Abstract
The liver kinase B1 (LKB1)/5′-adenosine monophosphate-activated protein kinase pathway has been reported to facilitate glioma cell growth by improving growth conditions. To investigate the clinical significance of LKB1 in human gliomas western blot analysis and quantitative polymerase chain reaction experiments were performed. The present study demonstrated that LKB1 expression was markedly decreased at the messenger RNA and protein levels in 30 freshly prepared glioma tissues, compared with non-neoplastic brain tissues (P<0.001). Subsequently, immunohistochemical analysis demonstrated that LKB1 immunostaining in 180 glioma tissues was significantly decreased compared with that in the corresponding non-neoplastic brain tissues (P<0.001). Notably, this downregulation frequently occurred in high-grade gliomas, and statistical analysis revealed that low LKB1 expression was significantly associated with large tumor size (P=0.02), advanced World Health Organization grade (P=0.006) and low Karnofsky performance scale (P=0.01). The prognostic value of LKB1 expression in patients with glioma was additionally evaluated using Kaplan-Meier survival curves and Cox proportional hazards regression models. As a result, the overall survival time of patients with glioma with low LKB1 expression was shorter compared with that of patients with high LKB1 expression (P<0.001), and low LKB1 expression also indicated decreased survival time in patients with high-grade glioma (P<0.001). Collectively, the present data indicated that the downregulation of LKB1 was closely associated with the malignant degree of human gliomas, exhibiting lower expression at a higher grade. Notably, LKB1 may serve as a potential prognostic biomarker for patients with glioma following surgery.
Keywords: liver kinase B1, glioma, clinicopathological features, prognosis
Introduction
Human glioma is located within the brain or spinal cord, and represents the most frequently occurring type of malignant central nervous system tumor (1). The annual incidence rate of gliomas has increased, and accounts for 1.9% of total tumor incidences in the world (2). Gliomas are characterized by rapid growth, high infiltration and difficulty in surgical resection, and the majority of patients with glioma are diagnosed at stage IV (3). Treatment strategies for human gliomas primarily consist of surgery, followed by radiotherapy and chemotherapy. However, this malignancy often has a high degree of malignancy, recurrence rate, multi-drug resistance and invasiveness, leading to poor efficacy and final clinical outcomes (3). The 5-year survival rate for patients with gliomas is poor. Particularly, the median survival of patients who have glioblastoma multiforme (GBM) ranges between 9 and 12 months (4). Previous studies on molecular biology of tumors have indicated that human gliomas may be complex diseases caused by interactions among various genes, a series of multiple oncogene activation and tumor-suppressor inactivation (5,6). Therefore, it is of great clinical significance to understand the molecular pathogenesis of gliomas in order to identify novel drug targets to improve treatment efficacy and enhance patients' survival.
Liver kinase B1 (LKB1), also termed serine/threonine protein kinase 11, is located on chromosome 19p13.3 and encodes a ~48 kDa serine/threonine protein kinase (7). LKB1 was originally observed to be mutated in Peutz-Jeghers syndrome, a rare cancer susceptibility syndrome featured by predisposition to gastrointestinal polyposis, mucocutaneous melanin pigmentation and multi-organ cancer susceptibility (8). Functionally, LKB1 serves roles in multiple cellular processes, including cell structure control, cell cycle regulation, apoptosis and cellular metabolism (9). As a multifunctional protein, LKB1 acts as a key metabolic enzyme in the 5′ adenosine monophosphate-activated protein kinase (AMPK) pathway, and its inactivation often leads to the activation of the mammalian target of rapamycin pathway, which is important for controlling cellular energy metabolism, cell survival and growth under metabolic stress such as nutrient deficiency (10). Growing evidence shows that LKB1 acts as a tumor-suppressor gene by activating AMPK or AMPK-related kinases, which in turn regulate cell cycle, cell apoptosis, cell polarity and metabolism (11). In human gliomas, Godlewski et al (12) identified microRNA (miR)-451 as a regulator of the LKB1/AMPK pathway, which may be a fundamental mechanism contributing to cellular adaptation in response to altered energy availability. Jiang et al (13) reported that probucol, which exerts antitumor activities at various stages of tumor initiation, promotion and progression, suppressed human glioma cell proliferation in vitro via reactive oxygen species production and LKB1-AMPK activation. Although these previous findings suggested that the LKB1-AMPK pathway facilitates glioma cell growth, the clinical significance of LKB1 expression in large numbers of glioma patients remains unclear.
