Abstract
Placenta-specific protein 1 (PLAC1) expression is co-opted in numerous human cancers. As a consequence of PLAC1 expression, tumor cells exhibit enhanced proliferation and invasiveness. This characteristic is associated with increased aggressiveness and worse patient outcomes. Recently, the presence of the tumor suppressor p53 was shown in vitro to inhibit PLAC1 transcription by compromising the P1, or distal/cancer, promoter. We sought to determine if this phenomenon occurs in primary patient tumors as well. Furthermore, we wanted to know if p53 mutation influenced PLAC1 expression as compared with wild-type. We chose to study serous ovarian tumors as they are well known to have a high rate of p53 mutation. We report herein that the phenomenon of PLAC1 transcription repression does occur in serous ovarian carcinomas but only when TP53 is wild-type. We find that mutant or absent p53 protein de-represses PLAC1 transcription. We further propose that the inability of mutant p53 to repress PLAC1 transcription is due to the fact that the altered TP53 protein is unable to occupy a putative p53 binding site in the PLAC1 P1 promoter thus allowing transcription to occur. Finally, we show that PLAC1 transcript number is significantly negatively correlated with patient survival in our samples. Thus, we suggest that characterizing tumors for TP53 mutation status, p53 protein status and PLAC1 transcription could be used to predict likely prognosis and inform treatment options in patients diagnosed with serous ovarian cancer.
Keywords: ovarian cancer, placenta-specific protein 1, p53
Introduction
Placenta-specific protein 1 (PLAC1), located at human chromosome Xq26, is a small (212 amino acid) secreted protein whose normal expression is limited almost exclusively to placental trophoblasts (1–3). It is unique to the Placentalia having first arisen some 165 million years ago and is conserved in placental mammal genomes by purifying selection (4). Since its discovery, PLAC1 has been shown to be a crucial element in the establishment and maintenance of normal placentation (5,6). In addition, beginning in 2006, PLAC1 expression has been detected in a wide range of cancers and cancer cell lines (7–16). Moreover, high PLAC1 expression is significantly associated with more advanced and/or aggressive disease leading to worse patient outcomes (13,15). Two crucial in vitro studies of PLAC1 demonstrated that knockdown in MCF-7 and BT-549 breast cancer cells (8) and in HTR8/SVneo trophoblast cells (6) significantly ablated both proliferation and invasiveness of the cells. Such results clearly provide an explanation as to why tumors induce PLAC1 expression.
PLAC1 mRNA transcription is initiated from two separate promoters, usually termed P1, or distal, and P2, or proximal. Both promoters appear to be active within cells that express PLAC1, but the latter promoter has been shown to be the primary transcription driver in placental tissues while the former is the primary transcription driver in cancers (14,17). A recent in vitro study of P1 transcription in transformed fibroblasts found that p53 protein can repress transcription by blocking binding of the RXRα and LXRβ transcription factors (18). We wanted to determine if this transcriptional repression mechanism was operating in patient tumors and what effect p53 mutation might have on PLAC1 transcription repression. In addition, we wanted to know whether or not the p53/PLAC1 relationship had an influence on patient outcomes, specifically survival. The presence of PLAC1 expression in ovarian cancers has been known for some time (12) and because serous ovarian cancers characteristically have high rates of p53 mutation (19) we considered it to be the ideal cancer on which to focus. To this end, we assembled a panel of 38 serous ovarian tumors in which we determined PLAC1 mRNA expression, the percentage of PLAC1 expression specifically driven from the P1 promoter, TP53 mutation status and p53 protein expression. Importantly, we obtained clinical information including stage, grade, treatment and survival to use in assessing the effects of p53 status and PLAC1 expression.
