Abstract
Long non-coding RNAs (lncRNAs) are key regulatory molecules in many fundamental cellular processes and their deregulation is assumed to contribute to carcinogenesis. Exosomal lncRNAs are thought to be involved in the dissemination of cell signals to control local cellular microenvironments. In the current study, exosomal expression of growth arrest specific 5 (GAS5), an inhibitor of cell proliferation and promoter of apoptosis, was evaluated in apoptotic processes initiated by different mechanisms. Therefore, MCF-7 and MDA-MB-231 breast cancer cells were treated with Taxol (2 and 10 nM) and bleomycin (2 and 10 ng/ml) for 24 h. Following cell viability determination and measurement of apoptosis, cellular and exosomal expression levels of GAS5 were investigated using a quantitative polymerase chain reaction assay. The findings indicate that Taxol is more toxic than bleomycin at the indicated doses and the effect was more evident in the MCF-7 cells. Despite varying toxicity rates, comparable levels of apoptotic nucleosomes were measured between Taxol- and bleomycin-treated cells. Upon drug treatment, cellular expression levels of GAS rose (≤1.5-fold) in the two cell lines. It appears that even a small increase in cellular expression leads to exosomal enrichment, as the accumulation of GAS5 in exosomes was marked in the MCF-7 cells (≤5.8-fold). Compared with the MCF-7 cells, the extent of GAS5 enrichment in the exosomes secreted from MDA-MB-231 cells was moderate (≤1.9-fold), potentially as a result of reduced cell death. The present study indicates that GAS5 accumulation in exosomes is a prevalent event in apoptotic processes that are initiated by different mechanisms.
Keywords: apoptosis, bleomycin, exosomes, GAS5, Taxol
Introduction
To facilitate next generation sequencing techniques, the discovery of pervasive transcription in the human genome was one of the major advances of genome research in the last decade (1,2). The vast majority (~98%) of the mammalian genome is pervasively transcribed into countless and multifunctional forms of RNA molecules termed non-coding RNAs (ncRNAs) (3,4). Based on the length of the functional transcripts, human ncRNAs have been largely classified into two classes: Small (20–30 nt.) and long (>200 nt.) non-coding transcripts. The small ncRNA class includes housekeeping and regulatory RNAs (tRNAs and rRNAs), microRNAs (miRNAs), as well as other small RNAs.
The long ncRNA (lncRNA) class includes an ambiguous, functionally distinct class of RNAs that encompasses thousands of lncRNA molecules with >15,000 encoded transcripts (5,6). Certain molecules have been reported to be similar to protein-coding genes with respect to splicing and polyadenylation. As a result of relatively high functional diversity, confusing and overlapping terminology has been used to name and classify lncRNAs (7). Various attributes, including transcript length, association with annotated protein-coding gene or other DNA elements/repeats, association with a biochemical pathway or stability, or sequence and structure conservation have been used for classification. Accumulating evidence reveals that lncRNAs are important in various biologic processes, including gene regulation, chromatin modification, immune surveillance, differentiation, cell cycle control and apoptosis, or act as nuclear architecture, subnuclear compartments, or enhancers and promoters in RNA processing (8–10).
The current study evaluates lncRNA, growth arrest specific 5 (GAS5). It is a recently identified tumor suppressor and displays reduced expression levels in various types of cancer, such as breast, prostate, lung and colorectal cancer. As GAS5 is involved in the regulation of cell proliferation, growth arrest and apoptosis (11), its low-expression pattern confers the elevated capacity of tumor cells for proliferation (12). GAS5, among others, was previously shown to be detectable in secreted exosomes (13). The existence of lncRNAs in exosomes may be of relevance in cancer, as exosomes are important mediators of intercellular communication, which regulates a diverse range of biologic processes (14), and their pathophysiologic roles in disease, including cancer are increasingly being recognized. As a promoter of apoptosis, exosomal GAS5 may be involved in the communication of dying tumor cells in their local microenvironment. The aim of the current study was to determine whether exosomal expression levels of GAS5 are adjusted during apoptosis induction, which is triggered by different mechanisms, in cultured breast cancer cells.
