Abstract
Biomarkers that facilitate the prediction of breast cancer prognosis can improve the quality of life in patients during the long period of illness and treatment. Particularly in recent years, with the advent of a more exhaustive analysis of genetic information and gene products, the molecular mechanisms at play during breast cancer have gradually become clearer. In the present study, a systematic review of the literature between 2009 and 2014 was conducted by searching for the keywords ‘breast cancer’, ‘biomarkers’, ‘diagnosis’, ‘prognosis’ and ‘drug response’ to clarify the present state of knowledge regarding biomarkers. In the final analysis, 16 studies on biomarkers for the breast cancer prognosis were retrieved. From these, 7 biomarkers in 9 studies were found to be strongly reliable predictors of prognosis and a further 7 biomarkers in 7 studies were poorly reliable. The use of these prognostic biomarkers should increase the options available for treatment algorithms.
Keywords: breast cancer, biomarkers, prognosis, diagnosis, drug response
1. Introduction
Currently, breast cancer treatment is progressing on a daily basis, and is divided into subtypes based on the hormone receptors human epidermal growth factor receptor type 2 (HER2) and Ki-67 (1). However, treatment algorithms do not necessarily result in satisfactory clinical outcomes. Therefore, to further improve the breast cancer prognosis, predictive factors are required in order to arrive at more accurate prognoses and improve treatment efficacy. If such biomarkers could be identified, it would be possible to provide appropriate treatments to relevant subjects, resulting in excellent clinical outcomes. To date, prognostic prediction has been based on older studies of morphological characteristics (2). More recent research has concentrated on molecular biomarkers (3). The present study reports the findings of a systematic review of prognosis for patients with breast cancer based on molecular biomarkers. The correlations between these biomarkers, prognosis and the treatment response may be useful for all breast cancer patients.
2. Literature search
A search of the PubMed database (National Center for Biotechnology Information, Bethesda, MD, UDA) using the key words ‘breast cancer,’ ‘biomarkers,’ ‘diagnosis,’ ‘prognosis’ and ‘drug response’ retrieved 1,689 potential studies. Subsequent to filtering for studies involving humans and written in English, 76 studies were excluded. When the remaining reports were limited to the period between 2009 and 2014, an additional 688 were excluded. Of the remainder, 520 studies were excluded, as they did not contain the full text. Finally, the abstracts of 405 studies were evaluated and those that contained insufficient descriptions of diagnostic performance, prognosis and drug response were excluded, resulting in a total of 16 studies for analysis (Fig. 1).
Figure 1.
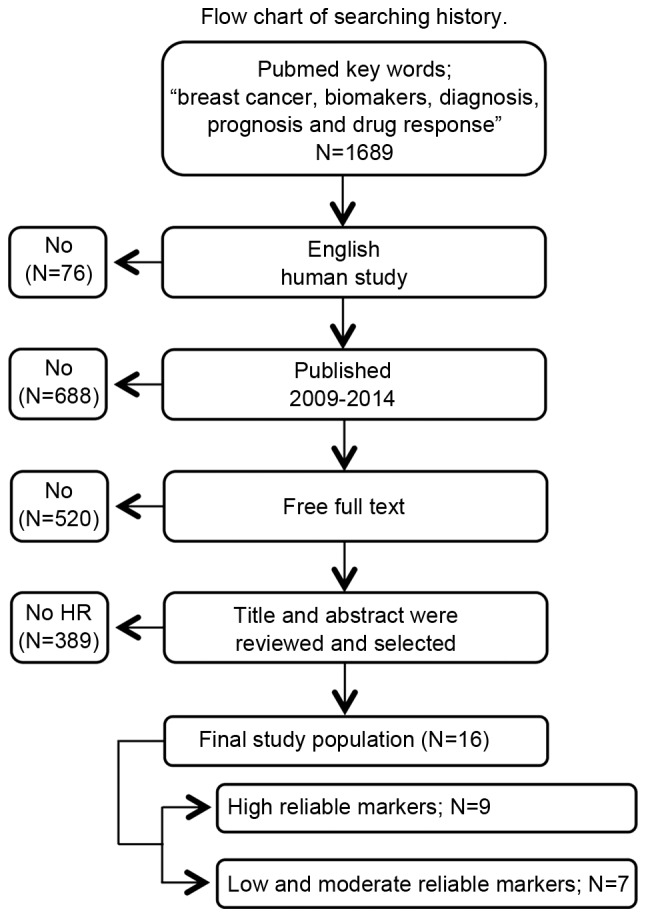
Flow chart of search history. PubMed searches were conducted. A total of 16 studies were selected according to the inclusion criteria of the present study.
3. Highly reliable biomarkers (Table I)
Table I.
List of biomarkers that presented ≥2 risk ratios compared with the control populations (high reliable markers).