We are able to report here that PLAC1 expression, and specifically that portion driven by the P1 promoter, is significantly determined by TP53 mutation status. We found that 13 of the 38 tumors were wild-type TP53 (34.2%) while 25 of 38 carried a p53 mutation (65.8%). Wild-type TP53 is significantly associated with PLAC1 transcription repression whereas most p53 mutants lead to de-repressed PLAC1 expression (p<0.01) and, in particular, the portion due to P1-driven transcription (p<0.001). We also find a significant negative relationship between PLAC1 transcript number and patient survival (log-rank test p<0.02). We suggest that patients with p53 compromised tumors experience a worse prognosis in part due to the positive effect that de-repressed PLAC1 expression has on cell growth and proliferation. This finding suggests the intriguing possibility that determining the combination of PLAC1 promoter-specific mRNA expression and p53 mutation status in a patient tumor could be useful as an aid in making therapeutic decisions in ovarian cancer in order to improve outcomes for those among whom there is evidence of p53 mutation and high PLAC1 expression.
Patients and methods
Patient tumor samples
Primary tumor tissues from 38 patients diagnosed with serous ovarian carcinoma and treated in the Department of Obstetrics and Gynecology of University of Iowa Hospitals and Clinics were obtained at the time of surgery under informed consent (IRB#201605841). Following histologic confirmation, tissues were flash frozen and archived in the Women's Health Tissue Repository housed within the University of Iowa Department of Obstetrics and Gynecology (20). Primary patient demographics and clinical data are presented in Table I.
Table I.
Patient demographics and clinical data.
| Characteristic | Category | No. | % |
|---|---|---|---|
| Age | 20–29 | 1 | 2.6 |
| 30–39 | 2 | 5.3 | |
| 40–49 | 5 | 13.2 | |
| 50–59 | 8 | 21.1 | |
| 60–69 | 12 | 31.6 | |
| 70–79 | 7 | 18.4 | |
| 80+ | 3 | 7.9 | |
| Stagea | R | 2 | 5.3 |
| IIIA | 3 | 7.9 | |
| IIIB | 1 | 2.6 | |
| IIIC | 24 | 63.2 | |
| IV | 8 | 21.1 | |
| Grade | ND | 4 | 10.5 |
| 2 | 3 | 7.9 | |
| 3 | 31 | 81.6 | |
| Responseb | Complete | 29 | 76.3 |
| Incomplete | 9 | 23.7 |
R, recurrent.
Complete response is defined as no evidence of disease for six months following a standard course of platinum plus Taxol chemotherapy.
Nucleic acid purifications
Total cellular RNA was purified from 0.1 g pieces of frozen tumor tissue using the mirVana RNA isolation kit according to the manufacturer's instructions (Thermo Fisher Scientific). High molecular weight genomic DNA (gDNA) was purified from adjacent tumor pieces using the DNeasy Blood and Tissue kit according to the manufacturer's instructions (Qiagen). RNA and DNA yield and purity were determined using a NanoDrop Model 100 spectrophotometer (Thermo Fisher Scientific).
Quantitative real-time PCR assay
Messenger RNA (mRNA) expression levels in each tumor was determined by SYBR Green qPCR assay. To begin, 500-ng aliquots of total RNA were reverse transcribed in the presence of SuperScript III reverse transcriptase under recommended conditions (Thermo Fisher Scientific). Resulting single stranded cDNAs were then amplified in the presence of SYBR Green on an Applied Biosystems Model 7900HT Real-Time PCR system in the Genomics Division of the University of Iowa Institute of Human Genetics (IIHG). The specific primer sequences used to amplify total PLAC1 message, promoter 1 (P1)-specific PLAC1 message, and the 18S rRNA endogenous normalizer are given in Table II.
Table II.