Materials and methods
Cell lines and cell culture
Two breast cancer cell lines (MCF-7 and MDA-MB-231) were used in the present study and purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA]. The MCF-7 cell line represents hormone-responsive human-invasive breast adenocarcinoma with receptor positivity resulting in a luminal A subtype. The MDA-MB-231 cells were of basal subtype, as these cells do not express hormone receptors and possess the mutant p53 protein (15). Cells were grown in Dulbecco's modified Eagle's medium (Biochrom GmbH, Berlin, Germany), including exosome-free serum, in standard culture conditions. To obtain exosome-free serum, fetal calf serum (Biochrom GmbH) was centrifuged at 120,000 × g for 2 h. Taxol and bleomycin (AppliChem GmbH, Darmstadt, Germany) served as apoptosis-inducer anticancer compounds. Taxol is an anti-mitotic that interferes with normal breakdown of microtubules during cell division and induces cell death. Bleomycin induces apoptotic cell death via DNA strand breaks.
Evaluation of cytotoxicity
Taxol- and bleomycin-induced cytotoxicity were evaluated using real-time cellular analysis in the microelectronic biosensor-based iCELLigence instrument (ACEA Biosciences, San Diego, CA, USA). In this system, numeric cell changes affect the local ionic environment at the electrode/solution interface, which leads to an increase or decrease in electrode impedance. Data are recorded as impedance change and expressed as the cell index (CI), as a mean of cell proliferation. For that analysis, 25,000 cells were plated and Taxol (2 and 10 nm) or bleomycin (2 and 10 nm/ml) (AppliChem GmbH) was added after 24 h; the cells were monitored for an additional 24 h. Untreated cells were used as control cells.
Measurement of apoptosis
For a quantitative analysis of apoptosis, cytoplasmic levels of mono- and oligonucleosomes released into the cytoplasm during apoptosis prior to membrane breakdown were measured using the Cell Death Detection ELISA kit (Roche Diagnostics GmbH, Mannheim, Germany). Detection of nucleosomal fragments as a result of DNA degradation has widely served as a hallmark of apoptotic cell death (16). The assay is based on an immunoassay using specific monoclonal antibodies against histones and DNA, allowing the determination of nucleosomes in the cytoplasmic fraction of cell lysates. In the present study, cells in culture suspension were removed by discarding the medium and only intact cells were used. For this assay, 150,000 cells were used and analyzed as previously described (17).
RNA isolation and gene expression analysis
Total cellular RNA was extracted using the RNA isolation solution, Tripure (Roche Diagnostics GmbH) according to the manufacturer's instructions. Exosome extraction from the culture medium was performed using the Total Exosome Isolation reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) as per the manufacturer's instructions. RNA was isolated from the extracted exosomes using the Total Exosome RNA & Protein Isolation kit (Thermo Fisher Scientific, Inc.). GAS5 expression levels were quantified as previously reported (13), using GAPDH as internal control. The results of three different experiments were used.
Statistical analysis
The present study evaluates whether GAS5 in exosomes is a marker of apoptosis. Results of three independent cell culture experiments were used. Cell viability in the drug-treated cells is expressed as the percentage of control cells while oligonucleosome-associated apoptosis and GAS5 expression levels are presented as fold-change relative to the control cells. Mean changes relative to basal values were statistically analyzed using the independent t-test where P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using SPSS version 15 (SPSS Inc., Chicago, IL, USA).
