Abstract
The DNA mismatch repair (MMR) gene MutL homolog 1 (MLH1) is critical for the maintenance of genomic integrity. Methylation of the MLH1 gene promoter was identified as a prognostic marker for numerous types of cancer including glioblastoma, colorectal, ovarian and gastric cancer. The present study aimed to determine whether MLH1 promoter methylation was associated with survival in male patients with esophageal squamous cell carcinoma (ESCC). Formalin-fixed, paraffin-embedded ESCC tissues were collected from 87 male patients. MLH1 promoter methylation was assessed using the methylation-specific polymerase chain reaction approach. Kaplan-Meier survival curves and log-rank tests were used to evaluate the association between MLH1 promoter methylation and overall survival (OS) in patients with ESCC. Cox regression analysis was used to obtain crude and multivariate hazard ratios (HR), and 95% confidence intervals (CI). The present study revealed that MLH1 promoter methylation was observed in 53/87 (60.9%) of male patients with ESCC. Kaplan-Meier survival analysis demonstrated that MLH1 promoter hypermethylation was significantly associated with poorer prognosis in patients with ESCC (P=0.048). Multivariate survival analysis revealed that MLH1 promoter hypermethylation was an independent predictor of poor OS in male patients with ESCC (HR=1.716; 95% CI=1.008–2.921). Therefore, MLH1 promoter hypermethylation may be a predictor of prognosis in male patients with ESCC.
Keywords: esophageal squamous cell carcinoma, MutL homolog 1, survival, methylation, methylation specific polymerase chain reaction
Introduction
Esophageal cancer (EC) is a lethal disease that presents a global health threat to humans (1) particularly in developing countries, including China (2). A major histological subtype of EC is esophageal squamous cell carcinoma (ESCC) (3). Despite medical and surgical advances, the prognosis for EC remains poor (4).
Esophageal cancer may be caused by various environmental factors, including chronic exposure to nitrosamines, obesity, smoking and alcohol (5). Tobacco and alcohol consumption are the predominant risk factors for ESCC (6,7). In addition, accumulating epidemiological studies indicated an association between obesity and EC (2,8–10).
Encompassing the genetic and environmental aspects of cancer development, epigenetic modification is considered to have a crucial role in the carcinogenesis of EC (11). DNA methylation is as a key mechanism in the inhibition of the expression of tumor suppressor genes (12). Aberrantly methylated genes have previously been identified as prognostic markers in EC (13,14).
DNA mismatch repair (MMR) genes, including MutL homolog (MLH)1, MLH3, MutS protein homolog (MSH)2 and MSH3, are critical for maintaining genomic integrity (15,16). The loss of MMR function is closely correlated with carcinogenesis (17–20). MLH1 encodes a protein that has been implicated in the maintenance of genome stability during DNA replication (21–23). A previous study indicated that MLH1 promoter hypermethylation was able to inactivate gene transcription, inducing defects in the function of the DNA repair system (24). Therefore, MLH1 contributes to the subsequent development of tumors including ESCC (24,25). MLH1 promoter methylation is significantly higher in EC tissues, as compared with paired adjacent tissues (15); however, the association between MLH1 promoter methylation and the prognosis of EC has yet to be elucidated.
In the current study, MLH1 methylation was investigated in a total of 87 male patients with ESCC, with the aim of determining the prognostic value of MLH1 methylation in this disease.
Materials and methods
Tissue samples
In a retrospective study, formalin-fixed paraffin-embedded (FFPE) tissues (4 µm in thickness) from the primary tumors of 87 male patients diagnosed with ESCC were selected from the archive at the Department of Thoracic Surgery, Shaoxing People's Hospital (Shaoxing, China) between October 1998 and June 2007. No preoperative radiotherapy or chemotherapy was administered prior to the collection of FFPE samples. The paraffin-embedded tissue block of ESCC was sliced transversely using Leica RM2245 Semi-Automated Rotary Microtome (Leica Microsystems GmbH, Wetzlar, Germany). All sections were reviewed with minimum of 75% malignant cells. Pathological parameters were defined according to the World Health Organization guidelines and Union for International Cancer Control tumor-node-metastasis classifications (26). Follow-up data was recorded for 70/87 patients until January 2010, and the maximum follow up period was 11 years and 3 months. The overall survival (OS) time was evaluated from the date of primary surgery to patient mortality, or to the date of the final follow-up (27). Information on clinicopathological characteristics was retrieved from the patient medical records and pathological diagnosis. The present study was approved by the Bioethics Committee of Shaoxing People's Hospital and written informed consent was obtained from all participants.
