Abstract
RAS protein activator like 2 (RASAL2) is a RAS-GTPase-activating protein and has recently been identified to be a tumor suppressor in various types of human cancer; however, the function of RASAL2 in colorectal carcinoma (CRC) remains unclear. In the present study, the function of RASAL2 in CRC cells was investigated using a RASAL2 loss-of-function cell model. RASAL2 short hairpin RNA was transfected into the human CRC cell lines LoVo, SW620 and HCT116, and the wild-type colon cell line NCM460. The subsequent downregulation of RASAL2 was evaluated using western blot and reverse transcription-quantitative polymerase chain reaction analyses. It was observed that RASAL2 expression was significantly decreased in human CRC tissues and cell lines (P<0.01). In the loss-of-function cell models, RASAL2 expression was decreased significantly, while cell proliferation, colony formation, migration and invasion were increased (all P<0.01). These effects were associated with the induction of epithelial-mesenchymal transition and Raf/mitogen-activated protein kinase hyperactivation. The results of the present study indicate that RASAL2 is a potential therapeutic target to inhibit CRC progression and metastasis.
Keywords: RAS protein activator like 2, colorectal cancer, proliferation, metastasis, epithelial-mesenchymal transition
Introduction
Colorectal carcinoma (CRC) is one of the most frequent and serious digestive tract malignancies worldwide, and the second leading cause of cancer-associated mortality (1,2). CRC is the fourth leading cause of cancer-associated mortality in China (3,4). The high mortality rate of CRC may be due to its high propensity for recurrence and metastasis (5). The liver is the most frequent site of CRC metastasis and ~50% of patients with CRC in China are diagnosed with liver metastases, either in the initial diagnosis or following primary tumor surgery, which makes treatment difficult (6,7). Identification of the cytokines regulating the proliferation and metastasis of CRC is required to understand the nature of CRC. In addition, investigation of the underlying mechanisms of CRC tumor progression may enable the development of potential therapeutic intervention strategies.
Ras-GTPase is an important regulator of the Ras signaling pathway, which is involved in a number of cellular activities. Aberrant activation of the Ras signaling pathway is common in the majority of human cancers, and is due to mutations in RAS genes, upstream inducers and downstream effectors (8,9). Ras proteins (H-Ras, N-Ras and K-Ras) are activated by GDP-GTP exchange (10).
RAS protein activator like 2 (RASAL2) functions as a GTPase activating protein (GAP) and was recently reported to be a tumor suppressor, and to mediate the growth and metastasis of various types of cancer (11). RASAL2 expression is suppressed by hypermethylation of its promoter, which leads to Ras signaling, and subsequent tumor growth and metastasis in human breast cancer (12,13). Decreased RASAL2 expression is correlated with metastasis, recurrence and poor disease outcome in luminal B tumors (14). However, RASAL2 is upregulated in a subset of triple-negative breast cancer (TNBC) and estrogen receptor (ER)-negative tumors, and is a direct target of microRNA 203, which has anti-invasive functions (13). RASAL2 is oncogenic in TNBC, where it drives mesenchymal invasion and metastasis, but not in luminal B ER-positive breast cancers. High RASAL2 expression is a predictor of poor disease outcome (13). In ovarian cancer, downregulation of RASAL2 promotes metastasis by increasing epithelial-mesenchymal transition (EMT) (15). Inactivation of RASAL2 promotes lung cancer metastasis through the induction of EMT via mitogen-activated protein kinase 1 (ERK) activation (16). RASAL2 was reported to be hypomethylated in hepatocellular carcinoma, suggesting that it functions as an oncogene in this cancer (12). However, to the best of our knowledge, no research focusing on the role of RASAL2 in CRC has been performed.
In the present study, tissue microarray immunochemistry was used to evaluate RASAL2 expression in CRC tissue samples at various stages of tumor development. In addition, a RASAL2 loss-of-function cell model was established to clarify the functional role of RASAL2 in CRC progression.
Materials and methods
Tumor tissue sample preparation and immunohistochemical (IHC) analysis
A total of 10 CRC samples were obtained during initial surgery in patients at the Hepatobiliary and Enteric Surgery Research Center of Xiangya Hospital (Central South University, Changsha, China) between December 2014 and June 2015. Samples included tumor, wild-type and metastatic tissue. The patients included 6 men and 4 women, with an age range of 32–72 years. All patients showed lymph node metastasis. The reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was used to analyze the expression of RASAL2 mRNA in the tissue samples. All experiments were approved by the Ethics Committee of Xiangya Hospital.