In the present study, western blotting and quantitative polymerase chain reaction (qPCR) were performed to examine the expression of LKB1 at the protein and messenger RNA (mRNA) levels in 30 pairs of freshly prepared glioma and non-neoplastic brain tissues. Subsequently, immunohistochemistry was used to validate the expression pattern of LKB1 protein in 180 patients with glioma. The associations between LKB1 immunoreactive scores and various clinicopathological characteristics were then statistically analyzed. Furthermore, the prognostic value of LKB1 expression in glioma patients was additionally evaluated using Kaplan-Meier survival curves and Cox proportional hazards regression models.
Materials and methods
Patients and tissue samples
The current study was approved by the Research Ethics Committee of The First Affiliated Hospital of Medical College, Shantou University (Shantou, China). All patients enrolled in the study provided written informed consent. All specimens were handled and made anonymous according to ethical and legal standards of The First Affiliated Hospital of Medical College, Shantou University, and were obtained under sterile conditions during surgery.
For western blotting and qPCR, 30 pairs of freshly prepared glioma and non-neoplastic brain tissues were obtained from the Department of Neurosurgery, The First Affiliated Hospital of Medical College, Shantou University (Shantou, China) between January 2010 and December 2014. The mean patient age was 50.6 years (range, 12–88 years), and 20 (66.67%) of them were male, while 10 (33.33%) were female. According to the clinicopathological criteria provided by the World Health Organization (WHO) (14): 3 patients were WHO grade I as pilocytic astrocytomas; 10 patients were grade II, including 5 patients with fibrillary astrocytoma, 3 patients with protoplasmic astrocytoma and 2 patients with oligodendroglioma; 12 patients were WHO grade III, including 8 patients with anaplastic astrocytoma and 4 patients with anaplastic oligodendroglioma; and 5 patients were WHO grade IV, all GBM.
For immunohistochemistry, 180 patients-derived paraffin-embedded glioma tissues were obtained from the Department of Neurosurgery, The First Affiliated Hospital of Medical College, Shantou University between January 2005 and December 2014. The mean patient age was 50.8 years (range, 10–86 years). Of these patients, 110 (61.11%) were male and 70 (38.89%) were female. According to the clinicopathological criteria provided by WHO (14), 15 patients were WHO grade I as pilocytic astrocytomas; 55 patients were WHO grade II, including 25 patients with fibrillary astrocytoma, 20 patients with protoplasmic astrocytoma and 10 patients with oligodendroglioma; 85 patients were WHO garde III, including 50 patients with anaplastic astrocytoma and 35 patients with anaplastic oligodendroglioma; and 25 patients were WHO grade IV, all GBM. Follow-up data were completed for all 180 patients, with a median follow-up time of 32 months (range, 2–118 months). The clinicopathological characteristics are summarized in Table I. All patients did not undergo any other treatments prior to surgery.
Table I.
Association of LKB1 expression with clinicopathological characteristics in human glioma.
| Clinicopathological characteristics | Patients, n (%) | LKB1-low, n (%) | P-value |
|---|---|---|---|
| Age, years | |||
| <50 | 88 (48.89) | 45 (51.14) | NS |
| ≥50 | 92 (51.11) | 47 (51.09) | |
| Gender | |||
| Male | 110 (61.11) | 56 (50.91) | NS |
| Female | 70 (38.89) | 36 (51.43) | |
| Tumor size, cm | |||
| <5 | 108 (60.00) | 40 (37.04) | 0.020 |
| ≥5 | 72 (40.00) | 52 (72.22) | |
| Tumor location | |||
| Supratentorial | 160 (88.89) | 82 (51.25) | NS |
| Subtentorial | 20 (11.11) | 10 (50.00) | |
| Karnofsky performance scale | |||
| <90 | 100 (60.00) | 72 (72.00) | 0.010 |
| ≥90 | 80 (40.00) | 20 (25.00) | |
| WHO grade | |||
| I | 15 (12.00) | 0 (0.00) | 0.006 |
| II | 55 (36.00) | 15 (27.27) | |
| III | 85 (42.00) | 52 (61.18) | |
| IV | 25 (10.00) | 25 (100.00) | |
| Tumor recurrence | |||
| Absent | 118 (65.56) | 62 (52.54) | NS |
| Present | 62 (34.44) | 30 (48.39) |
LKB1, liver kinase B1; NS, no significance difference; WHO, World Health Organization.