Primer sequences.
| Amplicon | Size (bp) | Sequencea |
|---|---|---|
| PLAC1 | 232 | F: 5′-CACCAGTGAGCACAAAGCCACATT-3′ |
| R: 5′-CCATGAACCAGTCTATGGAG-3′ | ||
| PLAC1P1 | 370 | F: 5′-AAACTTACACGAGGAGTCTGTC-3′ |
| R: 5′-CTGTGACCATGAACCAGTCTAT-3′ | ||
| 18S rRNA | 104 | F: 5′-AACTTTCGATGGTAGTCGCCG-3′ |
| R: 5′-CCTTGGATGTGGTAGCCGTTT-3′ | ||
| p53 block 1b | 908 | F: 5′-CAGACACTGGCATGGTGTT-3′ |
| R: 5′-ATATTCAACTTTGGGACAGGAGT-3′ | ||
| p53 block 2b | 661 | F: 5′-TGTAGACGCCAACTCTCTCTA-3′ |
| R: 5′-AACCCATTTACTTTGCACATCTC-3′ | ||
| p53 block 3b | 1,002 | F: 5′-TCATCTTGGGCCTGTGTTAT-3′ |
| R: 5′-AAAGCTGGTCTGGTCCTTTA | ||
| p53 block 4b | 1,412 | F: 5′-GGTACTTGAAGTGCAGTTTCTACT-3′ |
| R: 5′-GTGCAGGCCAACTTGTTCA-3′ |
All primers were designed using PrimerQuest software available on-line at Integrated DNA Technologies (www.idtdna.com). All primers were synthesized by IDT as well.
Block 1 contains exons 2–4, block 2 contains exons 5–6, block 3 contains exons 7–9 and block 4 contains exons 10–11.
Cycle threshold values (Ct) for each tumor were normalized (ΔCt) against the 18S rRNA endogenous control. Total PLAC1 transcripts and percent P1-specific message were determined for each tumor based upon a linear regression from total PLAC1- and PLAC1P1-specific standard curves. Standard curves consist of serial dilutions of PLAC1- and PLAC1P1-specific target clones ranging from 1012 to 106 copies. Total PLAC1- and PLAC1P1-specific transcripts were calculated as and . Percent P1 is then (PLAC1P1/PLAC1). Note that PLAC1 transcript numbers are per 500 ng of starting RNA for each tumor. Standard curve-derived primer efficiencies computed from the regression equations are 0.968 for total PLAC1 and 0.946 for PLAC1P1.
p53 sequencing
Tumor p53 sequencing was carried out on an Applied Biosystems Model 3730×l 96-capillary sequencer in the Genomics Division of the University of Iowa Institute of Human Genetics (IIHG). gDNA aliquots (100 ng) were PCR amplified in four blocks using primer sequences shown in Table II. Each block amplicon was visualized on a 1.3% agarose gel and then processed for sequencing using the QIAquick PCR Purification kit according to the manufacturer's instructions (Qiagen). Each purified amplicon was completely sequenced in both directions.
Protein purification and blots
Total protein was purified from the tumor tissues by homogenizing tumor tissue with a Fisher PowerGen 125 homogenizer in the presence of protease and phosphatase inhibitors. Final protein concentration was determined by BCA assay (21) (Thermo Fisher Scientific) read in duplicates on a BioRad xMark microplate spectrophotometer. Protein concentration was calculated against within-plate standard curves.
Protein expression was assessed via dot blot. A 40-µg aliquot of total protein from each tumor as determined by the BCA assay was spotted on a gridded nitrocellulose membrane. P53 expression was assessed using Santa Cruz antibody sc-126 and the β-actin control with Sigma-Aldrich A1978. Dot blots were analyzed in Image Studio ver. 5.2 (Licor). Dots were pseudo colored in Image Studio with red and blue corresponding to high and low expression respectively. To reduce the impact of outlier bright pixels the total protein signal was calculated by subtracting the local median background signal from the total pixel intensity for each dot. Protein loading was controlled by normalizing each dot to their respective β-actin protein levels.
Clinical variables and survival analysis
Univariate and multivariate association analyses via linear regressions were carried out for both total PLAC1 expression and % P1 contribution against numerous patient characteristics including age at diagnosis, BMI and treatment response as well as tumor variables including histologic type, stage, grade, TP53 mutation status and p53 protein expression. Significant variables in the univariate analyses were then included in a multivariate analysis to assess independent association. Survival analyses were performed using Cox proportional hazard ratios and comparisons between survival curves were calculated with long-rank test. Statistical significance was accepted at p<0.05.