A tissue microarray containing 80 CRC specimens was obtained from Auragene Bioscience (cat no. TC0217; Changsha, China). Tissues for microarray analysis were fixed using 3% methanol for 15 min and subsequently incubated at 104°C for 32 min in citrate buffer (pH 6.0). The microarray was left to cool for 10 min at room temperature prior to blocking with 10% goat serum (Auragene Bioscience) for 15 min at room temperature. The microarray was incubated with the RASAL2 primary antibody (1:200; cat. no. sc-390605; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C, prior to a 15 min incubation with horseradish peroxidase-labeled chain mildew avidin (cat. no. P003H-1; Auragene Bioscience) at room temperature. The antibody-labeled microarray was subsequently incubated with 3,3′-diaminobenzidine for 5 min at room temperature.
Cell culture and transfection
Human LoVo, HCT116 and SW620 CRC cell lines, and the wild-type colon cell line NCM460, were purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and 100 µg/ml streptomycin, and incubated at 37°C with 5% CO2. Transfection was carried out using Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RASAL2-short hairpin (sh)RNA (GCCTTCCACCTCTTCATAGTA) and control (Ct)-shRNA plasmids (pRNAT-U6.1-RASAL2) were obtained from Auragene Bioscience. The Ct-shRNA sequence was synthesized by scrambling the RASAL2-shRNA sequence. All subsequent experiments were carried out using transfected cells.
RT-qPCR
Total RNA was extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) was used to convert RNA into cDNA. cDNA was quantified via qPCR using the Power SYBR Green kit (Toyobo Co., Ltd., Osaka, Japan) and the Applied Biosystems® 7500 Real-Time PCR system (Thermo Fisher Scientific, Inc.). PCR thermocyling conditions were as follows: Denaturation at 95°C for 3 min, followed by 35 cycles of 95°C for 10 sec and 58°C for 30 sec. Primer sequences are illustrated in Table I. β-actin was used as an endogenous control and the 2−ΔΔCq method was used to quantify relative mRNA expression (17).
Table I.
PCR primer sequences.
| Gene | Primer sequences |
|---|---|
| RASAL2 | F: TCAACAAGGAGAAGGAGATACC |
| R: GGAAACTGACCCTCGGAAG | |
| E-cadherin | F: CTCGGCCTGAAGTGACTCGTAAC |
| R: CAGCAACGTGATTTCTGCATTTC | |
| Vimentin | F: GACGCCATCAACACCGAGTT |
| R: CTTTGTCGTTGGTTAGCTGGT | |
| N-Cadherin | F: CAGTATCCGGTCCGATCTGC |
| R: GTCCTGCTCACCACCACTAC | |
| β-actin | F: AGGGGCCGGACTCGTCATACT |
| R: GGCGGCACCACCATGTACCCT |
RASAL2, RAS protein activator-like 2F; E-cadherin, epithelial cadherin; N-cadherin, neural cadherin; F, forward; R, reverse.
Western blot analysis
The cells were lysed using 2X lysis buffer (cat. no. P002A; Auragene Bioscience) on ice for 15 min. Protein (20 µg) was separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. After blocking with 5% milk at room temperature for 40 min, the membrane was incubated with primary antibodies against Raf-1 serine/threonine kinase proto-oncogene (Raf-1; cat no. ab32025; 1:250), phosphorylated (p)-Raf-1 (ab130572; 1:250) (both Abcam, Cambridge, UK), ERK (cat no. Sc-292838; 1:500), p-ERK (cat no. sc-101761; 1:500), E-cadherin (cat no. sc-31020; 1:500), N-cadherin (cat no. sc-7939; 1:500), vimentin (YT4480; 1:1,000) (all Santa Cruz Biotechnology, Inc.) and β-actin (cat no. LCA01; 1:2,000; Auragene Biosciences) at 4°C overnight. Subsequently, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit (cat. no. 111-035-003) or goat anti-mouse (cat. no. 111-035-008) secondary antibodies (both 1:5,000; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA), which was detected using an enhanced chemiluminescence kit (Auragene Bioscience). β-actin was used as the endogenous control. Image-Pro Plus software (version 6.0; Media Cybernetics, Inc., Rockville, MD, USA) was used to quantify the intensity of the bands.