| Result | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Authors | Biomarker | Endpoint | Condition | HR | 95% CI | No. | Sample | Periods | (Refs.) |
| Johansson et al | RARA | RFS | Low RARA | 1.0 | – | 72 | Blood | 1991–1996 | (7) |
| High RARA | 4.1 | 1.55–11.0 (P=0.046) | |||||||
| Ellis et al | Aromatase | BCSS | Absent | 1.0 | – | 169 | Biopsy | 1998–1999 | (15) |
| Present | 3.76 | 1.4–10.0 (P=0.008) | |||||||
| Pang et al | Osteopontin | DFS | Low | 1.0 | – | 200 | Biopsy | 2002 | (18) |
| High | 3.094 | 1.229.7.789 (P=0.016) | |||||||
| OS | Low | 1.0 | – | ||||||
| High | 2.558 | 1.048–6.243 (P=0.039) | |||||||
| Ohno et al | Ki-67 | pCR | <10 | 1.0 | – | 504 | Biopsy | 2002–2004 | (21) |
| ≥10 | 2.718 | 1.331–5.549 (P=0.0061) | |||||||
| Denkert et al | Ki-67 | pCR | 0–15% | 1.0 | – | 1,166 | Biopsy | 2002–2005 | (22) |
| 15.1–35% | 3.3 2 | – | |||||||
| >35% | 9.2 | − (P=0.00005) | |||||||
| DeCensi et al | Ki-67 | OS | Pre-tamoxifen <20 | 1.0 | – | 86 | Biopsy | 2004–2007 | (24) |
| ≥20 | 3.66 | 0.81–16.47 (P=0.128) | |||||||
| Post-tamoxifen <20 | 1.0 | – | |||||||
| ≥20 | 5.50 | 1.26–23.16 (P=0.006) | |||||||
| Tsang et al | CEACAM6 | OS | CEACAM6 (−) | 1.0 | – | 312 | Biopsy | 2003–2009 | (28) |
| CEACAM6 (+) | 4.107 | 1.478–11.417 (P=0.007) | |||||||
| Cizkova et al | PIK3CA | DFS | Wild type | 1.0 | – | 80 | Biopsy | 2007 | (31) |
| Mutant type | 4.26 | 1.4–13 (P=0.0063) | |||||||
| Zhu et al | TIMP- | DFS | Low | 1.0 | – | 99 | Biopsy | 2008 | (36) |
| High | 3.085 | 1.096–8.680 (P=0.033) | |||||||
| OS | Low | 1.0 | – | ||||||
| High | 12.907 | 1.499–111.2 (P =0.020) | |||||||
RARA, Retinoic acid receptor α; RFS, recurrence-free survival; BCSS, breast cancer-specific survival; pCR, pathological complete response; CEACAM6, Carcinoembryonic antigen-related cell adhesion molecule 6; PIK3CA, Phosphatidylinositol-4,5-bisphosphate 3-kinase; catalytic subunit α; TIMP-1, Tissue inhibitor of metalloproteinases-1; DFS, disease free survival; OS, overall survival.
Retinoic acid receptor α (RARA)
Approximately 1/3 of estrogen receptor α (ERα)-positive breast cancer patients treated with tamoxifen experienced a relapse of the disease (4). RARA is a potential biomarker for tamoxifen resistance (5). The anti-tumor properties of RARA can be explained in association with the interaction of the receptor with ERα and their joint genomic binding site (6). The association between ERα resistance and RARA resistance was confirmed using tamoxifen-susceptible and -resistant cell lines (MCF7 and LCC2, respectively). The tamoxifen-resistant cells were found to express high levels of RARA (7).
Patients with ERα-positive breast cancer tumors with high internal levels of RARA protein that were treated with tamoxifen as adjuvant therapy exhibited shorter recurrence-free survival (RFS) than patients with low internal levels of RARA protein (7). Johansson et al (7) performed an investigation into serum RARA levels using ELISA, and found significantly higher RFS rates in patients with high RARA expression levels compared with patients with low levels: Hazard ratio (HR)=4.1; 95% confidence interval (CI)=1.55–11.0; P=0.0046. Therefore, RARA may potentially be a useful target of new treatment regimens and a biomarker to predict the effectiveness of tamoxifen adjuvant treatment in ERα-positive breast cancer.
Aromatase expression
Aromatase expression by breast cancer cells has been shown to influence the effectiveness of endocrine treatments for breast cancer (8,9). Ellis et al (10) conducted a study using a sample from a clinical trial comparing tamoxifen and letrozole as neoadjuvant endocrine therapies, and found that aromatase expression levels in tumor and somatic cells was correlated with treatment-induced changes in Ki-67, RFS, and breast cancer-specific survival (BCSS) (11–13).
Aromatase expression was correlated with a smaller tumor size (P=0.01), a higher Allred score of estrogen receptor (P=0.006) (14) and lower Ki-67 levels (P=0.003). In addition, aromatase expression by tumor cells was a significant prognostic factor of the independent variables RFS (HR=2.3; 95% CI=1.2–4.6; P=0.01) and BCSS (HR=3.76; 95% CI=1.4–10.0; P=0.008) (15). The aforementioned data supports the use of aromatase blockers as the first choice treatment for post-menopausal, hormone-positive breast cancer.
Osteopontin
Osteopontin is a secreted extracellular matrix adhesion protein associated with tumor cell invasion and metastasis (16,17). Pang et al (18) examined the clinical and pathological effects of the adhesion molecules osteopontin-c, E-cadherin and β-catenin in breast cancer, and found higher expression levels of all the aforementioned adhesion molecules in breast cancer compared with normal tissue. The expression of osteopontin-c was associated with lymph node metastasis, and higher tumor node-metastasis classification (19) and histological grade (19). In addition, high expression levels of osteopontin-c have been correlated with tumor recurrence and metastasis, as well as triple negative subtypes, which are predictive factors of the independent variables disease-free survival (DFS; HR=3.094; 95% CI, 1.229–7.789; P=0.016) and overall survival (OS; HR=2.558; 95% CI, 1.048–6.243; P=0.039) (20). Therefore, the development of treatments targeting osteopontin-c may be beneficial for the treatment of breast cancer.
Ki-67
Ohno et al (21) examined the role of Ki-67 as a predictive biomarker of treatment response in a randomized, multicenter study to compare the effectiveness of docetaxel subsequent to treatment with fluorouracil/epirubicin/cyclophosphamide with or without capecitabine, in patients with operable breast cancer. The endpoint was the rate of pathological complete response (pCR). Analysis of hormone receptors and the Ki-67 labeling index (Ki-67LI) by multivariate logistic regression analysis identified Ki-67 as an independent prognostic factor (HR=2.718; 95% CI=1.331–5.549; P=0.0061). In addition, the aforementioned results also suggest that the Ki-67LI prior to treatment was a predictor for the response to preoperative docetaxel treatment and preoperative capecitabine treatment in early-stage breast cancer.