qPCR
qPCR was performed to detect the expression of LKB1 mRNA in 30 pairs of freshly prepared glioma and non-neoplastic brain tissues. Total RNA was extracted from 30 pairs of freshly prepared glioma and non-neoplastic brain tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. The RNA was pretreated with DNase, and single-stranded complementary DNA was synthesized using the SuperScript First-Strand Synthesis System or RT-PCR (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Relative expression of LKB1 mRNA was determined using a SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and an ABI 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.), and normalized using GAPDH. The primer sequences used in the present study were as follows: LKB1 forward, 5′-AGGGATGCTTGAGTACGAACC-3′ and reverse, 5′-GTCCTCCAAGTACGGCACC-3′; GAPDH forward, 5′-TGAACGGGAAGCTCACTGG-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′. PCR amplifications for each gene were repeated three times. The fold-change of each gene was calculated using the 2−ΔΔCq method (15,16).
Western blot analysis
Western blot analysis was performed to detect the expression of LKB1 protein in 30 pairs of freshly prepared glioma and non-neoplastic brain tissues. Tissue samples were ground with liquid nitrogen and lysed at 48°C for 30 min in lysis buffer (50 mM Tris, pH 7.4, 100 mM NaCl2, 1 mM MgCl2, 2.5 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 2.5 mM EDTA, 0.5 % Triton X-100, 0.5% NP-40, and 5 mg/ml each of aprotinin, pepstatin A and leupeptin), and centrifuged at 10,000 × g for 30 min to collect the supernatant. Protein concentration was determined using the Bradford method. An equivalent amount of protein from each sample was separated by 10% SDS-PAGE and then transferred to a polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA, USA), followed by incubation in blocking buffer (PBS containing 5% nonfat milk) for 2 h at room temperature. Subsequently, the membrane was incubated with a goat polyclonal antibody against LKB1 (dilution, 1:200; catalogue no., sc-5638; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Next, the membrane was washed twice with PBS for 5 min and incubated with secondary horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin G (IgG) antibody (dilution, 1:1,000; catalogue no., sc-2949; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature. GAPDH was detected using a specific goat polyclonal antibody (dilution, 1:500; catalogue no., sc-20358; Santa Cruz Biotechnology, Inc.) as a loading control. Proteins were visualized using an enhanced chemiluminescence reagent (Santa Cruz Biotechnology, Inc.). Relative expression of LKB1 protein was normalized to GAPDH. The experiments were repeated three times.
Immunohistochemistry
Immunohistochemistry was performed to detect the expression pattern and subcellular localization of LKB1 protein in 180 pairs of paraffin-embedded glioma and non-neoplastic brain tissues. Tissue sections were dewaxed in xylene, rehydrated using graded alcohol and soaked in 0.3% hydrogen peroxide to block endogenous peroxidase activity. The sections were then rinsed in PBS, pH 7.2, and 10% goat serum (Gibco; Thermo Fisher Scientific, Inc.) was applied for 1.5 h at room temperature to block nonspecific reactions. The aforementioned goat polyclonal antibody against LKB1 was incubated with the tissue sections overnight at 4°C. Negative control slides were processed in parallel using a nonspecific IgG (dilution, 1:1,000; catalogue no., sc-2949; Santa Cruz Biotechnology, Inc.) at the same concentration as the primary antibody. Subsequent to being rinsed in PBS, the peroxidase reaction was visualized by incubating the sections with a solution containing 0.1% phosphate buffer, 0.02% 3,3′-diaminobenzidine tetrahydrochloride and 3% H2O2. Finally, the sections were counterstained with hematoxylin, dehydrated and mounted in mounting resin.
The immunohistochemical results were evaluated by two independent pathologists blinded to the patients' clinicopathological characteristics. The immunoreactive scoring (IRS) of LKB1 expression was calculated by combining the intensity and the percentage of positive cells. The percentage of positive cells was as follows: 0–5% scored 0; 6–35% scored 1; 36–70% scored 2; and >70% scored 3. The staining intensity was as follows: No staining scored 0; weakly staining scored 1; moderately staining scored 2; and strongly staining scored 3. The final IRS was designated as low or high expression using the percentage of positive cell score × staining intensity score.