MTT assay
The proliferation ability of cells was measured using the MTT cell viability assay. RASAL2-shRNA and Ct-shRNA plasmids were transfected into LoVo and HCT116 cells, and subsequently seeded into 96-well plates at a density of 3×103 cells/well and incubated for 3 days at 37°C. Every 24 h, 10 µl 0.5 mg/ml MTT solution (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) was added to each well and incubated for 4 h at 37°C. The absorbance values were measured at 490 nm using a microplate reader.
Wound healing assay
Scratch wounds were made using sterile 20 µl pipette tips when the cells had reached between 80 and 90% confluence. Cells were washed with serum-free PBS twice and cultured in fresh DMEM supplemented with 10% FBS for 24 h at 37°C. The medium was subsequently changed to DMEM supplemented with 3% FBS. Images were captured at 0 and 48 h using an inverted microscope.
Colony formation assay
A total of 48 h following transfection, 300 cells were plated into 6-well plates. After 10 days, visible colonies were fixed using 4% methanol for 15 min and stained with 1 ml Giemsa. The number of colonies was counted manually and normalized to the control group. Experiments were performed in triplicate.
Cell invasion assay
Cell invasion assays were performed in 24-well plates using an 8.0 µm BD Biocoat™ Matrigel™ Invasion Chamber (BD Biosciences, Franklin Lakes, NJ, USA). A total of 0.5 ml DMEM containing 10% FBS was added to the lower chamber at 37°C for 2 h and 0.2 ml (1×106 cells/ml) of cell solution was seeded into the upper chamber in serum-free culture medium. Cells were incubated for 24 h at 37°C. Cells were subsequently stained using crystal violet for 20 min and dissolved in 10% acetic acid. Absorbance at 570 nm was measured using a microplate reader. All experiments were performed independently in triplicate.
Cell adhesion assay
Cells were transfected with the RASAL2-shRNA or Ct-shRNA plasmids for 48 h at 37°C. A total of 100 µl cell solution was seeded into fibronectin-coated 96-well plates (5×105 cells/well) and incubated at 37°C for 2 h. Plates were washed with PBS four times to remove non-adherent cells, and the remaining cells were stained using crystal violet for 20 min and dissolved in 10% acetic acid. Absorbance at 570 nm was measured using a microplate reader. The percentage of cell adhesion was calculated relative to the original seeded cell solution's fluorescence at 570 nm (set to 100%).
Statistical analysis