DNA isolation and bisulfite conversion
Genomic DNA was extracted from the tissue samples using the E.Z.N.A® Tissue DNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the manufacturer's protocol. DNA concentrations were evaluated using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation-Gold™ kit (Zymo Research Corp., Irvine, CA, USA), which converted only unmethylated cytosine to uracil.
Methylation-specific polymerase chain reaction (MSP)
MSP was performed to qualitatively detect the methylation status of MLH1 (Fig. 1). The total amplification involved a reaction volume of 20 µl, containing 0.5 µl forward and reverse primers, 1.6 µl sodium bisulfate-modified DNA, 10 µl ZymoTaq™ PreMix (Zymo Research Corp.) and 7.4 µl DNase/RNase-free water. The applied MSP primers were as described previously (28), whereas the primer sequences for the MLH1 methylated (M) alleles were as follows: Forward, 5′-AACGAATTAATAGGAAGAGCGGATAGCG-3′ and reverse, 5′-CGTCCCTCCCTAAAACGACTACCC-3′. The primer sequences for MLH1 unmethylated (U) alleles were forward, 5′-TAAAAATGAATTAATAGGAAGAGTGGATAGTG-3′ and reverse, 5′-AATCTCTTCATCCCTCCCTAAAACA-3′. For PCR, the M and U primer pairs were initially denatured at 95°C for 10 min, followed by 35 cycles with 30 sec denaturation, 45 sec annealing at 55°C, and 1 min extension at 72°C. Following a subsequent 7 min extension at 72°C, the product was stored at 4°C. A number of DNA samples were also randomly sequenced to determine a complete bisulfite conversion, using the Applied Biosystems™ 3730 DNA Analyzer (Thermo Fisher Scientific, Inc., Waltham, MA, USA; Fig. 1).
Figure 1.
Representative results of sequencing validation and methylation-specific polymerase chain reaction. (A) Top row of nucleotides represents the original sequence of the gene, and the second row provides the converted sequences. (B) M and U represent methylated (91 bp) and unmethylated (102 bp) MutL homolog 1 products. MLH1, MutL homolog 1; MLHI-MF, represents the use of the forward methylated primer for sequencing; MLHI-UF, represents the use of the forward unmethylated primer for sequencing.
Statistical analysis
Statistical analysis was conducted with SPSS 18.0 software (SPSS Inc., Chicago, IL, USA). The Fisher's exact probability method or χ2 test was performed to compare the methylation frequencies between the groups. The Kaplan-Meier method was used to estimate the medium OS time for survival analyses. Log-rank tests were used for comparison of the survival curves. Potentially important factors in univariate analyses were then included in multivariate analyses. The Cox proportional hazards models was applied to calculate hazard ratios (HR) and the corresponding 95% confidence intervals (CI). P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
As presented in Table I, the median survival time of patients with ESCC was 74.0 months, ranging from 1.5-135 months. The mean age of patients with ESCC was 62.63±8.48 years. Of the 87 patients with ESCC, 34 (39.1%) and 53 (60.9%) cases were in the early-stage (I and II) and the advanced-stage (III and IV), respectively. In addition, 43 (49.4%) and 42 (48.3%) patients consumed alcohol and smoked cigarettes, respectively. Patients were classified as being alcohol consumers if they had consumed at least one alcoholic drink in the last 30 days (29). Smoking was classified based upon whether the patient had smoked continuously for >6 months during their lifetime (30).
Table I.