Statistical analysis
SPSS software version 11.0 for Windows (SPSS, Inc., Chicago, IL, USA) was used for the statistical analyses. All data obtained from experiments were expressed as the mean ± standard deviation. The differences of LKB1 mRNA and protein expression between glioma and non-neoplastic brain tissues were evaluated by Student's paired t-test. Spearman's rank correlation analysis was used to analyze the correlation between the level of LKB1 mRNA and protein expression. The associations between LKB1 expression and various clinicopathological features were analyzed by the χ2 test. Kaplan-Meier survival plots were constructed to analyze survival data. Univariate and multivariate analyses were performed using Cox's proportional hazards model. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of LKB1 mRNA and protein in glioma tissues
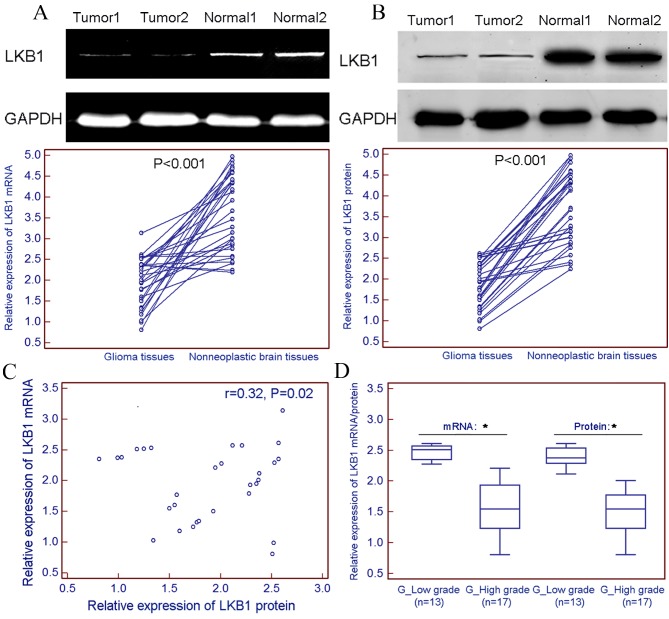
LKB1 expression was markedly decreased at the mRNA (tumor vs. normal, 1.96±0.59 vs. 3.66±0.92; P<0.001; Fig. 1A) and protein (tumor vs. normal, 1.89±0.54 vs. 3.71±0.87; P<0.001; Fig. 1B) levels in 30 freshly prepared glioma tissues, compared with non-neoplastic brain tissues. Spearman's rank correlation analysis revealed that the expression levels of LKB1 mRNA in glioma tissues were closely correlated with those of LKB1 protein in gliomas (r=0.32; P=0.02; Fig. 1C). Notably, the present results indicated a relatively lower expression level of LKB1 mRNA and protein in freshly prepared high-grade glioma tissue samples (WHO grades III–IV) compared with those in the low-grade glioma samples (WHO grades I–II; Fig. 1D).
Figure 1.
Downregulation of LKB1 mRNA and protein expression in glioma tissues. LKB1 expression was markedly decreased at (A) mRNA (tumor vs. normal, 1.96±0.59 vs. 3.66±0.92; P<0.001), and (B) protein (tumor vs. normal, 1.89±0.54 vs. 3.71±0.87; P<0.001) levels in 30 freshly prepared glioma tissues, compared with that in non-neoplastic brain tissues. (C) Spearman's rank correlation analysis revealed that the expression levels of LKB1 mRNA in glioma tissues were closely correlated with those of LKB1 protein in gliomas (r=0.32; P=0.02). (D) Relatively lower expression levels of LKB1 mRNA and protein were observed in freshly prepared high-grade glioma tissue samples (WHO grades III–IV) compared with those in low-grade glioma samples (WHO grades I–II). *P<0.05. LKB1, liver kinase B1; WHO, World Health Organization; mRNA, messenger RNA.