Denkert et al (22) obtained 1,166 breast cancer bioassay specimens from a large-scale cohort study established to investigate neoadjuvant treatment (the GeparTrio trial) and evaluated pre-treatment Ki-67 levels by immunohistochemical analysis. The study used the standardized, 3-endpoint, cut-off algorithm (pCR, DFS and OS) (23). The Ki-67 index and preoperative chemotherapy variables were divided into 3 subgroups each: ≤15, 15.1–35 and ≥35%, and pCR rates were 4.2, 12.8 and 29.0%, respectively (P<0.0005). The HR for prognosis also increased in response to Ki-67 (HR=1, 3.32 and 9.20, respectively), indicating that Ki-67 is a prognostic predictor for hormone receptor-positive, but not triple-negative, breast cancer. The aforementioned findings regarding Ki-67 may provide important information for the development of other quantitative biomarkers.
DeCensi et al (24) examined postoperative remission and prognosis in response to Ki-67 in early stage ERα-positive breast cancer patients treated with tamoxifen for 4 weeks as a short-term neo-adjuvant therapy and reported that post-treatment levels of Ki-67 in the second (14–19%), third (20–29%), and top (≥30%) quartiles had recurrence HRs of 2.92 (95% CI, 0.95–8.96), 4.37 (1.56–12.25) and 6.05 (2.07–17.65), respectively, compared with those in the bottom quartile (<14%; P=0.001). The mortality risks were 5.5-fold higher when Ki-67 levels were ≥20% (95% CI=1.26–23.16; P<0.006) when compared to those with Ki-67 levels <20% (P=0.006). The authors concluded that the level of Ki-67 subsequent to short-term neoadjuvant tamoxifen is a good predictor of RFS and OS, supporting the use of Ki-67 as a surrogate biomarker to personalize adjuvant treatment and to cost-effectively screen novel drugs.
Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6)
CEACAM6 is a human carcinoembryonic antigen that functions as a multi-functional regulatory protein and is overexpressed in various cell processes associated with cancer (25,26). CEACAM6 expression in atypical ductal hyperplasia has been suggested to serve an important role in the development of breast cancer (27). CEACAM6 has also been associated with invasive and treatment-resistant breast cancer (28). However, in a large-scale cohort study, CEACAM6 expression in luminal breast cancer exhibited no effect on OS or correlation with prognosis, although an association between CEACAM6 expression and prognosis in breast cancer overexpressing HER2 was revealed, as the high expression group tended to exhibit poorer OS (28). The aforementioned results appear to indicate that treatment is required for breast cancer patients with HER2 overexpression and the presence of CEACAM6.
Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α (PIK3CA)
PIK3CA is a cancer gene coding for 1 of 2 phosphoinositide 3-kinase (PI3 K) subunits (29), which is a gain-of-function mutation in certain types of cancer, and is present in 20–40% patients with breast cancer (30). Cizkova et al (31) identified PIK3CA mutations in 17 (21.3%) tumors among 80 HER2-positive patients treated with trastuzumab for 1 year. Patients exhibiting wild-type PIK3CA demonstrated an improved DFS compared with patients exhibiting the PIK3CA mutations. The prognosis for HER2-positive patients with PIK3CA mutations treated with trastuzumab was significantly worse than for patients exhibiting the wild-type variation, which is considered to occur since the P13K/protein kinase B pathway is adversely affected by PIK3CA mutations, resulting in the lower efficacy of trastuzumab. Thus, the detection of PIK3CA mutations is only required in HER2-positive patients.
Tissue inhibitor of metalloproteinases-1 (TIMP-1)
Paclitaxel is the first chemotherapy treatment of choice for patients with lymph node metastasis (32,33). However, there are currently no biomarkers to predict susceptibility to chemotherapy. TIMP-1 has been shown to protect cells from apoptosis (34). A previous epidemiological study demonstrated an association between high levels of TIMP-1 and reduced responsiveness to cyclophosphamide/methotrexate/5-fluorouracil and anthracycline-based chemotherapy regimens (35).
In a retrospective study of 99 breast cancer patients, Zhu et al (36) reported a correlation between TIMP-1 expression levels in primary tumors and improved responsiveness to paclitaxel-based chemotherapy. Kaplan-Meier survival analysis revealed that patients with high TIMP-1 levels had poorer 5-year DFS that those with lower TIMP-1 levels (71.1 vs. 88.5%, respectively; P=0.020). The 5-year OS was also lower (78.9 and 96.7%, respectively, P=0.004). The responsiveness to paclitaxel-based chemotherapy was significantly worse when the TIMP-1 expression levels were high. The aforementioned findings indicate that TIMP-1 may be a useful predictive biomarker for chemotherapy resistance.
4. Low and moderately reliable biomarkers (Table II)
Table II.
