwers GY. Esophageal squamous cell carcinoma with special reference to its early stage. Best Pract Res Clin Gastroenterol. 2013;27:171–186. doi: 10.1016/j.bpg.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Na YR, Jung D, Gu GJ, Seok SH. GM-CSF grown bone marrow derived cells are composed of phenotypically different dendritic cells and macrophages. Mol cells. 2016;39:734–741. doi: 10.14348/molcells.2016.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. 2004;10:301–313. doi: 10.1158/1078-0432.CCR-0999-3. [DOI] [PubMed] [Google Scholar]
- 34.Tseng SH, Yang CC, Yu EH, Chang C, Lee YS, Liu CJ, Chang KW, Lin SC. K14-EGFP-miR-31 transgenic mice have high susceptibility to chemical-induced squamous cell tumorigenesis that is associating with Ku80 repression. Int J Cancer. 2015;136:1263–1275. doi: 10.1002/ijc.29106. [DOI] [PubMed] [Google Scholar]
- 35.Kao JT, Feng CL, Yu CJ, Tsai SM, Hsu PN, Chen YL, Wu YY. IL-6, through p-STAT3 rather than p-STAT1, activates hepatocarcinogenesis and affects survival of hepatocellular carcinoma patients: A cohort study. BMC Gastroenterol. 2015;15:50. doi: 10.1186/s12876-015-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y, Song Q, Hu D, Zhuang X, Yu S. Tumor-infiltrating lymphocyte activity is enhanced in tumors with low IL-10 production in HBV-induced hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;461:109–114. doi: 10.1016/j.bbrc.2015.03.177. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Zhang J, Xia H, Zhang B, Jiang T, Wang J, Chen X, Wang Y. Stat3 is involved in the motality, metastasis and prognosis in lingual squamous cell carcinoma. Cell Biochem Funct. 2012;30:340–346. doi: 10.1002/cbf.2810. [DOI] [PubMed] [Google Scholar]
- 38.Hayakawa Y, Sato-Matsushita M, Takeda K, Iwakura Y, Tahara H, Irimura T. Early activation and interferon-γ production of tumor-infiltrating mature CD27 high natural killer cells. Cancer Sci. 2011;102:1967–1971. doi: 10.1111/j.1349-7006.2011.02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]