Results
Drug-induced cytotoxicity
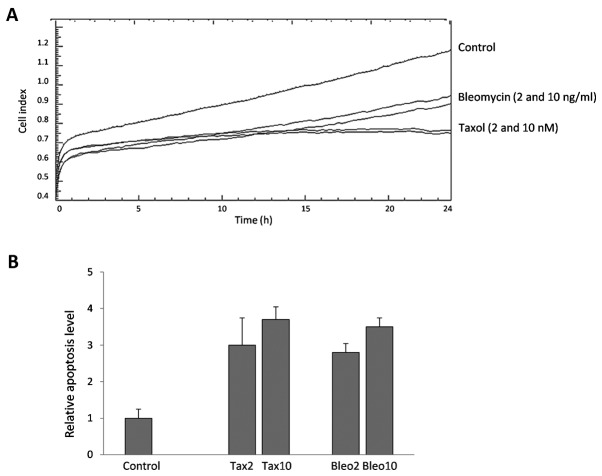
Taxol- and bleomycin-associated cytotoxicity in MCF-7 and MDA-MB-231 cells was determined. Cells were treated with each of the drugs for 24 h, and the anti-proliferative effect was measured using real-time cellular analysis. The CI, as a mean of cell proliferation, served as the measure of cytotoxicity. Different rates of cell viability were obtained, which indicated the variable sensitivity of the cell lines to those drugs. Fig. 1A representatively depicts the CI of the MCF-7 cells, and Table I shows the percentages of viable cells (% of control) in the MCF-7 and MDA-MB-231 cells following treatment. MCF-7 cells were more sensitive to the drugs than the MDA-MB-231 cells, and Taxol (2 and 10 nM) was identified to be more potent than bleomycin (2 and 10 ng/ml) at the applied doses.
Figure 1.
Cytotoxicity and apoptosis induction in breast cancer cells. MCF-7 and MDA-MB-231 breast cancer cells were cultured in exosome-free conditions for 24 h and treated with the indicated doses of Taxol (2 and 10 nM) and bleomycin (2 and 10 ng/ml) for a further 24 h. Antiproliferative effects and apoptosis induction were evaluated. (A) Cell index in control and drug-treated MCF-7 cells; (B) Apoptosis measurement based on cytoplasmic mono- and oligonucleosomes in control and drug-treated MCF-7 cells, expressed as fold-changes relative to control cells. The mean and maximum values are shown. Tax, Taxol; Bleo, bleomycin.
Table I.
Cell index and apoptosis levels in drug-treated breast cancer cells.
| Mean cell index (% of control) | Oligonucleosomes (−fold of control) | |||||||
|---|---|---|---|---|---|---|---|---|
| Cell line | Taxol 2 | Taxol 10 | Bleo 2 | Bleo 10 | Taxol 2 | Taxol 10 | Bleo 2 | Bleo 10 |
| MCF-7 | 69 | 67 | 91 | 87 | 3.0 | 3.7 | 2.8 | 3.5 |
| MDA-MB-231 | 80 | 75 | 98 | 97 | 1.8 | 2.1 | 1.5 | 1.9 |
Bleo, bleomycin.
Apoptosis induction
Taxol- and bleomycin-induced apoptosis was measured in breast cancer cells following 24 h of treatment. Despite differences in cell viability, considerable levels of cytoplasmic nucleosomes were detectable in the two cell lines. For example, similar quantities of nucleosomes were measured in the Taxol- and bleomycin-treated MCF-7 cells (Fig. 1B), although a difference of ~20% was noted in the mean CI between the two drugs. Similarly, although the toxicity of the two drugs was lower in the MDA-MB-231 cells, a considerable level of apoptosis was also detectable in these cells (Table I).