List of biomarkers that presented <2 risk ratios compared to the control populations (low and moderate reliable markers).
| Result | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Authors | Biomarker | Endpoint | Condition | HR | 95% CI | No. | Sample | Periods | (Refs.) |
| Jézéquel et al | FTL | MFS | <2.4 | 1.0 | – | 198 | Biopsy | 1980–2000 | (40) |
| >2.4 | 1.3 | 1.10–1.50 (P=0.001) | |||||||
| Harbeck et al | uPA/PAI-1 | DFS | Low | 1.0 | – | 409 | Blood | 1993–1998 | (43) |
| High | 1.84 | 1.1–3.0 (P=0.017) | |||||||
| OS | Low | 1.0 | – | ||||||
| High | 1.85 | 1.1–3.1 (P=0.02) | |||||||
| Villaseñor et al | CRP | OS | <1 mg/l | 1.0 | – | 2,919 | Blood | 1995–2000 | (46) |
| ≥10 mg/l | 1.96 | 1.22–3.13 (P<0.001) | |||||||
| Earl et al | Ch17CEP | OS | Normal | 1.0 | – | 1,762 | Biopsy | 1996–2001 | (50) |
| Duplication | 0.80 | 0.68–0.95 (P=0.009) | |||||||
| Moreno-Aspitia et al | sHER2 | DFS | <15 | 1.0 | – | 2,318 | Blood | 2004–2005 | (55) |
| ≥15 | 2.36 | 1.19–4.70 (P=0.01) | |||||||
| Sun et al | MAD1L1 | OS | Low | 1.0 | – | 461 | Biopsy | 2006 | (57) |
| High | 1.825 | −(P=0.027) | |||||||
| Hartmann et al | PITX2P2 Methylation | TDM | TDM | 1.66 | 1.21–2.28 (P=0.002) | 241 | Biopsy | NA | (66) |
MFS, metastasis-free survival; uPA/PAI-1, urokinase-type plasminogen activator and plasminogen activator type 1 inhibitor; CRP, C-reactive protein; Ch17CEP, Chromosome 17 centromere enumeration probe; sHER2, soluble human epidermal growth factor receptor 2; MAD1L1, mitotic arrest deficient like 1; PITX2P2, methylation of paired-like homeodomain 2; TDM, time to distant metastasis; DFS, disease free survival; OS, overall survival.
Ferritin light chain (FTL)
Ferritin is a ubiquitous iron-binding protein. In vertebrates, there are 2 types of apoferritin, which are assembled from 24 subunits including light and heavy chain types. The ratio between the ferritin heavy chain and FTL can vary greatly, depending on the tissue type and cellular conditions (37). The increase in ferritin from different cancer tissue samples exhibited a close correlation with disease onset (38). Ricolleau et al (39) investigated the utility of FTL as a prognostic marker for lymph node metastasis-positive breast cancer and determined an FTL cut-off level in tumors of 2.4. The high FTL level group had a significantly lower metastasis-free survival rate, indicating that FTL was an independent prognostic marker (HR=1.30; 95% CI=1.10–1.50; P=0.001) (40).
Urokinase-type plasminogen activator (uPA) and plasminogen activator type 1 inhibitor (PAI-1)
uPA, as a tumor-associated proteolytic factor, and PAI-1 serve important roles in tumor invasion and metastasis (41), and cell signaling, adhesion, migration and proliferation (42). In the final Chemo-N0 trial for the validation of The American Society of Clinical Oncology-recommended biomarkers (1993–1998; n=647; 12 centers; median follow-up period 113 months; range between 5 and 167 months), high uPA/PAI-1 levels were correlated with significantly lower DFS among breast cancer patients who did not receive adjuvant treatment (HR=1.84; 95% CI=1.1–3.0; P=0.017) and OS (HR=1.84; 95% CI=1.1–3.1; P=0.02). uPA/PAI-1 was also identified as a prognostic factor for breast cancer in other studies (43).
C-reactive protein (CRP)
Serum CRP is a marker of acute inflammatory response and is considered to be a prognostic indicator in breast cancer (44,45). The Women's Healthy Eating and Living study was a randomized comparative study examining the effect of a diet high in vegetables and low in fat on the prevention of premature mortality in women diagnosed with breast cancer. Serum protein analysis of 2,023 of 3,088 eligible women showed that acute inflammation (CRP ≥10 mg/l) was markedly suppressed by a high vegetable/low fat diet, resulting in improved long-term survival (46). Although the anti-oncological effects remain unclear, CRP was identified as an independent biomarker for prognosis of survival in breast cancer (HR=1.96; 95% CI=1.22–3.13).
Chromosome 17 centromere enumeration probe (Ch17CEP)
Chromosome 17 is the second densest chromosome in the human genome and codes for several genes, including BRCA1 and HER2 with important roles in breast cancer, as well as the housekeeping DNA repair genes TP53, RAD51C and RAD52B (47). Ch17 centromeric region duplication (Ch17CEP) is closely associated with HER2 amplification (48). Ch17CEP overlap is also a powerful marker for genome instability in breast cancer and is correlated with susceptibility to chemotherapy (48,49). In novel endovascular access trial/BR9601 clinical trials, prognostic factors were analyzed and categorized according to breast cancer subtype. Although numerous factors were not associated with subtype, Ch17CEP overlap was an independent prognostic factor for DFS and OS for all subtypes. Ch17CEP overlap was identified as a prognostic biomarker for breast cancer treated with cyclophosphamide, methotrexate and fluorouracil therapy in combination with epirubicin (HR=0.80; 95% CI=0.68–0.95; P=0.009) (50).
Soluble human epidermal growth factor receptor 2 (sHER2)
HER2 is a 185-kDa protein arising from the intracellular, transmembrane and extracellular domains (ECD) (51). The ECD is occasionally spliced by metalloprotease, resulting in the release of sHER2 into the peripheral circulation (52). sHER2 is an important biomarker for HER2-positive breast cancer at any stage (53). In the N9831 adjuvant breast cancer trials, early stage HER2-positivity was identified as a biomarker of clinical outcome and disease progression (54). Moreno-Aspitia et al (55) evaluated sHER2 levels at the time of relapse using a sample from the N9831 clinical trial and found that DFS was lower in patients with sHER2 levels >15 ng/ml, compared with patients with lower levels of sHER2 (HR=2.36; 95% CI=1.19–4.70; P=0.01). Therefore, in early stage HER2-positive breast cancer, sHER2 was found to be a suitable prognostic biomarker for relapse, and survival in relapsed patients.
