Abstract
Sphingosine kinase 1 (Sphk1) is an oncogenic kinase that is responsible for the phosphorylation of sphingosine to sphingosine-1-phosphate (S1P). Mounting evidence suggests that Sphk1 serves a crucial role in the proliferation and development of a variety of human cancer cells. However, the role of Sphk1 in hepatocellular carcinoma (HCC) has not been fully elucidated. Therefore, the expression of Sphk1 was examined in 127 formalin-fixed, paraffin-embedded HCC tissues using immunohistochemistry, and its clinical implications and prognostic significance were analyzed. As a result, the expression of Sphk1 in HCC tissue was revealed to be significantly higher than in normal tissue (P<0.01). In addition, Sphk1 expression was significantly associated with tumor size, tumor stage and histological differentiation (all P<0.05). The patients with low Sphk1 expression had higher overall survival and recurrence-free survival rates compared with patients with high Sphk1 expression. Furthermore, Sphk1-specific shRNA was used to downregulate the expression of Sphk1 in HCC cell lines, including hepatoblastoma G2 and HCC-9724. The CRISPR/Cas9 based transcription activation system was used to upregulate Sphk1 expression in the normal live cell, L02. Cell proliferation, mRNA expression and protein expression were measured using Cell Counting Kit-8, reverse transcription polymerase chain reaction and western blot analysis in the transfected cells. To the best of our knowledge, the present study provides the first evidence that Sphk1 promotes HCC cell proliferation and is involved in tumor progression. Notably, the data presented suggest that Sphk1 may be a potential independent prognosis biomarker for the treatment of HCC.
Keywords: hepatocellular carcinoma, sphingosine kinase 1, prognosis biomarker, synergistic activation mediator
Introduction
Hepatocellular carcinoma (HCC) ranked as the seventh most prevalent cancer and the second leading cause of cancer-associated morbidity and mortality worldwide in 2012 (1). Although new diagnostic and treatment methods have been developed, owing to the development of tumor biology over previous decades, live transplantation (LT) remains to be considered the most efficient therapeutic option for HCC at present. The incidence, recurrence and associated mortality of HCC are continuously increasing over years (2). The increase in mortality may be due to the inefficiency of early-stage diagnosis, resulting from the insidious onset of early-stage HCC (3). Therefore, there is increased interest in indentifying new indicators associated with the progression of HCC, with the aim to improve the overall survival time of patients with HCC.
Sphingosine-1-phosphate (S1P) is an important metabolite that serves a crucial role in intracellular and intercellular signal transduction (4). The formation of polar, bioactive S1P from the non-polar but bioactive sphingosine is catalyzed by sphingosine kinase 1 (Sphk1). Previous studies have demonstrated that S1P regulates multiple cellular processes including cell apoptosis inhibition and proliferation promotion, whereas sphingosine inhibits cell proliferation and stimulates apoptosis (5). Therefore, in previous years, numerous studies have focused their attentions on uncovering the role of Sphk1 in tumors. Sphk1 was firstly identified as a lipid kinase with its biological activity of balancing the expression and functions of substrate and product. Following that, it has been well established that Sphk1 is an oncogenic enzyme and its activation is associated with the cell transformation, survival and tumor microenvironment neovascularization (6). Accumulating evidence has demonstrated that Sphk1 could be activated by a number of reagents, including growth factors, cytokines and oncogenes. This activation is partly due to the phosphorylation of serine at residue 225 (S225), and appeared to be solely responsible for the Sphk1 oncogenic effect. Evidence also exists to show that the activation of Sphk1 is associated with relocalization of Sphk1 to the plasma membrane, which is considered to be critical in its transforming ability (7). Notably, downregulation of Sphk1 expression by Sphk1 short-hairpin RNA, or inactivation of Sphk1 function by specific inhibitors, suppressed prostate cancer cell and glioblastoma cell growth, indicating that Sphk1 may be a potential target for the treatment of human cancers (8).