Results
Promoter-specific PLAC1 expression
PLAC1 mRNA transcripts and the percent coming off the P1 promoter is shown for all 38 ovarian tumors in Table III. These data show there is a considerable range in both variables. As for total PLAC1 transcripts, the lowest value is 83.9×106 in OVC21 while the highest is 9.1×109 in OVC05 (median, 7.8×107). It is important to note that these values are relative to a starting mass of 500 ng of total cellular RNA. With regard to the percentage of transcript originating at the P1 promoter there is also a wide range with the lowest being 3% in OVC46 and the highest being 100% in OVC10, OVC24, OVC29, and OVC57 (median, 36%). Overall, there is a small but statistically significant Spearman rank correlation between total transcript number and % P1 transcription, r=0.363 (p<0.01).
Table III.
Clinical and molecular characteristics of the primary tumors in this study.
| Tumor ID | Age | Stage | Grade | Response | TP53 | p53 proteina | PLAC1 transcriptsb | % P1 |
|---|---|---|---|---|---|---|---|---|
| OVC02 | 71 | IIIC | 3 | CR | wt | 0.10 | 377870460 | 15 |
| OVC04 | 66 | IIIC | 3 | IR | wt | 0.04 | 218276486 | 07 |
| OVC05 | 81 | IIIC | 3 | CR | W91ter | 0.04 | 9068463009 | 05 |
| OVC06 | 44 | IV | 2 | CR | wt | 0.03 | 214248894 | 14 |
| OVC07 | 57 | IIIC | 3 | IR | wt | 0.00 | 5219928344 | 28 |
| OVC08 | 57 | R | CR | wt | 0.08 | 293903138 | 24 | |
| OVC10 | 55 | R | IR | R273H | 0.20 | 2203433716 | 100 | |
| OVC11 | 72 | IIIC | 3 | IR | F270S | 0.03 | 4483436342 | 33 |
| OVC12 | 68 | IIIC | 2 | CR | wt | 0.00 | 140271823 | 12 |
| OVC13 | 72 | IIIC | 3 | IR | R280G | 3.37 | 5534290300 | 42 |
| OVC14 | 58 | IV | 3 | CR | Y220C | 0.11 | 367149085 | 72 |
| OVC16 | 38 | IIIC | 3 | CR | R273H | 0.21 | 318066098 | 52 |
| OVC17 | 49 | IIIC | 2 | CR | R273C | 0.81 | 4497521474 | 97 |
| OVC18 | 59 | IIIC | 3 | IR | wt | 0.12 | 299093109 | 39 |
| OVC19 | 67 | IIIC | 3 | CR | E339ter | 0.29 | 3001202216 | 73 |
| OVC21 | 65 | IIIC | 3 | IR | wt | 0.01 | 83880665 | 18 |
| OVC23 | 70 | IIIC | 3 | CR | Y163C | 0.75 | 846229522 | 44 |
| OVC24 | 64 | IIIC | CR | C135R | 4.00 | 3254926969 | 100 | |
| OVC25 | 80 | IV | 3 | CR | wt | 0.01 | 565689548 | 07 |
| OVC26 | 64 | IIIC | 3 | CR | Y220H | 0.01 | 270915037 | 22 |
| OVC28 | 73 | IIIC | 3 | CR | E358D | 0.09 | 454113159 | 49 |
| OVC29 | 66 | IIIA | 3 | CR | Y126S | 1.15 | 781595028 | 100 |
| OVC32 | 61 | IV | 3 | CR | A276G | 1.66 | 2131955668 | 93 |
| OVC33 | 53 | IV | 3 | CR | del Y126-K132 | 0.83 | 2718172019 | 100 |
| OVC34 | 45 | IIIA | 3 | IR | del N131 | 1.68 | 4185362949 | 76 |
| OVC35 | 27 | IIIB | 3 | CR | wt | 0.01 | 237797036 | 09 |