Mitotic arrest deficient like 1 (MAD1L1)
MAD1L1 is a checkpoint gene associated with chromosomal instability. Abnormalities in MAD1L1 have been observed in a number of cancer types, including colon and lung cancer (56). Sun et al (57) analyzed MADL1 expression in breast cancer tissues from 461 patients and normal breast tissue to identify correlations between MAD1L1 expression and clinical pathological characteristics. The results of the aforementioned study revealed that OS was particularly worse in the high MAD1L1 expression group (HR=1.825; 95% CI=1.073–3.107; P=0.027). In patients with high nuclear MAD1L1 expression subsequent to taxol treatment, prognosis was poorer in the non-treated patients (P=0.026). Nuclear MAD1L1 expression therefore appears to have enhanced the treatment resistance and affected the prognosis of breast cancer, demonstrating that MAD1L1-positive breast cancer was not susceptible to taxol treatment.
Methylation of paired-like homeodomain 2 (PITX2P2)
C-phosphate-G islands located within the gene regulatory site are associated with the suppression of gene expression. The methylation of DNA dinucleotides in this gene is a common early event subsequent to the onset of cancer (58–60). Methylation patterns specific to tumor subtypes, including breast cancer, are reportedly associated with clinical outcomes (61–63). Several studies reported that PITX2 DNA methylation was associated with a high risk of relapse in lymph node metastasis-positive, hormone receptor-positive breast cancer patients undergoing whole body adjuvant tamoxifen therapy (64,65). In a cohort study of 241 lymph node metastasis-positive breast cancer patients with a history of anthracycline treatment, PITX2P2 methylation (a subtype of PITX2 methylation) was associated with an increased long-term relapse (HR=1.66; 95% CI=1.21–2.28; P=0.002) and reduced survival rates (HR=1.47; 95% CI=1.11–1.96; P=0.0084) (66).
5. Conclusions
In the present review of studies between 2009 and 2014, biomarkers were grouped according to reliability (high, medium, or low). A total of 3 studies were retrieved from the literature that classified Ki-67 as a high reliability biomarker. The utility of Ki-67 as a biomarker has been re-evaluated in the present study.
Of the high reliability biomarkers referred to in the 9 studies included in the present review, 0 were assessed by bioassays and only 1 mentioned biomarker measurement in peripheral blood. Although the evaluation of proteins in peripheral blood is relatively simple, in 2013, Johansson (7) reported that the identification of biomarkers is difficult, as measurements of biomarkers require too much quantification data. Therefore, the development of biomarkers from peripheral blood presents a challenge for future studies.
Although a number of molecules were identified in the present review, other markers, such as hormone receptors, were not widely evaluated. The ratio of ER-α/ER-β expression, βIII-tubulin and thyroid-stimulating hormone were identified as oncological indicators in breast cancer. Thus, additional studies of the correlations between the aforementioned biomarkers and prognosis and treatment response may be useful.
This study hypothesizes that the most important biomarkers of breast cancer are found in the blood, and that RARA, uPA/PAI-1, CRP, sHER2 are good biomarkers in routine examination. The authors highlight RARA in particular as an important biomarker in breast cancer. Future studies on biomarkers are likely to progress the understanding of the topic. For all biomarkers, reliability is important, but for the development of useful biomarkers, cost and ease of monitoring are crucial considerations.
Acknowledgements
The authors would like to thank Drs. Tousei Ohmura, Hidekazu Kameshima, and Yasuyo Suzuki for their help with preparing this study and participating in valuable discussions. The present study was partially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (grant nos. 26461921, 25293289, 24659592 and 50175634).
References
- 1.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ. Panel members: Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson IC, Patek AJ. The relationship between prognostic and predictive factors in the management of breast cancer. Breast Cancer Res Treat. 1998;52:261–288. doi: 10.1023/A:1006141703224. [DOI] [PubMed] [Google Scholar]
- 3.Lang JE, Wecsler JS, Press MF, Tripathy D. Molecular markers for breast cancer diagnosis, prognosis and targeted therapy. J Surg Oncol. 2015;111:81–90. doi: 10.1002/jso.23732. [DOI] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists' Collaborative Group (EBCTCG), corp-author Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 5.Niederreither K, Dollé P. Retinoic acid in development: Towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 6.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson HJ, Sanchez BC, Mundt F, Forshed J, Kovacs A, Panizza E, Hultin-Rosenberg L, Lundgren B, Martens U, Máthé G, et al. Retinoic acid receptor alpha is associated with tamoxifen resistance in breast cancer. Nat Commun. 2013;4:2175. doi: 10.1038/ncomms3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteban JM, Warsi Z, Haniu M, Hall P, Shively JE, Chen S. Detection of intratumoral aromatase in breast carcinomas. An immunohistochemical study with clinicopathologic correlation. Am J Pathol. 1992;140:337–343. [PMC free article] [PubMed] [Google Scholar]