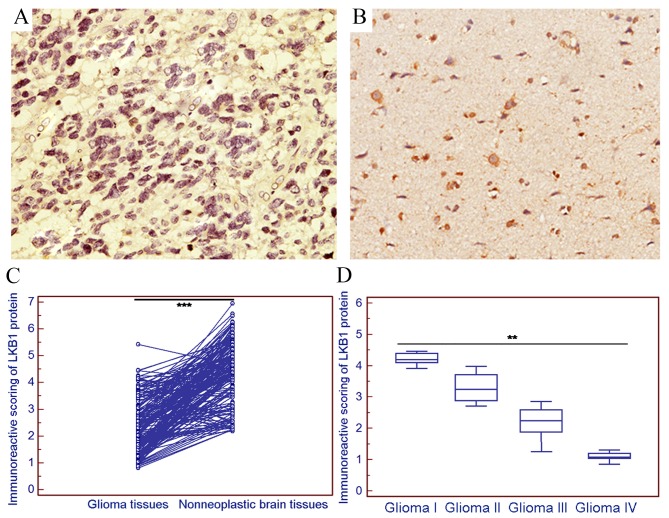
The positive expression of LKB1 protein was examined in 36/180 (20.00%) gliomas in the present cohort (Fig. 2A), and markedly strong LKB1 protein expression was observed in 132/180 (73.33%) non-neoplastic brain tissue samples (Fig. 2B). Statistically, the IRS of LKB1 protein expression in gliomas was significantly decreased compared with that in the corresponding non-neoplastic brain tissues (tumor vs. normal, 2.38±1.00 vs. 4.38±1.19; P<0.001; Fig. 2C). Notably, the expression of LKB1 in high-grade glioma was decreased compared with that in low-grade glioma (P<0.01; Fig. 2D).
Figure 2.
Immunostaining of LKB1 protein in glioma tissues. (A) Weak immunostaining of LKB1 protein in glioma tissues (magnification, ×200). (B) Strong immunostaining of LKB1 protein in non-neoplastic brain tissue samples (magnification, ×200). (C) Statistically, the IRS of LKB1 protein in gliomas was significantly decreased compared with that in the corresponding non-neoplastic brain tissues (tumor vs. normal, 2.38±1.00 vs. 4.38±1.19; ***P<0.001). (D) The expression of LKB1 in high-grade glioma was decreased compared with that in low-grade glioma (**P<0.01). LKB1, liver kinase B1; IRS, immunoreactive scoring.
Downregulation of LKB1 protein is associated with aggressive tumor progression of human gliomas
To explore the clinical significance of LKB1 downregulation in human gliomas, the associations between LKB1 and various clinicopathological parameters of glioma patients was statistically analyzed. The median value (2.31) of the IRS of LKB1 protein expression in glioma tissues was used as a cut-off point to divide all 180 patients with glioma into low LKB1 expression group (LKB1-low; n=98) and high LKB1 expression group (LKB1-high; n=82). Table I summarizes the associations between LKB1 expression and various clinicopathological parameters of glioma patients. As a result, low LKB1 expression was significantly associated with large tumor size (P=0.02; Table I), advanced WHO grade (P=0.006; Table I) and low Karnofsky performance scale (P=0.01; Table I). No significant associations of LKB1 with patients' age, gender, tumor location or recurrence were observed (Table I).
Downregulation of LKB1 protein predicts poor prognosis in patients with gliomas
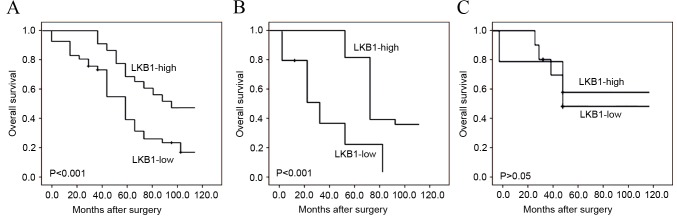
To additionally evaluate the prognostic implication of LKB1 downregulation in human gliomas, univariate and multivariate analyses using the Cox's proportional hazards model were constructed, including patients' age, gender, tumor size, tumor location, WHO grade, Karnofsky performance scale, tumor recurrence and LKB1 expression. Kaplan-Meier survival plots in Fig. 3A revealed that the overall survival of glioma patients with low LKB1 expression was clearly shorter compared with that of patients with high LKB1 expression (P<0.001). Notably, the subgroup analysis based on WHO classification demonstrated that low LKB1 expression indicated a poorer survival in high-grade glioma patients (P<0.001; Fig. 3B), but not in low-grade glioma patients (Fig. 3C). Additionally, the results of univariate and multivariate analyses revealed that LKB1 expression (P<0.001), WHO grade (P<0.001) and Karnofsky performance scale (P=0.01) were independent prognostic factors for overall survival of glioma patients (Table II).