The overexpression of Sphk1 has been well established in various human cancers, including head and neck (9), gastric (10), lung (11), colon (5) and ovarian cancers (12). Increasing evidence has demonstrated that Sphk1 is involved in tumorigenic processes through promoting survival and pro-growth signals, inhibiting cell apoptosis, inducing cell transformation and promoting angiogenesis (6). However, the role of Sphk1 in HCC has not been well studied. Therefore, the present study aimed to investigate the association between Sphk1 expression and clinicopathological characteristics of patients with HCC, and to evaluate the role of Sphk1 expression in the progression of HCC.
Materials and methods
Clinical specimens and tissues
A total of 127 patients (71 men and 56 women) that had undergone surgical liver resection at the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) from February 2008 to August 2010 were enrolled into the present study. None of these patients had ever received chemotherapy, radiotherapy or any other type of anti-cancer treatment prior to surgery. HCC tissues and adjacent normal tissues collected following surgical resection were immediately stored at −80°C for further research. All the clinicopathological characteristics were recorded and summarized in Table I. The stage and differentiation of patients were based on the TNM (Union for International Cancer Control) and Edmondson method. The present clinical study was approved and monitored by the ethics committee of the First Affiliated Hospital of Wenzhou Medical University. All the patients involved in the present study have signed the written informed consent as the requirement of Institutional Ethnic Committee.
Table I.
Association between Sphk1 expression and the clinicopathological features of HCC.
| Sphk1 expression | ||||
|---|---|---|---|---|
| Variable | No. of cases | High | Low | P-value |
| Gender | NS | |||
| Male | 71 | 52 | 19 | |
| Female | 56 | 41 | 15 | |
| Age | NS | |||
| ≥50 years | 66 | 53 | 13 | |
| <50 years | 61 | 40 | 21 | |
| Tumor size | 0.015 | |||
| ≥5 cm | 78 | 63 | 15 | |
| <5 cm | 49 | 30 | 19 | |
| HBsAg | NS | |||
| Positive | 62 | 48 | 13 | |
| Negative | 65 | 44 | 21 | |
| Histological differentiation | 0.018 | |||
| Well/Moderate | 81 | 65 | 16 | |
| Poor | 46 | 28 | 18 | |
| AFP | NS | |||
| >400 ng/ml | 76 | 56 | 20 | |
| ≤400 ng/ml | 51 | 37 | 14 | |
| Tumor stage | 0.009 | |||
| I–II | 58 | 36 | 22 | |
| III–IV | 69 | 57 | 12 | |
Sphk1, sphingosine kinase 1; NS, not significant; HBsAg, hepatitis B virus surface antigen; AFP, α-fetoprotein.
Cell lines
Human immortalized liver cell line L02 and two HCC cell lines (hepatoblastoma G2 and HCC-9724) were purchased from American Type Culture Collection (Manassas, VA, USA). All the cells were cultured in complete Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at a maintained humidified atmosphere (5% CO2, 37°C).
Silencing of Sphk1 expression using short-hairpin RNA (shRNA)
The Sphk1-specific shRNA and scramble shRNA as negative control were designed and synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). HepG2 and HCC-9724 cell lines stably expressing the Sphk1-specific shRNA or scramble shRNA were constructed by transfecting the shRNA expression plasmid (pENTR™/U6-shRNA; Invitrogen; Thermo Fisher Scientific, Inc.) using Lipofectamine (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Target sequences for specific shRNAs are: Sphk1 specific shRNA, 5′-GCAGCTTCCTTGAACCATTAT-3′; and scrambled shRNA, 5′-GGAATCTCATTCGATGCATAC-3′.
Upregulation of Sphk1 expression using synergistic activation mediator (SAM) constructs
The Sphk1-specific SAM guide RNA sequence was synthesized by GenScript (Nanjing, China) and cloned into lenti sgRNA (MS2)_zeo backbone. To activate the transcription of endogenous sphk1 gene, lenti sgRNA (MS2)_zeo backbone, dCas9-VP64_Blast and lenti MS2-P65-HSF1_Hygro, at the mass ratio of 1:1:1, were co-transfected into the L02 cells using Lipofectamine (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol and the description in a previous study (13). The gRNA sequence for Sphk1 activation is 5′-CCAGCTCGTGGCCCGGCCGT-3′.