Statistical analyses were performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Differences were evaluated using the Student t-test or one-way analysis of variance, followed by Fisher's least significant difference and Student-Newman-Keuls post hoc tests. Values are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulated RASAL2 expression is associated with CRC tumor grade and International Federation of Gynecology and Obstetrics (FIGO) stage
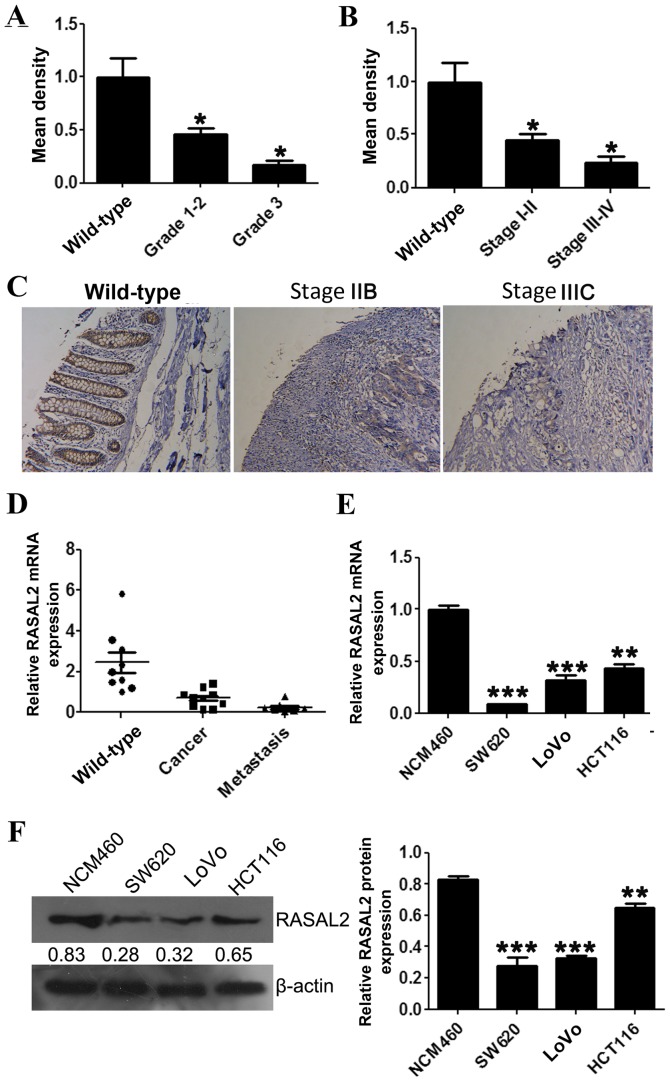
RASAL2 protein and mRNA expression levels in CRC tissues were evaluated using an IHC microarray and RT-qPCR, respectively. The IHC and RT-qPCR results confirmed that RASAL2 protein and mRNA levels were decreased in tumor tissue samples compared with wild-type colorectal tissues (Fig. 1A-D). Furthermore, significantly decreased RASAL2 protein expression was correlated with increased CRC tumor grade (P<0.05; Fig. 1A). Significantly decreased RASAL2 protein expression was also correlated with increased FIGO stage (18) (P<0.05; Fig. 1B).
Figure 1.
RASAL2 expression in CRC tissue samples and cell lines. RASAL2 expression was assessed using IHC staining of a human CRC tissue microarray. Relative RASAL2 protein expression in (A) grade 1–3 and (B) stage I–IV CRC tissue compared with wild-type colorectal tissue. (C) Representative images from the IHC stained tissue microarray. (D) RASAL2 mRNA expression in wild-type, cancerous and metastatic colorectal tissue samples relative to β-actin. RASAL2 (E) mRNA and (F) protein expression in the CRC cell lines SW620, LoVo and HCT116, relative to the wild-type colorectal cell line, NCM460. Values are presented as the mean ± standard deviation of triplicate data. *P<0.05, **P<0.01, ***P<0.001 vs. the wild-type tissue or cell line. RASAL2, RAS protein activator-like 2; CRC, colorectal carcinoma; IHC, immunohistochemistry.
RASAL2 mRNA and protein expression was evaluated in the CRC cell lines SW620, LoVo and HCT116, and the wild-type colon cell line NCM460, using RT-qPCR and western blotting, respectively. Consistent with the results observed in the tumor tissue samples, RASAL2 mRNA expression was significantly decreased in the CRC cell lines compared with the NCM460 cell line (all P<0.01; Fig. 1E), which was accompanied by a similar decrease in protein levels (all P<0.01; Fig. 1F). These results indicate that RASAL2 is downregulated in CRC tissues and cell lines.
Loss of RASAL2 promotes CRC cell proliferation
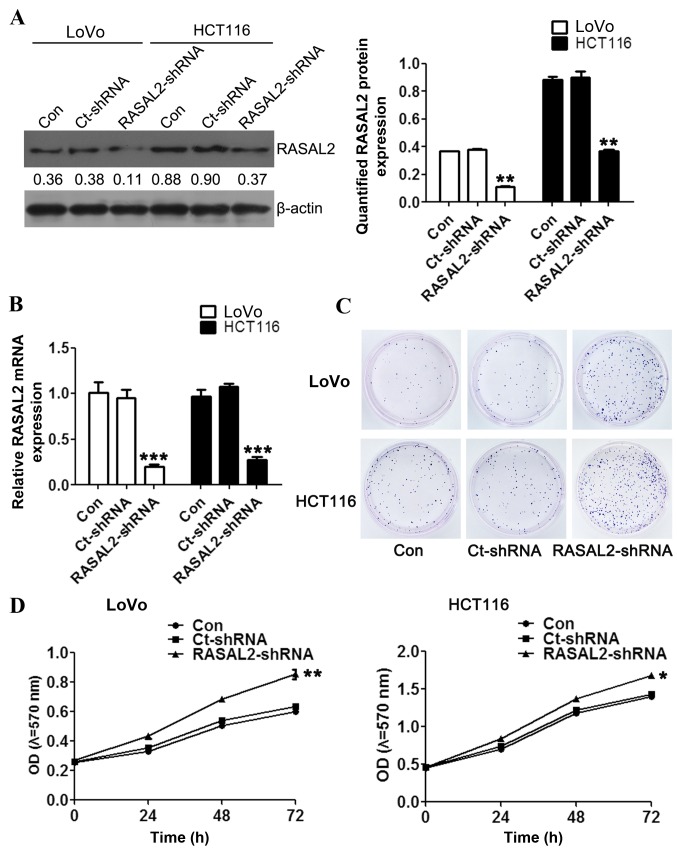