Figure 3.
Kaplan-Meier survival curves of glioma patients. (A) Overall survival of all 180 glioma patients with LKB1 protein expression: High vs. low. (B) Overall survival of high-grade glioma patients with LKB1 protein expression: High vs. low. (C) Overall survival of low-grade glioma patients with LKB1 protein expression: High vs. low. LKB1, liver kinase B1.
Table II.
Association of various candidate prognostic factors with overall survival of 180 glioma patients, as evaluated by Cox regression analysis.
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Variable | Groups | Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value |
| Age, years | <50 vs. ≥50 | 1.082 (0.568–1.966) | NS | – | – |
| Gender | Male vs. female | 1.021 (0.553–1.922) | NS | – | – |
| Tumor size, cm | <5 vs. ≥5 | 1.168 (0.689–2.139) | NS | – | – |
| Tumor location | Supratentorial vs. subtentorial | 1.026 (0.581–1.938) | NS | – | – |
| Karnofsky performance scale | <90 vs. ≥90 | 2.689 (1.291–5.693) | 0.010 | 2.043 (0.896–4.338) | 0.030 |
| WHO grade | I–II vs. III–IV | 3.625 (1.673–7.932) | <0.001 | 3.267 (1.392–6.581) | 0.001 |
| Tumor recurrence | Absent vs. present | 2.018 (0.819–3.983) | NS | – | – |
| LKB1 expression | Absent vs. present | 3.348 (1.397–7.026) | <0.001 | 3.022 (1.002–6.016) | 0.001 |
WHO, World Health Organization; LKB1, liver kinase B1; NS, no significance; CI, confidence interval.
Discussion
Human gliomas, as a group of highly aggressive, angiogenic and incurable malignancies, exhibit poor clinical outcomes (3). Thus, there is an urgent requirement to screen novel and efficient molecular markers for diagnosis and prognosis in patients with gliomas, and to develop new therapeutic strategies. In the current study, decreased expression of LKB1 mRNA and protein levels in glioma tissues were observed, compared with those in adjacent normal brain tissues. Subsequently, immunohistochemistry was performed to observe the expression of LKB1 protein in glioma tissues of different stage based on complete follow-up data and in the corresponding non-neoplastic brain tissue specimens. The present data demonstrated that the mean IRS of LKB1 protein expression in glioma tissues, particularly in high-grade glioma tissues, was significantly increased compared with that in the corresponding non-neoplastic brain tissues. These observations strongly suggest that the evaluation of LKB1 expression using immunohistochemistry discriminated between glioma and non-neoplastic brain tissues. In addition, LKB1 downregulation was significantly associated with larger tumor size, higher WHO grade and lower Karnofsky performance scale. Notably, the results revealed that LKB1 downregulation appeared to be an independent prognostic factor for overall survival in glioma patients, indicating that the detection of LKB1 may be valuable for the production of individual therapies and for the identification of patients who may or may not benefit from close monitoring subsequent to surgery. Additional research is required to validate these findings.
LKB1, a serine/threonine kinase, has been indicated to function as a tumor suppressor in various human cancers via regulating cancer cell growth, metabolism, survival and polarity (17). Mutations of LKB1 protein, such as K78I, D17GN, W308C and L67P, result in the loss of its kinase activity, and have been identified in different cancers, including malignant melanoma, head and neck squamous cell carcinoma, breast cancer, lung cancer, hepatocellular carcinoma, pancreatic and biliary carcinoma, colorectal cancer, cervical adenocarcinoma and testicular cancer (18–22). In addition, LKB1 expression has been documented to be regulated through epigenetic modification, such as hypermethylation in gene promoter CpG islands and global demethylation of the genome, transcriptional regulation and post-translational modification, such as phosphorylation, prenylation and ubiquitination (20). Aberrant expression of LKB1 gene and protein has been observed in multiple malignancies. For example, Huang et al (23) reported that LKB1 expression was decreased in hepatocellular carcinoma samples, and noticed that loss of LKB1 expression was significantly associated with aggressive clinical features and poor disease-free and overall survival. He et al (24) indicated that LKB1 loss at transcriptional level promoted tumor malignancy and poor patient outcomes in colorectal cancer. Yang et al (25) reported that the protein expression levels of LKB1 were significantly reduced in six pancreatic ductal adenocarcinoma cell lines and downregulated in 31.3% of pancreatic ductal adenocarcinoma lesions, compared with those in matched non-tumorous tissues. The authors also confirmed that decreased LKB1 expression predicted poor prognosis in pancreatic ductal adenocarcinoma. Shen et al (26) demonstrated that low LKB1 protein expression correlated with higher histological grade, larger tumor size, progesterone receptor status, presence of lymph node metastasis, higher relapse rate and poorer overall survival time. Jiang et al (27) revealed that the expression levels of LKB1 mRNA and protein were significantly reduced in non-small cell lung cancer tissues compared with those in the matched surrounding normal lung tissues, and determined that reduced expression of LKB1 was associated with poor survival of non-small cell lung cancer patients. Similar results were also obtained by the present analyses regarding the expression pattern and clinical implication of LKB1 in human gliomas.