| OVC43 | 60 | IV | 3 | CR | R248Q | 0.05 | 1652055526 | 74 |
| OVC45 | 62 | IV | 3 | CR | V173M | 0.79 | 2923011797 | 06 |
| OVC46 | 75 | IIIC | 3 | CR | wt | 1.16 | 722147991 | 08 |
| OVC50 | 67 | IIIC | 3 | CR | V173M | 1.44 | 1516484830 | 96 |
| OVC51 | 62 | IIIC | 3 | CR | R280G | 1.06 | 443139152 | 53 |
| OVC52 | 57 | IIIC | 3 | CR | R248Q | 0.36 | 769568136 | 60 |
| OVC53 | 45 | IV | 3 | CR | wt | 0.07 | 885711108 | 26 |
| OVC54 | 72 | IIIC | 3 | CR | A161ter | 0.33 | 779703161 | 05 |
| OVC56 | 86 | IIIA | 3 | CR | wt | 0.77 | 551335638 | 81 |
| OVC57 | 37 | IIIC | 3 | IR | D281H | 4.00 | 787667563 100 | |
| OVC59 | 49 | IIIC | 3 | CR | R248W | 0.29 | 4065716208 | 30 |
| OVC63 | 54 | IIIC | CR | R342ter | 0.18 | 928899559 | 15 |
Relative image intensity based upon dot blot scan.
per 500 ng of total protein.
p53 sequencing
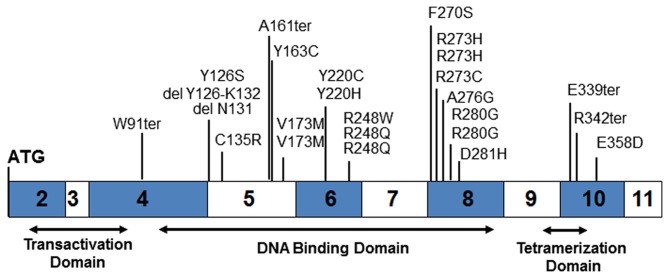
Sequencing of the p53 gene from tumor gDNA revealed 25 mutations (65.8%) and 15 wild-type (34.2%). Consistent with other p53 mutation studies, 21 of the 25 mutations are located in the DNA-binding domain (84.0%), one in the transactivation domain and three in the tetramerization domain (Fig. 1 and Table III). Four mutations result in the creation of premature stop codons and two others result in amino acid deletions. All of the remaining 19 mutants lead to single amino acid changes.
Figure 1.
Map of the twenty-five TP53 mutations found by sequencing all 38 tumors in this study. Organization of the p53 coding region including the three major functional domains is shown.
One of the two deletion mutants, OVC34, is loss of the codon for N131 while the other, OVC33, is a loss of the splice acceptor at exon 5 leading to skipping amino acids Y126 to K132. The K132 codon, AAG, places a splice acceptor in-frame at M133. Of the four termination mutants, three, OVC05, OVC19 and OVC63, are direct creation of stop codons. The fourth deletion mutant, OVC54, is a single base G loss at M160 or A161 resulting in a frame-shift ending with a stop codon at M169.
Twenty-one mutations occur in the DNA-binding domain, including the two deletion mutants and one of the four termination mutants. Several of these DNA-binding domain mutants are 'DNA contact' mutants involving R248, R273 or R280 (22). None of the observed mutations affect residues responsible for maintaining the structural integrity of the DNA binding surface such as R175, G245, R249 and R282 but mutants V173M (OVC45 and OVC50) and D281H (OVC57) are adjacent. Three mutants, F270S (OVC11), Y220C (OVC14) and Y220H (OVC26), are located in another tertiary structure, the β-sandwich, responsible for maintaining protein structural integrity (22).