Cell proliferation assay
The rate of cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8) purchased from Beyotime Institute of Biotechnology (Jiangsu, China), and followed the manufacturer's protocol. Cell lines transfected with shRNA or gRNA construct were seeded onto 96-well plates at a density of ~4,000 cells/well and incubated for 24, 48 and 72 h under the aforementioned culture conditions. A total of 10 µl of CCK-8 was added to each well 2 h prior to incubation completion. The absorbance of each well was measured using Thermo Multiskan® Spectrum (Thermo Fisher Scientific, Inc.) at a wavelength of 450 nm. All the experimental procedures were repeated at least three times independently.
Total RNA preparation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from human liver cell lines transfected with shRNA or SAM construct and the normal and tumor tissues were obtained from the patients with HCC using a TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's protocol. The concentration of RNA was then quantified (NanoDrop 3,000; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). cDNA chains were synthesized from total RNA (1 µg) using Moloney murine leukemia virus (M-MLV) RT (Promega Corporation, Madison, WI, USA) and an oligo-dT primer.
To quantify the mRNA expression of Sphk1 in the transfected cell lines, tumor tissues and normal tissues, iQ SYBRE Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was adopted and real-time RT-PCR was performed on a iCycle iQ Multicolor Real-Time RT-PCR Detection System (Bio-Rad Laboratories, Inc.). The optimized qPCR cycling conditions consisted of an initial step at 95°C for 10 min, followed by cycles (n=40) consisting of 10 sec at 95°C and 1 min annealing/extension at 55°C. All experiments were performed in triplicate. The data were analyzed with Q-Gene software (14) and expressed as the mean normalized expression (5). The expression level of Sphk1 was normalized to the reference gene, β-actin. The following primers were used for qPCR amplification: Sphk1 sense, 5′-ATGCACGAGGTGGTGAACG-3′ and antisense, 5′-GGAGGCAGGTGTCTTGGAAC-3′; and β-actin sense, 5′-TCCTCCCTGGAGAAGAGCTA-3′ and antisense, 5′-CCAGACAGCACTGTGTTGGC-3′.
Western blot analysis
To quantify the protein expression of Sphk1 in transfected cell lines and tissues, western blot analysis was used in the present study. The preparation of lysates from cultured cells or tissues was performed as previously described (15). The supernatant was collected, and the concentration of protein was then determined using the Bradford colorimetric protein assay. Following denaturation, equal amounts of protein were separated on a 12% SDS-PAGE gel and then blotted onto a nitrocellulose (NC) membrane in a humid environment. The NC membrane was blocked with 5% non-fat milk in TBST [10 mM Tris (pH 7.4), 100 mM NaCl and 0.05% Tween-20]. Following blocking, the NC membrane was incubated with 1:1,000 dilution of anti-Sphk1 antibody (catalog no. 12071; Cell Signaling Technology, Inc., Danvers, MA, USA) or 1:3,000 dilution of anti-β-actin antibody (catalog no. sc-4778; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at room temperature for 1 h, and then labeled with 1:3,000 dilution of horseradish peroxidase-conjugated anti-rabbit antibody (catalog no. RPN4301; GE Healthcare Life Sciences, Shanghai, China) at 1 h. Bands were visualized with enhanced chemiluminescence reaction western blotting substrate (Pierce; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. Autoradiography signals were analyzed using a gel imaging system (GE Healthcare Life Sciences).
Immunohistochemistry assay
All immunohistochemistry was analyzed on formalin-fixed, paraffin-embedded tissues with the avidin-biotin immunoperoxidase technique. Tissue sections were deparaffinized by xylene, and rehydrated with graded ethanol. Following blocking of endogenous peroxidase activity with 3% hydrogen peroxide in methanol for 10 min, the tissue sections were washed with 10 mM citrate buffer and then incubated with 1:200 dilution of rabbit polyclonal anti-Sphk1 antibody (cat. no. 3297; Cell Signaling Technology, Inc.) for 1 h at room temperature. The tissue sections were then incubated with biotinylated goat anti-rabbit immunoglobulin G and then horseradish peroxidase-conjugated streptavidin. Subsequent to washing with 0.01 M PBS (pH 7.34), the antibody complexes were colored with 3,3′-diaminobenzidine solution (cat. no. D8001; Sigma-Aldrich; Merck Millipore, Darmstadt, Gemany). The Olympus microscope was used to observe the stained tissues (BX63; Olympus Corp., Tokyo, Japan).