Baseline clinical phenotypes and MLH1 promoter methylation in patients with ESCC.
| Variables | N | MLH1 promoter methylation | MLH1 promoter unmethylation | P-value |
|---|---|---|---|---|
| Age at surgery, years | 0.810 | |||
| ≤60 | 37 | 22 | 15 | |
| >60 | 50 | 31 | 19 | |
| Tumor location | 0.600a | |||
| Upper | 4 | 2 | 2 | |
| Middle | 39 | 22 | 17 | |
| Lower | 44 | 29 | 15 | |
| Differentiation | 0.439a | |||
| Well differentiated | 8 | 4 | 4 | |
| Moderately differentiated | 46 | 26 | 20 | |
| Poorly differentiated | 33 | 23 | 10 | |
| Lymph node metastasis | 0.737 | |||
| Negative | 39 | 23 | 16 | |
| Positive | 48 | 30 | 18 | |
| Disease stage | 0.441 | |||
| I and II | 34 | 19 | 15 | |
| III and IV | 53 | 34 | 19 | |
| Smoking history | 0.486 | |||
| Positive | 42 | 24 | 18 | |
| Negative | 45 | 29 | 16 | |
| Drinking history | 0.335 | |||
| Positive | 43 | 24 | 19 | |
| Negative | 44 | 29 | 15 | |
| Surgical margin | 0.616 | |||
| Negative | 72 | 43 | 29 | |
| Positive | 15 | 10 | 5 | |
| Vascular invasion | 0.536 | |||
| Negative | 67 | 42 | 25 | |
| Positive | 20 | 11 | 9 |
Fisher's exact test, for all other P-values a Chi-square test was performed. MLH1, MutL homolog 1; N, number of patients; ESCC, esophageal squamous cell carcinoma.
MLH1 promoter methylation in ESCC tissues
As presented in Table I, MLH1 promoter methylation was observed in 53/87 (60.92%) ESCC tissue samples. No significant differences were observed in further correlation analyses between MLH1 methylation and clinical phenotypes (All P>0.05). The evaluated clinical phenotypes were as follows: Age at diagnosis, smoking behavior, history of alcohol consumption and clinicopathological characteristics, including tumor location, histological differentiation, lymph node metastasis, clinical stage, surgical margin status and vascular invasion.
Survival analysis
As presented in Table II, the univariate survival analysis demonstrated that the OS of patients with ESCC was associated with age at the time of surgery (P=0.028), history of alcohol consumption (P=0.015) and vascular invasion (P=0.015). Meanwhile, MLH1 promoter methylation was significantly associated with poor OS in patients with ESCC (P=0.048; Fig. 2). Following adjustments for these potential confounding factors in a multivariate Cox proportional hazards model, the OS of patients with ESCC was observed to be significantly associated with MLH1 promoter hypermethylation (HR=1.716; 95% CI=1.008–2.921), age ≤60 years old (HR=0.486, 95%; CI=0.279–0.849), alcohol consumption (HR=1.968; 95% CI=1.157–3.348) and vascular invasion (HR=1.791; 95% CI=1.007–3.185).
Table II.
Survival analysis of 87 patients with esophageal cancer with MSP detection of MLH1 promoter methylation.
| Variables | Median OS, (months) | χ2 | Univariate P-value | HR (95% CI) | Multivariate P-value |
|---|---|---|---|---|---|
| Age at surgery, years | 0.028a | 0.011a | |||
| ≤60 | 31.5 | 4.812 | 0.486 (0.279–0.849) | ||
| >60 | 14.0 | ||||
| Tumor location | 0.835 | ||||
| Upper | 31.5 | 0.360 | |||
| Middle | 19.0 | ||||
| Lower | 17.0 | ||||
| Differentiation | 0.138 | ||||
| Well differentiated | 50.0 | 3.965 | |||
| Moderately differentiated | 21.0 | ||||
| Poorly differentiated | 14.0 | ||||
| Lymph node metastasis | 0.211 | ||||
| Negative | 24.5 | 1.566 | |||
| Positive | 14.5 | ||||
| Disease stage | 0.213 | ||||
| I and II | 21.0 | 1.551 | |||
| III and IV | 14.5 | ||||
| Smoking history | 0.307 | ||||
| Positive | 17.0 | 1.042 | |||
| Negative | 20.0 | ||||
| Drinking history | 0.015a | 0.013a | |||
| Positive | 13.0 | 5.880 | 1.968 (1.157–3.348) | ||
| Negative | 22.0 | ||||
| Surgical margin | 0.421 | ||||
| Negative | 19.0 | 0.647 | |||
| Positive | 14.5 | ||||
| Vascular invasion | 0.015a | 0.047a | |||
| Positive | 12.5 | 5.903 | 1.791 (1.007–3.185) | ||
| Negative | 22.5 | ||||
| Methylated status of MLH1 promoter | 0.048a | 0.047a | |||
| Methylation | 16.5 | 3.902 | 1.716 (1.008–2.921) | ||
| Unmethylation | 27.0 |
P<0.05. MHL1, MutL homolog 1; OS, overall survival; HR, hazard rate; MSP, methylation-specific polymerase chain reaction.