p53 and PLAC1 transcription
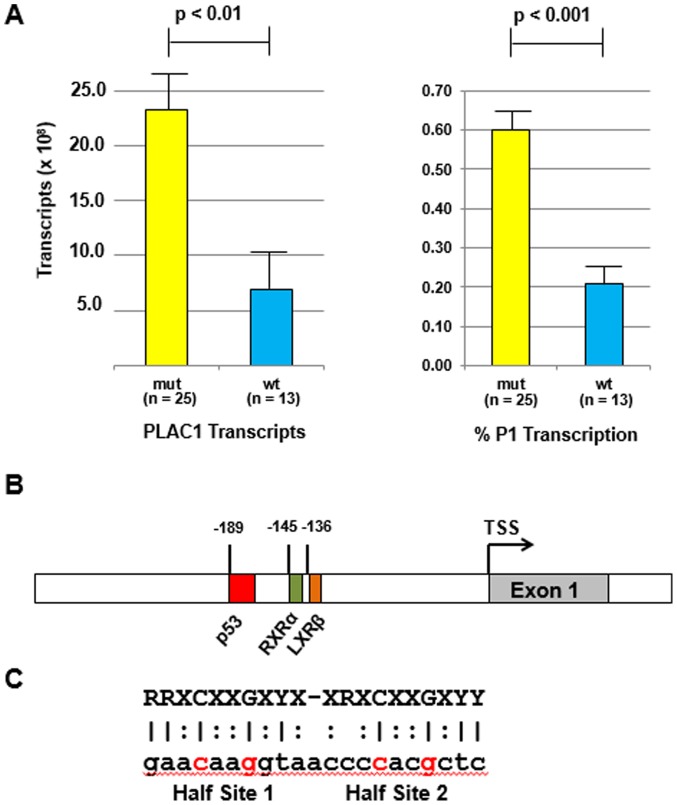
The relationship between PLAC1 transcription and p53 mutation status is shown in Fig. 2A. There is a clear association with p53 mutation status with both total PLAC1 transcripts as well as for % P1 transcripts. For total PLAC1 mRNA transcripts, tumors containing a p53 mutant present significantly more message than do tumors with wild-type p53 (p<0.01). Similarly, tumors containing a p53 mutant have a significantly higher percentage of that PLAC1 message coming from the P1 promoter (p<0.001). These results are consistent with the finding in vitro that PLAC1 transcription from the P1 promoter is ablated in the presence of wild-type p53 (18).
Figure 2.
(A) PLAC1 transcript number (left) and % P1 promoter-specific transcription (right) in tumors harboring a TP53 mutation versus tumors with wild-type TP53. (B) The genomic region around the PLAC1P1 transcription start site (TSS) showing the location of the RXRα and LXRβ transcription factor binding sites (17) as well as the putative p53 binding site. (C) Sequence of the putative p53 binding site aligned with a consensus transcription repressor sequence (23).
Examination of human PLAC1 genomic sequence upstream of the P1 transcription start site includes the previously reported binding sites for RXRα and LXRβ transcription factors (17) and a possible p53 response element (Fig. 2B). While this p53RE is not canonical for p53 transcription initiation sites, it is consistent with the more relaxed structures associated with p53 transcription repression (Fig. 2C) (23).
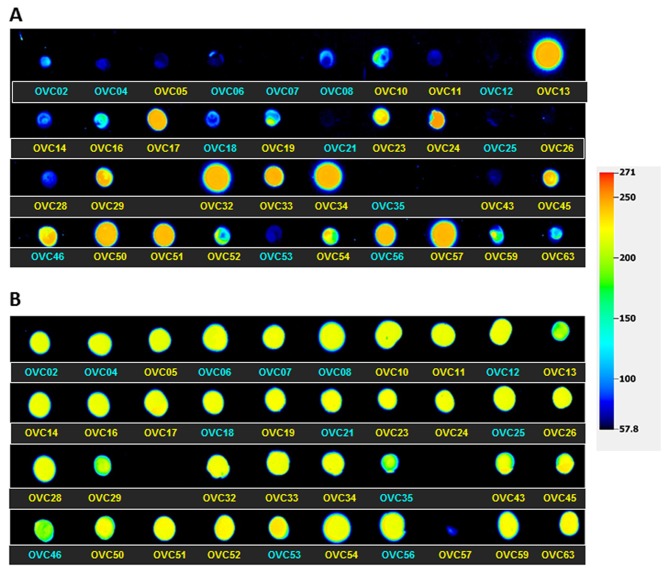
p53 protein expression
Recognizing that a p53 null tumor would likely de-repress PLAC1 transcription in the same way that TP53 mutants would, we purified protein from each of the 38 tumors and spotted a dot blot. The blot was then scanned and a relative signal intensity value assigned to each tumor (Table III). A false-color image of p53 protein expression levels in all 38 tumors is shown in Fig. 3. These data show measurable protein detected in all but two of the tumors, OVC07 and OVC12. Several of the other wild-type tumors display very low but detectable p53 protein levels. It can also be seen that it is the mutant p53 proteins that are present in significantly higher levels versus wild-type protein (p<0.01).