Statistical analysis
Data were presented as the mean ± standard deviation. The χ2 test was performed to determine whether Sphk1 expression in HCC was associated with clinicopathological characteristics. The significance of difference among groups was evaluated by Student's two-tailed t-test. The Kaplan-Meier method was used for survival analysis, and differences in survival were estimated by long-rank test. Multivariate analysis was performed on parameters with P<0.05 in the univariate analysis using Cox proportional hazards model (95% confidence interval). P<0.05 was considered to indicate a statistically significant difference.
Results
Sphk1 expression in hepatocellular carcinoma (HCC) samples
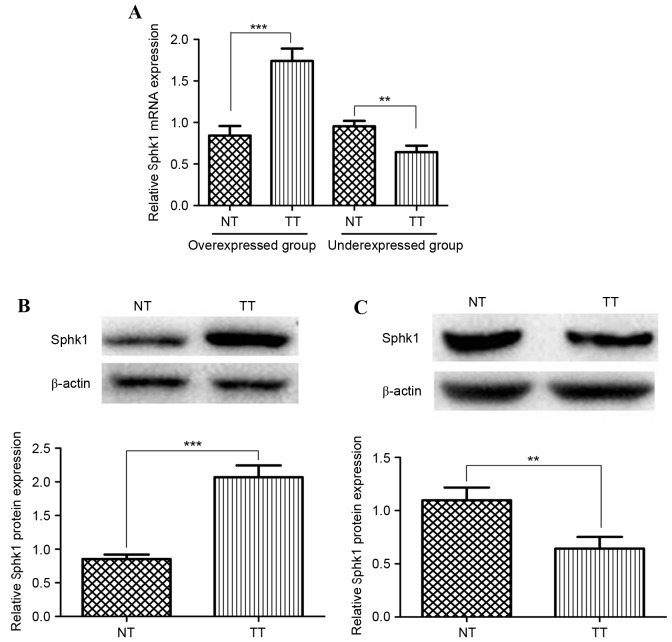
To observe the expression of Sphk1 in HCC tissues, the HCC and matched adjacent noncancerous tissues obtained from 127 patients were examined at the mRNA and protein level, using RT-qPCR and western blot analysis. The mRNA expression analysis result is shown in Fig. 1A. In total, 93/127 (73.23%) of the cancer tissue samples were revealed to have a higher Sphk1 expression compared with adjacent normal tissues (overexpressed group). The other 26.77% demonstrated lower expression (Fig. 1A, under-expressed group). The results revealed that Sphk1 overexpression is universal in HCC tissues. Furthermore, western blotting was performed to quantify the Sphk1 protein expression of HCC tissues in these two groups. As depicted in Fig. 1B, the Sphk1 expression level in HCC tissue is significantly higher than that of normal tissue in the overexpressed group, which is consistent with the result obtained from RT-qPCR (P<0.001). The same result could be drawn from the under-expressed group (Fig. 1C). Therefore, Sphk1 may be important in HCC.
Figure 1.
Analysis of Sphk1 expression in hepatocellular carcinoma tissues and adjacent normal tissues. (A) Quantitative reverse transcription polymerase chain reaction analysis of Sphk1 mRNA expression level. The patients with increased sphk1 mRNA expression were classified as the overexpressed group. Otherwise, patients were classified as the underexpressed group. (B) Western blot analysis of the protein expression of Sphk1 as the overexpressed group. (C) Western blot analysis of the protein expression of Sphk1 in the underexpressed group (***P<0.001; **P<0.01). NT, normal liver tissues; TT, liver tumor tissues; NS, not significant; Sphk1, sphingosine kinase 1.