Figure 2.
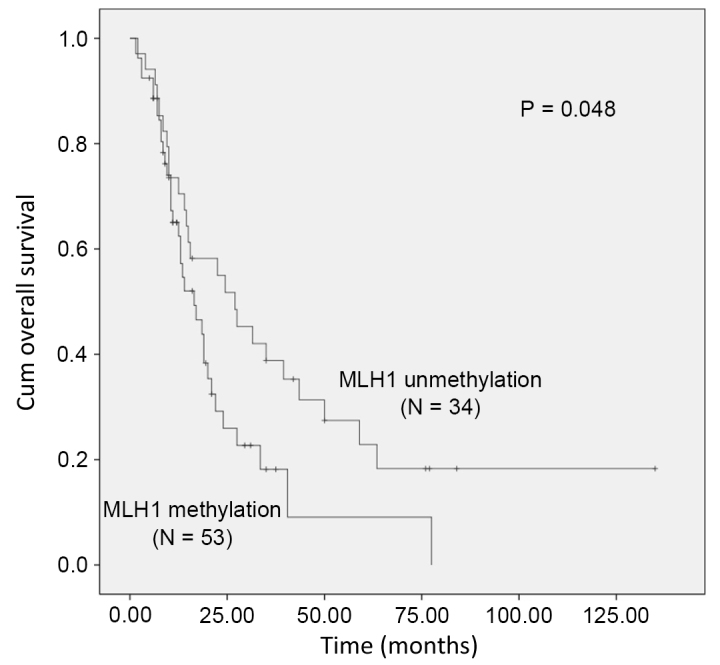
Kaplan-Meier survival curves comparing the survival (months) of patients with esophageal cancer and methylated or unmethylated MLH1 promoter regions. MLH1, MutL homolog 1; cum, cumulative.
Discussion
In the current study, MLH1 promoter methylation and OS was investigated in male patients with ESCC. MLH1 promoter hypermethylation was significantly associated with poor OS in male patients with ESCC. Furthermore, diagnostic age, alcohol consumption and vascular invasion were also independent factors for the OS of patients with ESCC.
DNA mismatch repair is a conserved process that is critical for correcting any generated mismatches that escape proofreading during DNA replication (25). MLH1 is one of the major genes involved in this process (25). The loss of the MLH1 protein may be detected in ~72% of ESCC tumors, concordant with MLH1 promoter hypermethylation, which is correlated with reduced gene expression levels (31,32). MLH1 promoter hypermethylation has been reported in numerous types of tumors, including colorectal (33), gastric cancer (34) and EC (35). Previous studies have also reported that MLH1 hypermethylation may be associated with poor prognosis for patients with gastric (22), ovarian (36) and colorectal cancer (37), whilst studies on the prognostic value of MLH1 methylation in Han Chinese patients with EC or ESCC are scarce. The present study demonstrated that MLH1 promoter hypermethylation is a prognostic marker of ESCC in Chinese males.
The epidemiology of ESCC demonstrates that there is a high gender bias, evidenced by a gender ratio of 8–9:1 in favor of males (38). In addition, the consumption of alcohol and cigarettes occurs more frequently in males, as compared with in females in China (39,40). Although the present study did not examine whether alcohol and cigarette consumption were risk factors for ESCC, as investigated in previous studies (41,42), the results demonstrated that the consumption of alcohol was significantly associated with poor OS in ESCC. Vascular invasion has previously been significantly correlated with ESCC (43). In the present study, patients with vascular invasion had a significantly shorter survival time, compared with patients without, suggesting a role for vascular invasion in the progression and prognosis of ESCC.