Figure 3.
(A) False color image of p53 protein expression intensity on a dot blot containing all 38 ovarian cancers. (B) False color image of β-actin loading control expression on the same dot blot as shown in (A). Tumors containing a wild-type TP53 gene are in blue, tumors containing a TP53 mutation are yellow. Blue images indicate low expression levels and red images indicate high expression levels. Tumor ID numbers correspond to those listed in Table III. Two tumors, OVC30 and OVC39, were removed from this figure and from the rest of the study as it was determined on final review that neither was a serous ovarian cancer.
Patient outcome
We and others have clearly associated high levels of PLAC1 in cancers with poor patient outcomes (24). Moreover, mutations in p53, especially a group of mutants collectively referred to as oncomorphic (25), have also been linked to poor outcomes in ovarian cancers. One of the selection criteria applied to the tumor panel used in the present study is that there must be complete follow-up on the patients. Using these data we analyzed tumor characteristics against patient survival. We found a significant negative relationship between PLAC1 transcript number (in 108 copies per 500 ng total RNA) and patient survival (in months from surgery) (log-rank test p<0.02) (Fig. 4).
Figure 4.
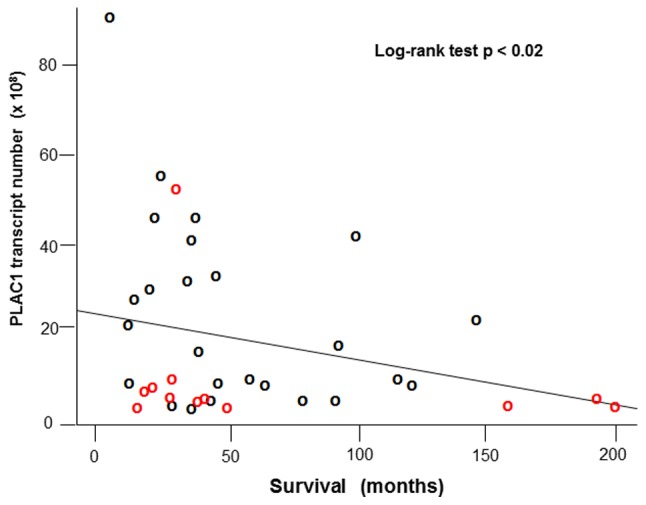
Log-rank regression of PLAC1 transcript number (x108 per 500 ng of total RNA) on patient survival (in months from diagnosis). Tumors with wild-type TP53 are indicated with red circles while tumors bearing TP53 mutations are indicated with black circles.
Discussion
We have presented evidence from a panel of 38 serous ovarian tumors that expression of placenta-specific protein 1 (PLAC1) as well as the proportion of that expression emanating from the P1, or distal/cancer, promoter is significantly determined by the presence of the tumor suppressor p53 and its mutation status. We have shown that whether p53 protein is mutated or absent the effect is to de-repress PLAC1 transcription. Also, consistent with studies in other cancers (13,15), high PLAC1 expression in serous ovarian cancer is associated with more aggressive disease and worse patient outcomes.