In conclusion, the current data indicated that downregulation of LKB1 was closely associated with the malignant degree of human gliomas, displaying lower expression at a higher grade. Notably, LKB1 may serve as a potential prognostic biomarker for glioma patients following surgery.
References
- 1.Shapiro WR, Shapiro JR. Biology and treatment of malignant glioma. Oncology (Williston Park) 1998;12:233–240. [PubMed] [Google Scholar]
- 2.Preusser M, Haberler C, Hainfellner JA. Malignant glioma: Neuropathology and neurobiology. Wien Med Wochenschr. 2006;156:332–337. doi: 10.1007/s10354-006-0304-7. [DOI] [PubMed] [Google Scholar]
- 3.Wang B, Li M, Wu Z, Li X, Li Y, Shi X, Cheng W. Associations between SOX2 and miR-200b expression with the clinicopathological characteristics and prognosis of patients with glioma. Exp Ther Med. 2015;10:88–96. doi: 10.3892/etm.2015.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong ML, Kaye AH, Hovens CM. Targeting malignant glioma survival signalling to improve clinical outcomes. J Clin Neurosci. 2007;14:301–308. doi: 10.1016/j.jocn.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Qi J, Yang H, Wang X, Tu Y. The progress in molecular biomarkers of gliomas. Cancer Transl Med. 2016;2:125–129. doi: 10.4103/2395-3977.189305. [DOI] [Google Scholar]
- 6.Rizzo D, Ruggiero A, Martini M, Rizzo V, Maurizi P, Riccardi R. Molecular biology in pediatric high-grade glioma: Impact on prognosis and treatment. Biomed Res Int. 2015;2015:215135. doi: 10.1155/2015/215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun R, Li J, Wang B, Guo Y, Ma L, Quan X, Chu Z, Li T. Liver kinase B1 promoter CpG island methylation is related to lung cancer and smoking. Int J Clin Exp Med. 2015;8:14070–14074. [PMC free article] [PubMed] [Google Scholar]
- 8.Rhodes LV, Tate CR, Hoang VT, Burks HE, Gilliam D, Martin EC, Elliott S, Miller DB, Buechlein A, Rusch D, et al. Regulation of triple-negative breast cancer cell metastasis by the tumor-suppressor liver kinase B1. Oncogenesis. 2015;4:e168. doi: 10.1038/oncsis.2015.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng DY, Song H, Liu LB. Resveratrol-downregulated phosphorylated liver kinase B1 is involved in senescence of acute myeloid leukemia stem cells. J Huazhong Univ Sci Technolog Med Sci. 2015;35:485–489. doi: 10.1007/s11596-015-1457-7. [DOI] [PubMed] [Google Scholar]
- 10.Rao F, Xu J, Fu C, Cha JY, Gadalla MM, Xu R, Barrow JC, Snyder SH. Inositol pyrophosphates promote tumor growth and metastasis by antagonizing liver kinase B1. Proc Natl Acad Sci USA. 2015;112:1773–1778. doi: 10.1073/pnas.1424642112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia C, Ye F, Hu X, Li Z, Jiang B, Fu Y, Cheng X, Shao Z, Zhuang Z. Liver kinase B1 enhances chemoresistance to gemcitabine in breast cancer MDA-MB-231 cells. Oncol Lett. 2014;8:2086–2092. doi: 10.3892/ol.2014.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godlewski J, Bronisz A, Nowicki MO, Chiocca EA, Lawler S. microRNA-451: A conditional switch controlling glioma cell proliferation and migration. Cell Cycle. 2010;9:2742–2748. doi: 10.4161/cc.9.14.12248. [DOI] [PubMed] [Google Scholar]