Cellular and exosomal GAS5 expression levels
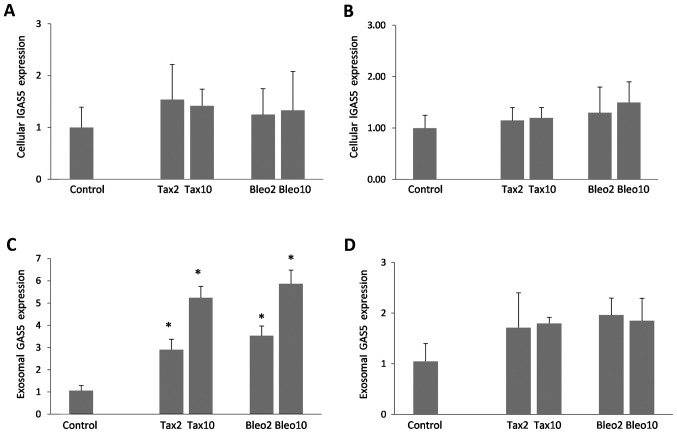
Subsequently, GAS5 expression levels in Taxol- and bleomycin-treated cells, and exosomes secreted from the drug-treated cells were investigated. In the two cell lines, an increase of up to 1.5-fold was detected in the cellular expression levels of GAS5 (Fig. 2A and B, P>0.05). Compared with cellular GAS5, exosomal expression levels of GAS5 appeared to be more responsive to apoptosis induction, as the extent of accumulation in secreted exosomes was marked, particularly in the MCF-7 cells (Fig. 2C and D). As seen in Fig. 2C and D, dose-dependent increases of ≤5.8-fold (P<0.001) and ≤1.9-fold (P>0.05) were detected in the MCF-7 and MDA-MB-231 cells, respectively.
Figure 2.
Cellular and exosomal expression of GAS5. Expression levels of GAS5 relative to GAPDH in (A) MCF-7 and (B) MDA-MB-231 cells treated with Taxol (2 and 10 nM) and bleomycin (2 and 10 ng/ml), and (C and D, respectively) exosomes secreted from those cells. Expression levels in control cells were taken as 1 and changes in drug-treated cells were expressed as fold-changes. Mean and maximum values are shown. *P<0.05 vs. control. GAS5, growth arrest specific 5; Tax, Taxol; Bleo, bleomycin.
Discussion
As important mediators of intercellular communication, exosomes have been implicated in cancer development and progression (18). Their involvement in malignant development is associated with their cargo in the form of proteins and nucleic acids of different types, including lncRNAs. In this respect, lncRNAs and exosomes may function together to disseminate cell signals that alter and/or control local cellular microenvironments (19). Such an intercellular communication via RNA molecules may also occur within tumors with high cell turnover. The present study focused on clarifying whether GAS5 is enriched in exosomes during the apoptotic processes. To accomplish this goal, cellular and exosomal accumulation of GAS5 was investigated in relation to Taxol- and bleomycin-induced apoptosis in two breast cancer cell lines that differ in hormone receptor expression and p53 mutational status.
At the administered doses, Taxol was identified to be more potent than bleomycin in the MCF-7 and MDA-MB-231 cells. This is consistent with expectations, as Taxol directly interferes with cell division, whereas bleomycin-associated toxicity occurs via DNA damage. Similarly, Taxol-associated toxicity was stronger in MCF-7 cells when compared with MDA-MB-231 cells, which indicates the involvement of the hormone receptor and p53 status in MD-MB-231 cells. However, a previous study reported that Taxol-induced apoptosis in MCF-7 and MDA-MB-231 cells is estrogen receptor- and p53-independent (20). Despite variances in toxicity rates, similar levels of cell death-associated nucleosomes were measured following treatment with the two drugs, thereby indicating the varying efficacy in blocking cell proliferation and apoptosis induction. In addition, the current findings confirm that cytoplasmic nucleosomal fragments are useful tools for evaluating apoptosis in intact cells, as DNA degradation occurs several hours before plasma membrane breakdown during the apoptotic process (21).