To investigate the function of RASAL2 in CRC pathogenesis, RASAL2 loss-of-function models were established using shRNA interference in the CRC cell lines LoVo and HCT116. Significantly decreased RASAL2 mRNA expression was detected in cells following transfection with the RASAL2- or Ct-shRNA (all P<0.01 vs. the control group; Fig. 2A), which was accompanied by a similar decrease in protein levels (both P<0.01; Fig. 2B). In order to investigate the effect of RASAL2 knockdown on tumorigenesis, cell proliferation was measured following transfection using the colony formation and MTT assays, respectively. The colony formation assay demonstrated increased colony formation in the LoVo and HCT116 cells following RASAL2-shRNA transfection compared with the untransfected and Ct-shRNA-transfected cells (Fig. 2C). Furthermore, RASAL2-shRNA transfection significantly enhanced the viability of the CRC cells compared with the untransfected cells (both P<0.05; Fig. 2D). These results indicate that RASAL2 suppresses CRC cell proliferation in vitro.
Figure 2.
RASAL2 knockdown promotes LoVo and HCT116 cell proliferation in vitro. (A) Decreased RASAL2 protein expression was validated using western blotting and quantified relative to β-actin. (B) Decreased RASAL2 mRNA expression was validated using the reverse transcription-quantitative polymerase chain reaction. (C) Representative images of the colony formation assay following RASAL2 shRNA knockdown. (D) Evaluation of cell viability following RASAL2-shRNA transfection using the MTT assay. Values are presented as the mean ± standard deviation of triplicate data. *P<0.05, **P<0.01, ***P<0.001 vs. the Con cells. RASAL2, RAS protein activator-like 2; shRNA, short hairpin RNA; Con, untransfected; Ct, control; OD, optical density.
RASAL2 suppresses CRC cell migration and invasion
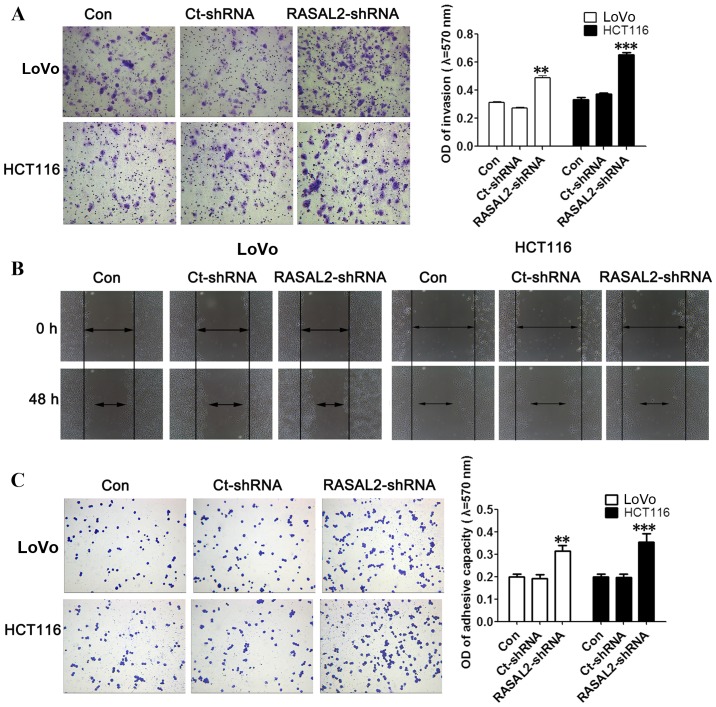
The effect of RASAL2 knockdown on the invasion and migration of LoVo and HCT116 cells was investigated using the cell invasion and wound healing assays, respectively. RASAL2 knockdown was demonstrated to significantly promote invasion (both P<0.01; Fig. 3A) and markedly increase migration (Fig. 3B) in LoVo and HCT116 cells compared with the untransfected control cells. In addition, the effect of RASAL2 on cell adhesion was evaluated using a cell adherence assay. RASAL2 knockdown significantly enhanced the adhesive ability of LoVo and HCT116 cells compared with the untransfected control cells (both P<0.01; Fig. 3C). These results demonstrate that RASAL2 knockdown promotes CRC cell invasion, migration and adhesion in vitro.