- 9.Miki Y, Suzuki T, Sasano H. Controversies of aromatase localization in human breast cancer-stromal versus parenchymal cells. J Steroid Biochem Mol Biol. 2007;106:97–101. doi: 10.1016/j.jsbmb.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Ellis MJ, Coop A, Singh B, Mauriac L, Llombert-Cussac A, Jänicke F, Miller WR, Evans DB, Dugan M, Brady C, et al. Letrozole is more effective neoadjuvant endocrine therapy than tamoxifen for ErbB-1- and/or ErbB-2-positive, estrogen receptor-positive primary breast cancer: Evidence from a phase III randomized trial. J Clin Oncol. 2001;19:3808–3816. doi: 10.1200/JCO.2001.19.18.3808. [DOI] [PubMed] [Google Scholar]
- 11.Eiermann W, Paepke S, Appfelstaedt J, Llombart-Cussac A, Eremin J, Vinholes J, Mauriac L, Ellis M, Lassus M, Chaudri-Ross HA, et al. Preoperative treatment of postmenopausal breast cancer patients with letrozole: A randomized double-blind multicenter study. Ann Oncol. 2001;12:1527–1532. doi: 10.1023/A:1013128213451. [DOI] [PubMed] [Google Scholar]
- 12.Ellis MJ, Tao Y, Luo J, A'Hern R, Evans DB, Bhatnagar AS, Ross HA Chaudri, von Kameke A, Miller WR, Smith I, et al. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100:1380–1388. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis MJ, Coop A, Singh B, Tao Y, Llombart-Cussac A, Jänicke F, Mauriac L, Quebe-Fehling E, Chaudri-Ross HA, Evans DB, Miller WR. Letrozole inhibits tumor proliferation more effectively than tamoxifen independent of HER1/2 expression status. Cancer Res. 2003;63:6523–6531. [PubMed] [Google Scholar]
- 14.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 15.Ellis MJ, Miller WR, Tao Y, Evans DB, Ross HA Chaudri, Miki Y, Suzuki T, Sasano H. Aromatase expression and outcomes in the P024 neoadjuvant endocrine therapy trial. Breast Cancer Res Treat. 2009;116:371–378. doi: 10.1007/s10549-008-0161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senger DR, Wirth DF, Hynes RO. Transformed mammalian cells secrete specific proteins and phosphoproteins. Cell. 1979;16:885–893. doi: 10.1016/0092-8674(79)90103-X. [DOI] [PubMed] [Google Scholar]
- 17.Brown LF, Papadopoulos-Sergiou A, Berse B, Manseau EJ, Tognazzi K, Perruzzi CA, Dvorak HF, Senger DR. Osteopontin expression and distribution in human carcinomas. Am J Pathol. 1994;145:610–623. [PMC free article] [PubMed] [Google Scholar]
- 18.Pang H, Lu H, Song H, Meng Q, Zhao Y, Liu N, Lan F, Liu Y, Yan S, Dong X, Cai L. Prognostic values of osteopontin-c, E-cadherin and β-catenin in breast cancer. Cancer Epidemiol. 2013;37:985–992. doi: 10.1016/j.canep.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Ortiz-Martínez F, Perez-Balaguer A, Ciprián D, Andrés L, Ponce J, Adrover E, Sánchez-Payá J, Aranda FI, Lerma E, Peiró G. Association of increased osteopontin and splice variant-c mRNA expression with HER2 and triple-negative/basal-like breast carcinomas subtypes and recurrence. Hum Pathol. 2014;45:504–512. doi: 10.1016/j.humpath.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Bramwell VH, Tuck AB, Chapman JA, Anborgh PH, Postenka CO, Al-Katib W, Shepherd LE, Han L, Wilson CF, Pritchard KI, et al. Assessment of osteopontin in early breast cancer: Correlative study in a randomised clinical trial. Breast Cancer Res. 2014;16:R8. doi: 10.1186/bcr3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno S, Chow LW, Sato N, Masuda N, Sasano H, Takahashi F, Bando H, Iwata H, Morimoto T, Kamigaki S, et al. Randomized trial of preoperative docetaxel with or without capecitabine after 4 cycles of 5-fluorouracil-epirubicin-cyclophosphamide (FEC) in early-stage breast cancer: Exploratory analyses identify Ki67 as a predictive biomarker for response to neoadjuvant chemotherapy. Breast Cancer Res Treat. 2013;142:69–80. doi: 10.1007/s10549-013-2691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denkert C, Loibl S, Muller BM, Eidtmann H, Schmitt WD, Eiermann W, Gerber B, Tesch H, Hilfrich J, Huober J, et al. Ki67 levels as predictive and prognostic parameters in pretherapeutic breast cancer core biopsies: A translational investigation in the neoadjuvant GeparTrio trial. Ann Oncol. 2013;24:2786–2793. doi: 10.1093/annonc/mdt350. [DOI] [PubMed] [Google Scholar]
- 23.Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, Denkert C. Cutoff finder: A comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeCensi A, Guerrieri-Gonzaga A, Gandini S, Serrano D, Cazzaniga M, Mora S, Johansson H, Lien EA, Pruneri G, Viale G, Bonanni B. Prognostic significance of Ki-67 labeling index after short-term presurgical tamoxifen in women with ER-positive breast cancer. Ann Oncol. 2011;22:582–587. doi: 10.1093/annonc/mdq427. [DOI] [PubMed] [Google Scholar]
- 25.Kuroki M, Matsuo Y, Kinugasa T, Matsuoka Y. Three different NCA species, CGM6/CD67, NCA-95 and NCA-90, are comprised in the major 90 to 100-kDa band of granulocyte NCA detectable upon SDS-polyacrylamide gel electrophoresis. Biochem Biophys Res Commun. 1992;182:501–506. doi: 10.1016/0006-291X(92)91760-N. [DOI] [PubMed] [Google Scholar]