- 13.Jiang YS, Lei JA, Feng F, Liang QM, Wang FR. Probucol suppresses human glioma cell proliferation in vitro via ROS production and LKB1-AMPK activation. Acta Pharmacol Sin. 2014;35:1556–1565. doi: 10.1038/aps.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Vigneswaran K, Neill S, Hadjipanayis CG. Beyond the world health organization grading of infiltrating gliomas: Advances in the molecular genetics of glioma classification. Ann Transl Med. 2015;3:95. doi: 10.3978/j.issn.2305-5839.2015.03.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballester M, Castelló A, Ibáñez E, Sánchez A, Folch JM. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. Biotechniques. 2004;37:610–613. doi: 10.2144/04374ST06. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Lu J, Sun P, Sun B, Wang C. Low LKB1 expression results in unfavorable prognosis in prostate cancer patients. Med Sci Monit. 2015;21:3722–3727. doi: 10.12659/MSM.894847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Momcilovic M, McMickle R, Abt E, Seki A, Simko SA, Magyar C, Stout DB, Fishbein MC, Walser TC, Dubinett SM, Shackelford DB. Heightening energetic stress selectively targets LKB1-deficient non-small cell lung cancers. Cancer Res. 2015;75:4910–4922. doi: 10.1158/0008-5472.CAN-15-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan RY, Li HB. Recent progress on liver kinase B1 (LKB1): Expression, regulation, downstream signaling and cancer suppressive function. Int J Mol Sci. 2014;15:16698–16718. doi: 10.3390/ijms150916698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Momcilovic M, Shackelford DB. Targeting LKB1 in cancer-exposing and exploiting vulnerabilities. Br J Cancer. 2015;113:574–584. doi: 10.1038/bjc.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, Zhang J, Marcus AI. LKB1 Tumor Suppressor: Therapeutic opportunities knock when LKB1 is inactivated. Genes Dis. 2014;1:64–74. doi: 10.1016/j.gendis.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monteverde T, Muthalagu N, Port J, Murphy DJ. Evidence of cancer-promoting roles for AMPK and related kinases. FEBS J. 2015;282:4658–4651. doi: 10.1111/febs.13534. [DOI] [PubMed] [Google Scholar]
- 23.Huang YH, Chen ZK, Huang KT, Li P, He B, Guo X, Zhong JQ, Zhang QY, Shi HQ, Song QT, et al. Decreased expression of LKB1 correlates with poor prognosis in hepatocellular carcinoma patients undergoing hepatectomy. Asian Pac J Cancer Prev. 2013;14:1985–1988. doi: 10.7314/APJCP.2013.14.3.1985. [DOI] [PubMed] [Google Scholar]
- 24.He TY, Tsai LH, Huang CC, Chou MC, Lee H. LKB1 loss at transcriptional level promotes tumor malignancy and poor patient outcomes in colorectal cancer. Ann Surg Oncol. 2014;21 doi: 10.1245/s10434-014-3824-1. (Suppl 4) S703-S710. [DOI] [PubMed] [Google Scholar]
- 25.Yang JY, Jiang SH, Liu DJ, Yang XM, Huo YM, Li J, Hua R, Zhang ZG, Sun YW. Decreased LKB1 predicts poor prognosis in Pancreatic Ductal Adenocarcinoma. Sci Rep. 2015;5:10575. doi: 10.1038/srep10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Z, Wen XF, Lan F, Shen ZZ, Shao ZM. The tumor suppressor gene LKB1 is associated with prognosis in human breast carcinoma. Clin Cancer Res. 2002;8:2085–2090. [PubMed] [Google Scholar]
- 27.Jiang L, Liang X, Liu M, Wang W, Ma J, Guo Q, Han L, Yang C, Nan K. Reduced expression of liver kinase B1 and Beclin1 is associated with the poor survival of patients with non-small cell lung cancer. Oncol Rep. 2014;32:1931–1958. doi: 10.3892/or.2014.3432. [DOI] [PubMed] [Google Scholar]