Figure 3.
RASAL2 is involved in the motility and invasion of LoVo and HCT116 cells. (A) The cell invasion assay demonstrated that RASAL2 knockdown significantly promoted cell invasion. (B) The wound healing assay demonstrated that RASAL2 knockdown promoted cell migration. (C) The cell adhesion assay demonstrated that RASAL2 knockdown significantly promoted cell adherence. Values are presented as the mean ± standard deviation of triplicate data. **P<0.01, ***P<0.001 vs. the Con cells. RASAL2, RAS protein activator-like 2; Con, untransfected; Ct, control; shRNA, short hairpin RNA; OD, optical density.
RASAL2 functions upstream of the ERK and EMT signaling pathways
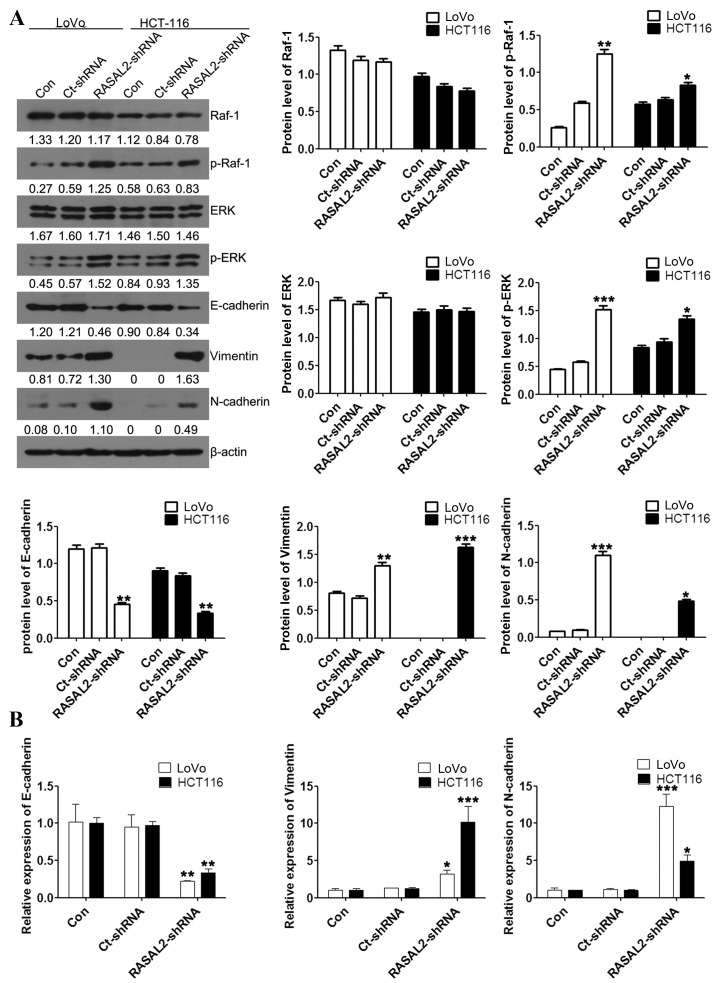
EMT is a critical event in the progression and metastasis of cancer (19). To investigate whether the effect of RASAL2 knockdown on CRC cell migration and invasion was EMT-mediated, the expression of several EMT markers was assessed using western blotting and RT-qPCR analyses. RASAL2 knockdown significantly increased the protein and mRNA levels of the mesenchymal markers vimentin and N-cadherin, while significantly decreasing the protein and mRNA expression of the epithelial marker E-cadherin, compared with the untransfected control cells (all P<0.05; Fig. 4). Furthermore, activation of the ERK and Raf-1 signaling pathways was evaluated using western blot analysis. This identified no significant difference in the protein expression of Raf-1 and ERK between the cell lines (all P>0.05; Fig. 4A). However, RASAL2 knockdown led to significantly increased expression of p-Raf-1 and p-ERK compared with the untransfected control cells (all P<0.05; Fig. 4A.) These results indicate that RASAL2 knockdown activates the ERK signaling pathway and promotes EMT in CRC cells in vitro.
Figure 4.