- 26.Blumenthal RD, Leon E, Hansen HJ, Goldenberg DM. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poola I, Shokrani B, Bhatnagar R, DeWitty RL, Yue Q, Bonney G. Expression of carcinoembryonic antigen cell adhesion molecule 6 oncoprotein in atypical ductal hyperplastic tissues is associated with the development of invasive breast cancer. Clin Cancer Res. 2006;12:4773–4783. doi: 10.1158/1078-0432.CCR-05-2286. [DOI] [PubMed] [Google Scholar]
- 28.Tsang JY, Kwok YK, Chan KW, Ni YB, Chow WN, Lau KF, Shao MM, Chan SK, Tan PH, Tse GM. Expression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in breast cancers. Breast Cancer Res Treat. 2013;142:311–322. doi: 10.1007/s10549-013-2756-y. [DOI] [PubMed] [Google Scholar]
- 29.German S, Aslam HM, Saleem S, Raees A, Anum T, Alvi AA, Haseeb A. Carcinogenesis of PIK3CA. Hered Cancer Clin Pract. 2013;11:5. doi: 10.1186/1897-4287-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanker AB, Pfefferle AD, Balko JM, Kuba MG, Young CD, Sánchez V, Sutton CR, Cheng H, Perou CM, Zhao JJ, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci USA. 2013;110:14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cizkova M, Dujaric ME, Lehmann-Che J, Scott V, Tembo O, Asselain B, Pierga JY, Marty M, de Cremoux P, Spyratos F, Bieche I. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br J Cancer. 2013;108:1807–1809. doi: 10.1038/bjc.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hortobagyi GN, Holmes FA. Single-agent paclitaxel for the treatment of breast cancer: An overview. Semin Oncol. 1996;23(1):S4–S9. Suppl 1. [PubMed] [Google Scholar]
- 33.Bergh J, Jönsson PE, Glimelius B, Nygren P. SBU-group. Swedish Council of Technology Assessment in Health Care. A systematic overview of chemotherapy effects in breast cancer. Acta Oncol. 2001;40:253–281. doi: 10.1080/02841860120784. [DOI] [PubMed] [Google Scholar]
- 34.Chromek M, Tullus K, Lundahl J, Brauner A. Tissue inhibitor of metalloproteinase 1 activates normal human granulocytes, protects them from apoptosis, and blocks their transmigration during inflammation. Infect Immun. 2004;72:82–88. doi: 10.1128/IAI.72.1.82-88.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrohl AS, Meijer-van Gelder ME, Holten-Andersen MN, Christensen IJ, Look MP, Mouridsen HT, Brünner N, Foekens JA. Primary tumor levels of tissue inhibitor of metalloproteinases-1 are predictive of resistance to chemotherapy in patients with metastatic breast cancer. Clin Cancer Res. 2006;12:7054–7058. doi: 10.1158/1078-0432.CCR-06-0950. [DOI] [PubMed] [Google Scholar]
- 36.Zhu D, Zha X, Hu M, Tao A, Zhou H, Zhou X, Sun Y. High expression of TIMP-1 in human breast cancer tissues is a predictive of resistance to paclitaxel-based chemotherapy. Med Oncol. 2012;29:3207–3215. doi: 10.1007/s12032-012-0239-3. [DOI] [PubMed] [Google Scholar]
- 37.Arosio P, Yokota M, Drysdale JW. Structural and immunological relationships of isoferritins in normal and malignant cells. Cancer Res. 1976;36:1735–1739. [PubMed] [Google Scholar]
- 38.Levi S, Yewdall SJ, Harrison PM, Santambrogio P, Cozzi A, Rovida E, Albertini A, Arosio P. Evidence of H- and L-chains have co-operative roles in the iron-uptake mechanism of human ferritin. Biochem J. 1992;288:591–596. doi: 10.1042/bj2880591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricolleau G, Charbonnel C, Lodé L, Loussouarn D, Joalland MP, Bogumil R, Jourdain S, Minvielle S, Campone M, Déporte-Fety R, et al. Surface-enhanced laser desorption/ionization time of flight mass spectrometry protein profiling identifies ubiquitin and ferritin light chain as prognostic biomarkers in node-negative breast cancer tumors. Proteomics. 2006;6:1963–1975. doi: 10.1002/pmic.200500283. [DOI] [PubMed] [Google Scholar]
- 40.Jézéquel P, Campion L, Spyratos F, Loussouarn D, Campone M, Guérin-Charbonnel C, Joalland MP, André J, Descotes F, Grenot C, et al. Validation of tumor-associated macrophage ferritin light chain as a prognostic biomarker in node-negative breast cancer tumors: A multicentric 2004 national PHRC study. Int J Cancer. 2012;131:426–437. doi: 10.1002/ijc.26397. [DOI] [PubMed] [Google Scholar]
- 41.Duffy MJ. Urokinase plasminogen activator and its inhibitor, PAI-1, as prognostic markers in breast cancer: From pilot to level 1 evidence studies. Clin Chem. 2002;48:1194–1197. [PubMed] [Google Scholar]
- 42.Mengele K, Napieralski R, Magdolen V, Reuning U, Gkazepis A, Sweep F, Brünner N, Foekens J, Harbeck N, Schmitt M. Characteristics of the level-of-evedence-1 disease forcast cancer biomarkers uPA and its inhibitor PAI-1. Expert Rev Mol Diagn. 2010;10:947–962. doi: 10.1586/erm.10.73. [DOI] [PubMed] [Google Scholar]
- 43.Harbeck N, Schmitt M, Meisner C, Friedel C, Untch M, Schmidt M, Sweep CG, Lisboa BW, Lux MP, Beck T, et al. Ten-year analysis of the prospective multicentre Chemo-N0 trial validates American society of clinical oncology (ASCO)-recommended biomarkers uPA and PAI-1 for therapy decision making in node-negative breast cancer patients. Eur J Cancer. 2013;49:1825–1835. doi: 10.1016/j.ejca.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Favaro E, Amadori A, Indraccolo S. Cellular interactions in the vascular niche: Implications in the regulation of tumor dormancy. APMIS. 2008;116:648–659. doi: 10.1111/j.1600-0463.2008.01025.x. [DOI] [PubMed] [Google Scholar]