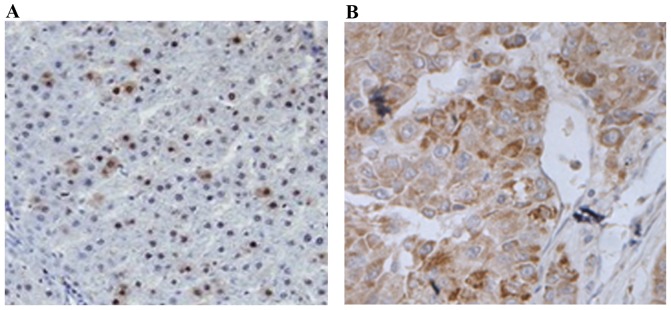
To analyze the in situ expression of Sphk1 in HCC tissues, immunohistochemistry (IHC) staining was performed in the present study. IHC analysis revealed that the expression of Sphk1 in the tissues obtained from the overexpressed group was significantly higher than those from the under-expressed group (P<0.001). This is evidence of universally higher Sphk1 expression in patients with HCC, and also verifies the results drawn from RT-qPCR and western blot analysis (Fig. 2). In summary, the present study demonstrated that Sphk1 was overexpressed in HCC patients through RT-qPCR, western blotting and IHC analysis.
Figure 2.
Immunohistochemistry analysis of Sphk1 expression in hepatocellular carcinoma patients. (A) High Sphk1 expression. (B) Low Sphk1 expression. Sphk1, sphingosine kinase 1. Magnification, ×200.
Sphk1 overexpression promotes tumor cell proliferation
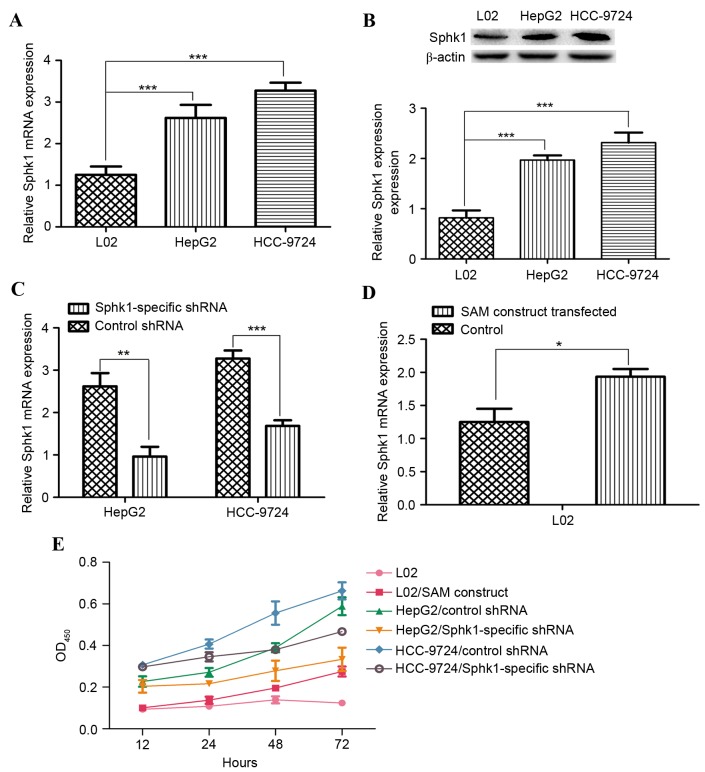
Overexpression of Sphk1 has been well established in the present study through analyzing the mRNA and protein expression in HCC tissues. Therefore, to reveal the role of Sphk1 expression in HCC, the effect of Sphk1 on cell proliferation was examined. The normal liver cell line (L02) and two HCC cell lines (HepG2 and HCC-9724) were selected in the present study. Firstly, Sphk1 mRNA and protein expression in L02, HepG2 and HCC-9724 was analyzed through RT-qPCR and western blot. As shown in Fig. 3A and B, Sphk1 expression in HCC cell lines was markedly higher than in the normal liver cell lines, which is consistent with the results drawn from HCC tissue analysis (all P<0.001). Next, the Sphk1-specific shRNA and negative control shRNA were used to downregulate the expression of Sphk1 in HepG2 and HCC-9724. The comparison of Sphk1 expression in Sphk1-specific shRNA and negative control shRNA transfected cells shows that sphk1 expression in the former cell is lower than in the latter cell and is statistically significant (all P<0.001; Fig. 3C), proving the effectiveness of Sphk1-specific shRNA. Lastly, upregulation of Sphk1 expression was attempted using the CRISPR/Cas9 based transcription activation system. The result was presented in Fig. 3D. The Sphk1 expression level in L02 cells with SAM construct transfection was higher than in normal L02 cells, meaning that Sphk1 expression was successfully activated by the SAM construct (P<0.05). In summary, through shRNA or SAM construct transfection, it is possible to regulate Sphk1 expression in normal liver and HCC cell lines.