The primary goal of the present study was to investigate GAS5 accumulation in exosomes during apoptotic induction triggered by two different mechanisms, e.g., microtubule stabilization and DNA strand breaks. Upon apoptosis induction, cellular expression of GAS5 was induced; however, to a limited extent (≤1.5-fold) whereas exosomal enrichment was more pronounced, particularly in MCF-7 cells. These findings indicate that even a small increase of cellular GAS5 expression leads to exosomal accumulation. Comparable levels of GAS5 accumulation were found in exosomes that were released from Taxol- and bleomycin-treated MCF-7 cells, suggesting that GAS5 enrichment in exosomes is associated with apoptosis initiation rather than the antiproliferative effect of anticancer agents (Table I). Accordingly, the extent of exosomal GAS5 enrichment was lower in MDA-MB-231 cells, possibly as a result of lower cell death rates in these cells.
In conclusion, the current study provides evidence that GAS5 accumulation in exosomes is a marker of apoptotic induction. Therefore, it is plausible to hypothesize that GAS5 is involved in communication of tumor cells upon cell death-promoting signals. Tracking GAS5 in secreted exosomes may be useful for evaluating apoptosis and assessing the efficacy of therapeutic interventions in cancer, as radiotherapy and many chemotherapeutic agents usually depend upon the efficient engagement of the apoptotic machinery for their action (22). However, any possible involvement of GAS5 in exosomal communication of dying tumor cells requires further research.
Acknowledgements
The present study is part of MSc Thesis of Oguz Koldemir and was supported by the Istanbul University Scientific Projects Coordination Unit (grant no. 56669). The authors would like to thank Mr. David F. Chapman for language editing of the manuscript.
Glossary
Abbreviations
- lncRNAs
long non-coding RNAs
- ncRNAs
non-coding RNAs
References
- 1.Jacquier A. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 2.Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Mol Cell. 2013;52:473–484. doi: 10.1016/j.molcel.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 3.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2011;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickard MR, Williams GT. Molecular and cellular mechanisms of action of tumour suppressor GAS5 lncRNA. Genes (Basel) 2015;6:484–499. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, Song Y. The growth arrest-specific transcript 5 (GAS5): A pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37:1437–1444. doi: 10.1007/s13277-015-4521-9. [DOI] [PubMed] [Google Scholar]
- 13.Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- 14.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 15.Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG, et al. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer (Auckl) 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Xu M. Apoptotic DNA fragmentation and tissue homeostasis. Trends Cell Biol. 2002;12:84–89. doi: 10.1016/S0962-8924(01)02206-1. [DOI] [PubMed] [Google Scholar]
- 17.Özgür E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin Exp Med. 2013;13:119–126. doi: 10.1007/s10238-012-0181-x. [DOI] [PubMed] [Google Scholar]
- 18.Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewson C, Morris KV. Form and function of exosome-associated long non-coding RNAs in cancer. Curr Top Microbiol Immunol. 2016;394:41–56. doi: 10.1007/82_2015_486. [DOI] [PubMed] [Google Scholar]
- 20.Choi YH, Yoo YH. Taxol-induced growth arrest and apoptosis is associated with the upregulation of the Cdk inhibitor, p21WAF1/CIP1, in human breast cancer cells. Oncol Rep. 2012;28:2163–2169. doi: 10.3892/or.2012.2060. [DOI] [PubMed] [Google Scholar]
- 21.Neukirchen J, Meier A, Rohrbeck A, Garcia-Pardillos G, Steidl U, Fenk R, Haas R, Kronenwett R, Rohr UP. The proteasome inhibitor bortezomib acts differently in combination with p53 gene transfer or cytotoxic chemotherapy on NSCLC cells. Cancer Gene Ther. 2007;14:431–439. doi: 10.1038/sj.cgt.7701029. [DOI] [PubMed] [Google Scholar]
- 22.Indran IR, Tufo G, Pervaiz S, Brenner C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim Biophys Acta. 2011;1807:735–745. doi: 10.1016/j.bbabio.2011.03.010. [DOI] [PubMed] [Google Scholar]