RASAL2 regulates EMT by activating the ERK signaling pathway in LoVo and HCT116 cells. (A) Raf-1, p-Raf-1, ERK, p-ERK, E-cadherin, vimentin and N-cadherin protein expression levels relative to β-actin were assessed using western blotting. (B) E-cadherin, vimentin and N-cadherin mRNA expression levels relative to β-actin were assessed using the reverse transcription-quantitative polymerase chain reaction. Values are presented as the mean ± standard deviation of triplicate data. *P<0.05, **P<0.01, ***P<0.001 vs. the Con cells. RASAL2, RAS protein activator-like 2; EMT, epithelial-mesenchymal transition; ERK, mitogen-activated protein kinase 1; Raf-1, Raf-1 serine/threonine kinase proto-oncogene; p, phosphorylated; Con, untransfected; Ct, control; shRNA, short hairpin RNA.
Discussion
The RAS gene family contains a number of proto-oncogenes involved in the proliferation, differentiation, apoptosis, cytoskeletal organization and motility of tumor cells. Mutations in RAS genes frequently lead to permanently activated RAS and thus its downstream effectors, which causes aberrant cell proliferation and potential tumorigenesis (14). Previous studies have demonstrated that mutation of RAS genes is the primary mechanism of aberrant RAS activation (8,20,21). However, other underlying mechanisms for RAS activation exist, which lead to abnormal activation of the RAS signaling pathway, including deregulation of RAS GTPase-activating proteins (RAS-GAPs) or RAS guanine nucleotide exchange factors (22).
A novel mechanism for the permanent activation of the RAS signaling pathway is the knockdown of RAS-GAPs. RASAL2 was reported to be mutated or suppressed in RAS- and ERK-hyperactivated breast cancer, where RASAL2 downregulation promoted tumor growth, progression and metastasis in two distinct human xenograft tumor models (14). Silencing of the RAS-GAPs RASAL1, DAB2 interacting protein (DAB2IP) and neurofibromin 1 leads to aberrant RAS activation in HCC cells (23). In a previous study, reactivation of RASAL1, DAB2IP and paired like homeodomain 1 inhibited cell growth, and induced apoptosis, while their silencing increased proliferation and inhibited apoptosis (23). In the present study, RASAL2 expression was downregulated in CRC tissue samples and cells. RASAL2 knockdown in CRC cells led to significantly increased expression of p-Raf-1 and p-ERK, indicating hyperactivation of the RAS signaling pathway.
Epithelial cell adhesion and communication, with the extracellular matrix and neighbouring cells, serves a fundamental role in tumour differentiation and progression (24,25). In addition, EMT is a critical event in cancer metastasis, during which differentiated epithelial cells obtain the characteristics of mesenchymal cells and subsequently migrate (26). During this process, epithelial marker expression is reduced and mesenchymal marker expression is increased (26). EMT in mammary epithelial cells typically facilitates tumor cell migration and invasion (27). Previously, RASAL2 was identified as a mediator of EMT and metastasis in ovarian cancer, lung adenocarcinoma and breast cancer (14–16). In the present study, RASAL2 was demonstrated to be a mediator of EMT, migration and invasion in CRC cells.
In conclusion, the present study confirmed that RASAL2 knockdown promotes proliferation and metastasis in CRC cells, through the activation of the RAS/ERK signaling pathway and subsequent induction of EMT in vitro. Therefore, further investigations of therapeutic strategies targeting RASAL2 for the treatment of CRC are warranted.
Acknowledgements
The present study was supported by the National and International Cooperation in Science and Technology Special Project (grant no. 2013DFA32150) and the Young Teachers Booster Project (grant no. 502010334).