In conclusion, the present study identified MLH1 promoter methylation as a prognostic marker of ESCC. It was also demonstrated that patients with ESCC significantly benefited from early-stage diagnosis. Alcohol consumption, smoking and vascular invasion may be involved in the progression of ESCC; however, further studies are required to investigate these findings.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 31100919 and 81371469), Zhejiang Provincial Natural Science Foundation (grant no. LR13H020003), the Clinical Research Foundation of Zhejiang Province Medical Association (grant no. 2015ZYC-A77) and Science and Technology Plan Project of Shaoxing (grant no. 2015B70069).
References
- 1.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu M, Zaninotto G, Nagata K, Graham DY, Lauwers GY. Esophageal squamous cell carcinoma with special reference to its early stage. Best Pract Res Clin Gastroenterol. 2013;27:171–186. doi: 10.1016/j.bpg.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Conteduca V, Sansonno D, Ingravallo G, Marangi S, Russi S, Lauletta G, Dammacco F. Barrett's esophagus and esophageal cancer: An overview. Int J Oncol. 2012;41:414–424. doi: 10.3892/ijo.2012.1481. [DOI] [PubMed] [Google Scholar]
- 5.Du Z, Ma K, Sun X, et al. Methylation of RASSF1A gene promoter and the correlation with DNMT1 expression that may contribute to esophageal squamous cell carcinoma. World J Surg Oncol. 2015;13:141. doi: 10.1186/s12957-015-0557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang AH, Liu Y, Wang B, et al. Epidemiological studies of esophageal cancer in the era of genome-wide association studies. World J Gastrointest Pathophysiol. 2014;5:335–343. doi: 10.4291/wjgp.v5.i3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma G, Zhang X, Ma Q, et al. A novel multivariate scoring system for determining the prognosis of lymph node-negative esophageal squamous cell carcinoma following surgical therapy: An observational study. Eur J Surg Oncol. 2015;41:541–547. doi: 10.1016/j.ejso.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Arnold M, Colquhoun A, Cook MB, Ferlay J, Forman D, Soerjomataram I. Obesity and the incidence of upper gastrointestinal cancers: An ecological approach to examine differences across age and sex. Cancer Epidemiol Biomarkers Prev. 2016;25:90–97. doi: 10.1158/1055-9965.EPI-15-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thrift AP. The epidemic of oesophageal carcinoma: Where are we now? Cancer Epidemiol. 2016;41:88–95. doi: 10.1016/j.canep.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Trevellin E, Scarpa M, Carraro A, Lunardi F, Kotsafti A, Porzionato A, Saadeh L, Cagol M, Alfieri R, Tedeschi U, et al. Esophageal adenocarcinoma and obesity: Peritumoral adipose tissue plays a role in lymph node invasion. Oncotarget. 2015;6:11203–11215. doi: 10.18632/oncotarget.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahrens TD, Werner M, Lassmann S. Epigenetics in esophageal cancers. Cell Tissue Res. 2014;356:643–655. doi: 10.1007/s00441-014-1876-y. [DOI] [PubMed] [Google Scholar]
- 12.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 13.Kuo IY, Chang JM, Jiang SS, Chen CH, Chang IS, Sheu BS, Lu PJ, Chang WL, Lai WW, Wang YC. Prognostic CpG methylation biomarkers identified by methylation array in esophageal squamous cell carcinoma patients. Int J Med Sci. 2014;11:779–787. doi: 10.7150/ijms.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murata A, Baba Y, Watanabe M, Shigaki H, Miyake K, Ishimoto T, Iwatsuki M, Iwagami S, Yoshida N, Oki E, et al. IGF2 DMR0 methylation, loss of imprinting, and patient prognosis in esophageal squamous cell carcinoma. Ann Surg Oncol. 2014;21:1166–1174. doi: 10.1245/s10434-013-3414-7. [DOI] [PubMed] [Google Scholar]