Figure 3.
Analysis of Sphk1 expression in normal liver and HCC cell lines. (A) Sphk1 mRNA expression in normal (L02) and tumor (HepG2 and HCC-9724) cell lines. (B) Sphk1 protein expression in normal and tumor cell lines. (C) Sphk1 mRNA expression analysis of the HCC cell lines with sphk1-specific shRNA transfection. (D) Sphk1 mRNA expression analysis of the normal liver cell line with CRISPR/Cas9-based SAM construct transfection. (E) Cell Counting Kit-8 assay to determine the cell proliferation rate of cell lines with and without shRNA or SAM construct transfection (***P<0.001; **P<0.01; *P<0.05). Sphk1, sphingosine kinase 1; HCC, hepatocellular carcinoma; SAM, synergistic activation mediator; shRNA, short-hairpin RNA; NS, not significant; HepG2, hepatoblastoma G2; OD450, optical density at 450 nm.
Furthermore, the proliferation rate of cells with shRNA or SAM construct transfection was examined. The proliferation rate of HCC cells with Sphk1-specific shRNA transfection was significantly lower than cells with negative control shRNA transfection (all P<0.05). In addition, the proliferation rate of normal liver cells with SAM construct transfection was notably higher than the cell without transfection (P<0.05; Fig. 3E). In general, the aforementioned results demonstrate that Sphk1 overexpression may promote cell proliferation, which may be one of the mechanisms that Sphk1 is involved in in tumor progression.
Clinical significance of Sphk1 expression in hepatocellular carcinoma
The clinicopathological data of the 127 patients enrolled in the present study was collected. The clinical significance of Sphk1 overexpression was then determined by investigating the association between Sphk1 overexpression and clinicopathological characteristics of HCC patients. The results are summarized in Table I. Sphk1 expression status was significantly associated with tumor size (P=0.015), histological differentiation (P=0.018) and tumor stage (P=0.009), while no significant association was observed between Sphk1 expression level and age, gender, hepatitis B virus surface antigen (HBsAg) and α-fetoprotein (AFP) (all P>0.05).
Sphk1 expression is an independent prognostic factor for recurrence-free survival in patients with hepatocellular carcinoma
To confirm the role of Sphk1 as a predictor of recurrence-free survival, survival analysis was performed in the HCC patients enrolled in the present study. As shown in Fig. 4, the patients with HCC that had high Sphk1 expression had shorter 5-year survival rates compared with patients with low Sphk1 expression. In addition, univariate analysis demonstrated that Sphk1 expression (P=0.019), tumor size (P=0.036), tumor stage (P=0.026) and histological differentiation (P=0.038) were significantly associated with the overall survival of patients with HCC (Table II). No significant association was observed for age, gender, HBsAg, AFP and outcome of patients (all P>0.05). Furthermore, multivariate analysis was conducted using Cox proportional hazards model to analyze the significant variables in the univariate analysis. The results showed that Sphk1 expression was one of the independent prognostic factors for HCC patients, along with tumor size, tumor stage and histological differentiation.
Figure 4.
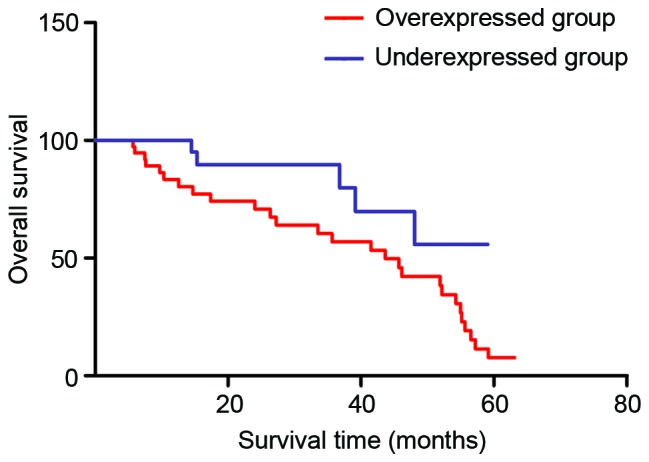
Overall survival rate assessed by Kaplan-Meier curves. Patients with Sphk1 overexpression in hepatocellular carcinoma cells have shorter 5-year survival rates compared with patients that have Sphk1 underexpression.