- 45.Allin KH, Nordestgaard BG. Elevated C-reactive protein in the diagnosis, prognosis, and cause of cancer. Crit Rev Clin Lab Sci. 2011;48:155–170. doi: 10.3109/10408363.2011.599831. [DOI] [PubMed] [Google Scholar]
- 46.Villaseñor A, Flatt SW, Marinac C, Natarajan L, Pierce JP, Patterson RE. Postdiagnosis C-reactive protein and breast cancer survivorship: Findings from the WHEL study. Cancer Epidemiol Biomarkers Prev. 2014;23:189–199. doi: 10.1158/1055-9965.EPI-13-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, et al. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature. 2006;440:1045–1049. doi: 10.1038/nature04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watters AD, Going JJ, Cooke TG, Bartlett JM. Chromosome 17 aneusomy is associated with poor prognostic factors in invasive breast carcinoma. Breast Cancer Res Treat. 2003;77:109–114. doi: 10.1023/A:1021399923825. [DOI] [PubMed] [Google Scholar]
- 49.Corzo C, Bellosillo B, Corominas JM, Salido M, Coll MD, Serrano S, Albanell J, Solé F, Tusquets I. Does polysomy of chromosome 17 have a role in ERBB2 and topoisomerase IIalpha expression? Gene, mRNA and protein expression: A comprehensive analysis. Tumor Biol. 2007;28:221–228. doi: 10.1159/000107583. [DOI] [PubMed] [Google Scholar]
- 50.Earl HM, Hiller L, Dunn JA, Vallier AL, Bowden SJ, Jordan SD, Blows F, Munro A, Bathers S, Grieve R, et al. Adjuvant epirubicin followed by cyclophosphamide, methotrexate and fluorouracil (CMF) vs CMF in early breast cancer: Results with over 7 years median follow-up from the randomised phase III NEAT/BR9601 trials. Br J Cancer. 2012;107:1257–1267. doi: 10.1038/bjc.2012.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sias PE, Kotts CE, Vetterlein D, Shepard M, Wong WL. ELISA for quantitation of the extracellular domain of p185HER2 in biological fluids. J Immunol Methods. 1990;132:73–80. doi: 10.1016/0022-1759(90)90400-P. [DOI] [PubMed] [Google Scholar]
- 52.Codony-Servat J, Albanell J, Lopez-Talavera JC, Arribas J, Baselga J. Cleavage of the HER2 ectodomain is a pervanadate-activable process that is inhibited by the tissue inhibitor of metalloproteases-1 in breast cancer cells. Cancer Res. 1999;59:1196–1201. [PubMed] [Google Scholar]
- 53.Tse C, Gauchez AS, Jacot W, Lamy PJ. HER2 shedding and serum HER2 extracellular domain: Biology and clinical utility in breast cancer. Cancer Treat Rev. 2012;38:133–142. doi: 10.1016/j.ctrv.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 55.Moreno-Aspitia A, Hillman DW, Dyar SH, Tenner KS, Gralow J, Kaufman PA, Davidson NE, Lafky JM, Reinholz MM, Lingle WL, et al. Soluble human epidermal growth factor receptor 2 (HER2) levels in patients with HER2-positive breast cancer receiving chemotherapy with or without trastuzumab: Results from north central cancer treatment group adjuvant trial N9831. Cancer. 2013;119:2675–2682. doi: 10.1002/cncr.28130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukasaki K, Miller CW, Greenspun E, Eshaghian S, Kawabata H, Fujimoto T, Tomonaga M, Sawyers C, Said JW, Koeffler HP. Mutations in the mitotic check point gene, MAD1L1, in human cancers. Oncogene. 2001;20:3301–3305. doi: 10.1038/sj.onc.1204421. [DOI] [PubMed] [Google Scholar]
- 57.Sun Q, Zhang X, Liu T, Liu X, Geng J, He X, Liu Y, Pang D. Increased expression of mitotic arrest deficient-like 1 (MAD1L1) is associated with poor prognosis and insensitive to Taxol treatment in breast cancer. Breast Cancer Res Treat. 2013;140:323–330. doi: 10.1007/s10549-013-2633-8. [DOI] [PubMed] [Google Scholar]
- 58.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- 59.Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 60.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 61.Frühwald MC. DNA methylation patterns in cancer: Novel prognostic indicators? Am J Pharmacogenomics. 2003;3:245–260. doi: 10.2165/00129785-200303040-00003. [DOI] [PubMed] [Google Scholar]
- 62.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 63.Maier S, Nimmrich I, Koenig T, Eppenberger-Castori S, Bohlmann I, Paradiso A, Spyratos F, Thomssen C, Mueller V, Nährig J, et al. DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients-Technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer. 2007;43:1679–1686. doi: 10.1016/j.ejca.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 64.Martens JW, Nimmrich I, Koenig T, Look MP, Harbeck N, Model F, Kluth A, Bolt-de Vries J, Sieuwerts AM, Portengen H, et al. Association of DNA methylation of phosphoserine aminotransferase with response to endocrine therapy in patients with recurrent breast cancer. Cancer Res. 2005;65:4101–4117. doi: 10.1158/0008-5472.CAN-05-0064. [DOI] [PubMed] [Google Scholar]
- 65.Widschwendter M, Siegmund KD, Müller HM, Fiegl H, Marth C, Müller-Holzner E, Jones PA, Laird PW. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 66.Hartmann O, Spyratos F, Harbeck N, Dietrich D, Fassbender A, Schmitt M, Eppenberger-Castori S, Vuaroqueaux V, Lerebours F, Welzel K, et al. DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res. 2009;15:315–323. doi: 10.1158/1078-0432.CCR-08-0166. [DOI] [PubMed] [Google Scholar]