- 15.Vasavi M, Ponnala S, Gujjari K, Boddu P, Bharatula RS, Prasad R, Ahuja YR, Hasan Q. DNA methylation in esophageal diseases including cancer: Special reference to hMLH1 gene promoter status. Tumori. 2006;92:155–162. doi: 10.1177/030089160609200212. [DOI] [PubMed] [Google Scholar]
- 16.De Oliveira SR, Da Silva IC, Mariz BA, Pereira AM, De Oliveira NF. DNA methylation analysis of cancer-related genes in oral epithelial cells of healthy smokers. Arch Oral Biol. 2015;60:825–833. doi: 10.1016/j.archoralbio.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Sun MZ, Ju HX, Zhou ZW, Jin H, Zhu R. Single nucleotide polymorphisms of DNA mismatch repair genes MSH2 and MLH1 confer susceptibility to esophageal cancer. Int J Clin Exp Med. 2014;7:2329–2333. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Xie XJ, Piao YS, Liu B, Wang LD. Methylation of p16 and hMLH1 genes in esophageal squamous cell carcinoma and reflux esophagitis. Zhonghua Bing Li Xue Za Zhi. 2011;40:537–541. (In Chinese) [PubMed] [Google Scholar]
- 19.Uehara H, Miyamoto M, Kato K, Cho Y, Kurokawa T, Murakami S, Fukunaga A, Ebihara Y, Kaneko H, Hashimoto H, et al. Deficiency of hMLH1 and hMSH2 expression is a poor prognostic factor in esophageal squamous cell carcinoma. J Surg Oncol. 2005;92:109–115. doi: 10.1002/jso.20332. [DOI] [PubMed] [Google Scholar]
- 20.Vogelsang M, Wang Y, Veber N, Mwapagha LM, Parker MI. The cumulative effects of polymorphisms in the DNA mismatch repair genes and tobacco smoking in oesophageal cancer risk. PLoS One. 2012;7:e36962. doi: 10.1371/journal.pone.0036962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, Xu X, Chen H, Wang J, Xiong W, Xu Y, Liu J. The hMLH1 promoter polymorphisms and cancer susceptibility in Asian populations: A meta-analysis. Gene. 2013;523:199–204. doi: 10.1016/j.gene.2013.03.138. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Yang Y, Lu Y, Herman JG, Brock MV, Zhao P, Guo M. Predictive value of CHFR and MLH1 methylation in human gastric cancer. Gastric Cancer. 2015;18:280–287. doi: 10.1007/s10120-014-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veganzones S, Maestro ML, Rafael S, de la Orden V, Vidaurreta M, Mediero B, Espantaleón M, Cerdán J, Díaz-Rubio E. Combined methylation of p16 and hMLH1 (CMETH2) discriminates a subpopulation with better prognosis in colorectal cancer patients with microsatellite instability tumors. Tumour Biol. 2015;36:3853–3861. doi: 10.1007/s13277-014-3027-1. [DOI] [PubMed] [Google Scholar]
- 24.Ling ZQ, Li P, Ge MH, Hu FJ, Fang XH, Dong ZM, Mao WM. Aberrant methylation of different DNA repair genes demonstrates distinct prognostic value for esophageal cancer. Dig Dis Sci. 2011;56:2992–3004. doi: 10.1007/s10620-011-1774-z. [DOI] [PubMed] [Google Scholar]
- 25.Baba Y, Watanabe M, Baba H. Review of the alterations in DNA methylation in esophageal squamous cell carcinoma. Surgery Today. 2013;43:1355–1364. doi: 10.1007/s00595-012-0451-y. [DOI] [PubMed] [Google Scholar]
- 26.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 27.Wu D, Xu J, Yu G, Zhang B, Wang H, Wang C, Ru G, Sun A, Shen L, Wei Q. Expression status of fatty acid synthase (FAS) but not HER2 is correlated with the differentiation grade and prognosis of esophageal carcinoma. Hepatogastroenterology. 2013;60:99–106. doi: 10.5754/hge12415. [DOI] [PubMed] [Google Scholar]
- 28.Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61:900–902. [PubMed] [Google Scholar]
- 29.Mu L, Mukamal KJ. Alcohol consumption and rates of cancer screening: Is cancer risk overestimated? Cancer Causes Control. 2016;27:281–289. doi: 10.1007/s10552-015-0692-3. [DOI] [PubMed] [Google Scholar]