Table II.
Univariate analysis and multivariate analysis of overall survival.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sphk1 | 2.150 | 1.051–4.395 | 0.019 | 2.021 | 1.011–4.037 | 0.017 |
| Age | 1.025 | 0.522–2.014 | 0.424 | – | – | – |
| Gender | 1.116 | 0.563–2.212 | 0.375 | – | – | – |
| Tumor size | 2.204 | 1.026–4.737 | 0.036 | 1.883 | 0.960–3.693 | 0.031 |
| HBsAg | 1.583 | 0.765–3.275 | 0.071 | – | – | – |
| Histological differentiation | 2.011 | 0.960–4.213 | 0.038 | 1.851 | 0.936–3.659 | 0.033 |
| AFP | 1.316 | 0.651–2.664 | 0.120 | – | – | – |
| Tumor stage | 2.087 | 1.009–4.316 | 0.026 | 1.848 | 0.935–3.654 | 0.034 |
Sphk1, sphingosine kinase 1; HR, hazard ratio; CI, confidence index; HBsAg, hepatitis B virus surface antigen; AFP, α-fetoprotein.
Discussion
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-associated morbidity and mortality worldwide. The lack of an efficient early diagnosis method may be responsible for that. Therefore, studies have focused their attention on molecular genetics and pathogenesis of HCC (16,17). Notably, rapid expansion of information was obtained from these studies, and new diagnostic markers were developed owing to this progression. However, the overall survival quality of patients with HCC remains unsatisfactory. Therefore, the molecular genetics of HCC and the molecular mechanism of hepatocarcinogenesis remain to be revealed.
Increasing studies have showed that Sphk1 is overexpressed in a number of human cancers, including head and neck, gastric, lung, colon and ovarian cancers. The link between Sphk1 overexpression and carcinogenesis has been well established in the aforementioned cancers (5,9–12). However, the role of Sphk1 overexpression in HCC remains elusive. Therefore, Sphk1 expression status of tissues obtained from patients with HCC was examined at the mRNA and protein level simultaneously. In the present study, the results revealed that Sphk1 was overexpressed in 73.23% patients enrolled in the present study. Furthermore, immunohistochemistry analysis demonstrated that Sphk1 overexpression is associated with tumor differentiation. Notably, overexpression of Sphk1 was linked with well-known cancer progression-associated clinicopathological parameters, including tumor size, tumor stage and histological differentiation, which implies that Sphk1 overexpression may be a valuable contributing factor in HCC progression.
In order to evaluate the mechanism of Sphk1 overexpression in HCC, Sphk1 expression was examined in HCC cell lines and normal liver cell lines. Consistent with the previous results obtained from human cancers and our result from HCC tissues, the present study demonstrated that the mRNA and protein level of Sphk1 are significantly elevated in HCC cell lines (all P<0.001). In addition, sphk1 expression in normal liver cell lines was upregulated by CRISPR/Cas9 based synergistic activation mediator construct, while sphk1 expression in HCC cell lines was downregulated through sphk1-specific shRNA. Based on these findings, the cell proliferation rate of the normal liver cell line, HCC cell lines and engineered cell lines constructed in the present study were further assessed. The data suggested that sphk1 overexpression could prompt cell proliferation and this effect may explain the role of sphk1 overexpression in HCC. Notably, the 5-year survival rates of HCC patients with high sphk1 overexpression were low. In the end, the association of clinicopathological parameters and survival rates of patients with HCC were analyzed using univariate and multivariate analysis methods. Sphk1 overexpression was revealed to be an independent indicator for poor prognosis of HCC patients.