References
- 1.Mao JD, Wu P, Huang JX, Wu J, Yang G. Role of ERK-MAPK signaling pathway in pentagastrin-regulated growth of large intestinal carcinoma. World J Gastroenterol. 2014;20:12542–12550. doi: 10.3748/wjg.v20.i35.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin HR, Carlos MC, Varghese C. Cancer control in the Asia Pacific region: Current status and concerns. Jpn J Clin Oncol. 2012;42:867–881. doi: 10.1093/jjco/hys077. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. Report of incidence and mortality in China cancer registries, 2009. Chin J Cancer Res. 2013;25:10–21. doi: 10.3978/j.issn.1000-9604.2012.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Song ZF, Xie RM, Pei J, Xiang MF, Wang H. Analysis of death causes of in-patients with malignant tumors in sichuan cancer hospital of China from 2002 to 2012. Asian Pac J Cancer Prev. 2013;14:4399–4402. doi: 10.7314/APJCP.2013.14.7.4399. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Lv Z, He G, Wang J, Zhang X, Lu G, Ren X, Wang F, Zhu X, Ding Y, et al. The SOX17/miR-371-5p/SOX2 axis inhibits EMT, stem cell properties and metastasis in colorectal cancer. Oncotarget. 2015;6:9099–9112. doi: 10.18632/oncotarget.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi H, Beppu T, Sakamoto Y, Miyamoto Y, Yokoyama N, Higashi T, Nitta H, Hashimoto D, Chikamoto A, Baba H. Prognostic value of Ki-67 expression in conversion therapy for colorectal liver-limited metastases. Am J Cancer Res. 2015;5:1225–1233. [PMC free article] [PubMed] [Google Scholar]
- 7.Ruers T, Bleichrodt RP. Treatment of liver metastases, an update on the possibilities and results. Eur J Cancer. 2002;38:1023–1033. doi: 10.1016/S0959-8049(02)00059-X. [DOI] [PubMed] [Google Scholar]
- 8.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 9.Malumbres M, Barbacid M. RAS oncogenes: The first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 10.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Deng Y, Ji Z, Liu H, Liu Y, Peng H, Wu J, Fan J. Identification of thyroid carcinoma related genes with mRMR and shortest path approaches. PLoS one. 2014;9:e94022. doi: 10.1371/journal.pone.0094022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefanska B, Cheishvili D, Suderman M, Arakelian A, Huang J, Hallett M, Han ZG, Al-Mahtab M, Akbar SM, Khan WA, et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin Cancer Res. 2014;20:3118–3132. doi: 10.1158/1078-0432.CCR-13-0283. [DOI] [PubMed] [Google Scholar]
- 13.Feng M, Bao Y, Li Z, Li J, Gong M, Lam S, Wang J, Marzese DM, Donovan N, Tan EY, et al. RASAL2 activates RAC1 to promote triple-negative breast cancer progression. J Clin Invest. 2014;124:5291–5304. doi: 10.1172/JCI76711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, Beroukhim R, Polyak K, Brown M, Kuperwasser C, Cichowski K. The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell. 2013;24:365–378. doi: 10.1016/j.ccr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Zhao M, Xu H, Wang K, Fu Z, Jiang Y, Yao Z. RASAL2 down-regulation in ovarian cancer promotes epithelial-mesenchymal transition and metastasis. Oncotarget. 2014;5:6734–6745. doi: 10.18632/oncotarget.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, Li S. RASAL2 promotes lung cancer metastasis through epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2014;455:358–362. doi: 10.1016/j.bbrc.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Kamata S, Uchida M. Relationship between TNM-classification and survival rate in cases of carcinoma of the oral cavity: A comparison between UICC and AJCC TNM staging systems. Gan No Rinsho. 1986;32:1339–1343. (In Japanese) [PubMed] [Google Scholar]
- 19.Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol Res Pract. 2015;211:557–569. doi: 10.1016/j.prp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Karnoub AE, Weinberg RA. Ras oncogenes: Split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Zhao JY, Qian XC, Cheng ZY, Liu Y, Wang Z. RASAL1 attenuates gastric carcinogenesis in nude mice by blocking RAS/ERK signaling. Asian Pac J Cancer Prev. 2015;16:1077–1082. doi: 10.7314/APJCP.2015.16.3.1077. [DOI] [PubMed] [Google Scholar]
- 23.Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An J, Lv J, Li A, Qiao J, Fang L, Li Z, Li B, Zhao W, Chen H, Wang L. Constitutive expression of Bcl-2 induces epithelial-Mesenchymal transition in mammary epithelial cells. BMC Cancer. 2015;15:476. doi: 10.1186/s12885-015-1485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Shi X, Peng Y, Wu M, Zhang P, Xie R, Wu Y, Yan Q, Liu S, Wang J. HIF-1α promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS One. 2015;10:e0129603. doi: 10.1371/journal.pone.0129603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keleg S, Büchler P, Ludwig R, Büchler MW, Friess H. Invasion and metastasis in pancreatic cancer. Mol Cancer. 2003;2:14. doi: 10.1186/1476-4598-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]