- 30.Organization WH. Guidelines for controlling and monitoring the tobacco epidemic. World Health Organization. 1998;119:63–64. [Google Scholar]
- 31.Tzao C, Hsu HS, Sun GH, Lai HL, Wang YC, Tung HJ, Yu CP, Cheng YL, Lee SC. Promoter methylation of the hMLH1 gene and protein expression of human mutL homolog 1 and human mutS homolog 2 in resected esophageal squamous cell carcinoma. J Thorac Cardiovasc Surg. 2005;130:1371. doi: 10.1016/j.jtcvs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Hernàndez I, Garcia JL, Santos-Briz A, Hernández-Laín A, González-Valero JM, Gómez-Moreta JA, Toldos-González O, Cruz JJ, Martin-Vallejo J, González-Sarmiento R. Integrated analysis of mismatch repair system in malignant astrocytomas. PLoS One. 2013;8:e76401. doi: 10.1371/journal.pone.0076401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Wang X, Li X, Wang M, Chen X, Wu X, Wang Y, Fan Y, Jin H. The specific methylation characteristics of cancer related genes in Chinese colorectal cancer patients. Tumour Biol. 2014;35:8267–8279. doi: 10.1007/s13277-014-2100-0. [DOI] [PubMed] [Google Scholar]
- 34.Jin J, Xie L, Xie CH, Zhou YF. Aberrant DNA methylation of MGMT and hMLH1 genes in prediction of gastric cancer. Genet Mol Res. 2014;13:4140–4145. doi: 10.4238/2014.May.30.9. [DOI] [PubMed] [Google Scholar]
- 35.Su Y, Yin L, Liu R, Sheng J, Yang M, Wang Y, Pan E, Guo W, Pu Y, Zhang J, Liang G. Promoter methylation status of MGMT, hMSH2, and hMLH1 and its relationship to corresponding protein expression and TP53 mutations in human esophageal squamous cell carcinoma. Med Oncol. 2014;31:784. doi: 10.1007/s12032-013-0784-4. [DOI] [PubMed] [Google Scholar]
- 36.Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, Khabir A, Frikha F, Kallel L, Frikha M, Sellami-Boudawara T, Gargouri A, Mokdad-Gargouri R. Aberrant methylation of hMLH1 and p16INK4a in Tunisian patients with sporadic colorectal adenocarcinoma. Biosci Rep. 2011;31:257–264. doi: 10.1042/BSR20100023. [DOI] [PubMed] [Google Scholar]
- 38.Sukocheva OA, Wee C, Ansar A, Hussey DJ, Watson DI. Effect of estrogen on growth and apoptosis in esophageal adenocarcinoma cells. Dis Esophagus. 2013;26:628–635. doi: 10.1111/dote.12000. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Liu S, Wang L, Zhou M. Burden of disease attributable to main behavioral risk factor of chronic disease inactivity in China, 1990 and 2010. Zhonghua Yu Fang Yi Xue Za Zhi. 2015;49:333–338. (In Chinese) [PubMed] [Google Scholar]
- 40.Lepage C, Drouillard A, Jouve JL, Faivre J. Epidemiology and risk factors for oesophageal adenocarcinoma. Dig Liver Dis. 2013;45:625–629. doi: 10.1016/j.dld.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Y, Cao X, Wen J, Yang H, Luo K, Liu Q, Huang Q, Chen J, Fu J. Smoking affects treatment outcome in patients with resected esophageal squamous cell carcinoma who received chemotherapy. PLoS One. 2015;10:e0123246. doi: 10.1371/journal.pone.0123246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumagai N, Wakai T, Akazawa K, Ling Y, Wang S, Shan B, Okuhara Y, Hatakeyama Y, Kataoka H. Heavy alcohol intake is a risk factor for esophageal squamous cell carcinoma among middle-aged men: A case-control and simulation study. Mol Clin Oncol. 2013;1:811–816. doi: 10.3892/mco.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang CC, Mao WM, Ling ZQ. DNA methylation status of RARβ2 and p16 (INK4α) in peripheral blood and tumor tissue in patients with esophageal squamous cell carcinoma. Zhonghua Zhong Liu Za Zhi. 2012;34:441–445. (In Chinese) [PubMed] [Google Scholar]