In conclusion, results drawn from the present study are consistent with previous studies demonstrating an association between increased sphk1 expression and aggressive oncogenic behaviors including tumor size and histological differentiation. To the best of our knowledge, this is the first study to reveal the expression status and clinical significance of sphk1 in HCC. Therefore, a valuable molecular target for the diagnosis and treatment of HCC has been provided.
References
- 1.McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: An emphasis on demographic and regional variability. Clin Liver Dis. 2015;19:223–238. doi: 10.1016/j.cld.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayo J, Fiore E, Aquino JB, Malvicini M, Rizzo M, Peixoto E, Alaniz L, Piccioni F, Bolontrade M, Podhajcer O, et al. Human umbilical cord perivascular cells exhibited enhanced migration capacity towards hepatocellular carcinoma in comparison with bone marrow mesenchymal stromal cells: A role for autocrine motility factor receptor. Biomed Res Int. 2014;2014:837420. doi: 10.1155/2014/837420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chessum N, Jones K, Pasqua E, Tucker M. Recent advances in cancer therapeutics. Prog Med Chem. 2015;54:1–63. doi: 10.1016/bs.pmch.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 5.Kawamori T, Kaneshiro T, Okumura M, Maalouf S, Uflacker A, Bielawski J, Hannun YA, Obeid LM. Role of sphingosine kinase 1 in colon carcinogenesis. FASEB J. 2009;23:405–414. doi: 10.1096/fj.08-117572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyne NJ, Tonelli F, Lim KG, Long JS, Edward J, Pyne S. Sphingosine 1-phosphate signaling in cancer. Biochem Soc Trans. 2012;40:94–100. doi: 10.1042/BST20110602. [DOI] [PubMed] [Google Scholar]
- 7.Pitson SM, Xia P, Leclercq TM, Moretti PA, Zebol JR, Lynn HE, Wattenberg BW, Vadas MA. Phosphorylation-dependent translocation of sphingosine kinase to the plasma membrane drives its oncogenic signaling. J Exp Med. 2005;201:49–54. doi: 10.1084/jem.20040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gault CR, Obeid LM. Still benched on its way to the bedside: Sphingosine kinase 1 as an emerging target in cancer chemotherapy. Crit Rev Biochem Mol Biol. 2011;46:342–351. doi: 10.3109/10409238.2011.597737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Facchinetti MM, Gandini NA, Fermento ME, Sterin-Speziale NB, Ji Y, Patel V, Gutkind JS, Rivadulla MG, Curino AC. The expression of sphingosine kinase-1 in head and neck carcinoma. Cells Tissues Organs. 2010;192:314–324. doi: 10.1159/000318173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Yu CP, Xia JT, Zhang L, Weng GX, Zheng HQ, Kong QL, Hu LJ, Zeng MS, Zeng YX, et al. Sphingosine kinase 1 is associated with gastric cancer progression and poor survival of patients. Clin Cancer Res. 2009;15:1393–1399. doi: 10.1158/1078-0432.CCR-08-1158. [DOI] [PubMed] [Google Scholar]
- 11.Johnson KR, Johnson KY, Crellin HG, Ogretmen B, Boylan AM, Harley RA, Obeid LM. Immunohistochemistry distribution of sphingosine kinase 1 in normal and tumor lung tissues. J Histochem Cytochem. 2005;53:1159–1166. doi: 10.1369/jhc.4A6606.2005. [DOI] [PubMed] [Google Scholar]
- 12.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- 13.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genoma-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. [PubMed] [Google Scholar]
- 15.Liu SQ, Huang JA, Qin MB, Su YJ, Lai MY, Jiang HX, Tang GD. Sphingosine kinase 1 enhances colon cancer cell proliferation and invasion by upregulating the production of MMP-2/9 and uPA via MAPK pathways. Int J Colorectal Dis. 2012;27:1569–1578. doi: 10.1007/s00384-012-1510-y. [DOI] [PubMed] [Google Scholar]
- 16.Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1:593–598. doi: 10.3892/mco.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu L, Zhang Y, Zhu Y, Cong Q, Xiang Y, Fu L. The effect of LOXL2 in hepatocellular carcinoma. Mol Med Rep. 2016;14:1923–1932. doi: 10.3892/mmr.2016.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]