The previously reported in vitro study of the role of TP53 in PLAC1 P1 transcriptional repression also examined common p53 DNA contact mutants R175H, R248W, R249S and R273H and showed that PLAC1P1 transcription is substantially increased (18). It is further commented on that, among human cancer cell lines, those with p53 mutations tend to have higher PLAC1 expression levels (8,9,13,18). The results reported herein further support and extend the in vitro data to primary serous ovarian tumors. Taking % P1 transcription ≤25% as an operational definition of repression, nearly all p53 wild-type tumors repress PLAC1 transcription and nearly all p53 mutant tumors de-repress PLAC1 transcription (Table III). However, there are exceptions. For example, tumors OVC45 and OVC50 both carry a V173M mutation and display relatively similar levels of PLAC1 transcript. While OVC50 is clearly de-repressed at 96% PLAC1P1, OVC45 is only 6% PLAC1P1. These data would suggest that more than just TP53 status is contributing to PLAC1 regulation in a subset of tumors and those nuances will require further study. Howeever, for the greater number of tumors we studied, TP53 status appears to be a primary determinant of PLAC1 expression.
Another phenomenon appearing in the present data is that while the wild-type TP53 tumors in which protein is detectable successfully appear to repress PLAC1P1 transcription, the two presumably null tumors act very differently. OVC07 is a clear example of de-repression while OVC12 performs as a good repressor with low PLAC1 transcript number and only 12% PLAC1P1 transcription. On the other hand, OVC56 p53, though wild-type, is highly expressed but PLAC1P1 is 81%. In general though, mutant p53 proteins are highly expressed in our tumors and PLAC1 expression is de-repressed in spite of protein levels. On the contrary, it appears that only a relative small amount of wild-type p53 is sufficient to repress PLAC1 mRNA transcription. Mutant p53 protein overexpression is termed the dominant-negative effect (22,26) and it appears that cells are trying to compensate for the defective protein by making a great deal more of it. We observe that this cellular response is in vain, at least as regards PLAC1 mRNA transcription repression.
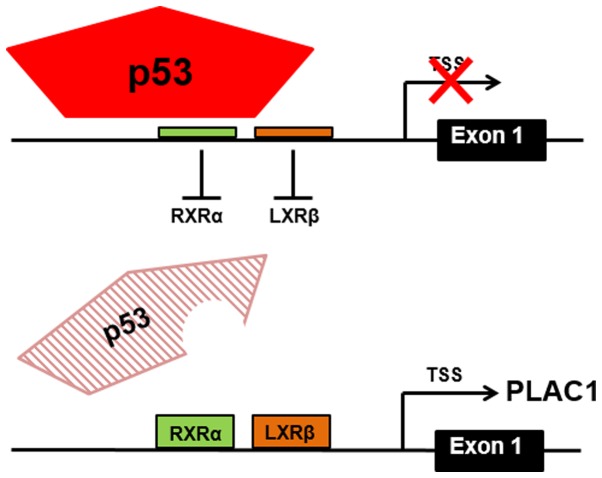
Deservedly known as the guardian of the genome (27), p53 carries out its role as a major transcriptional regulator by activating or repressing transcription of a host of target genes (28–30). Historically, the transcriptional activation function of p53 has been the most studied and best understood of the two functions but evidence is accumulating regarding the transcription repression function that is in play here (31). Among the mechanisms of transcriptional repression exerted by p53, it appears that simple direct competition with other transcription factors is the most likely means by which p53 exerts its influence on PLAC1 expression. That is, p53 occupies a binding site in the PLAC1P1 promoter through which it prevents binding of the usual transcription factors RXRα and LXRβ (18) (Fig. 5). Excellent in vitro experimental evidence supporting a more sophisticated model of PLAC1P1 transcriptional regulation has been presented elsewhere (18). This model involves the interplay of numerous components including p53, RB, the nuclear receptor co-activator NCOA2 and histone modulation. However, for the majority of the serous ovarian tumors in this study the simple assessment of whether or not p53 protein is present and whether or not it is mutated suffices to understand PLAC1 expression. By extension then, determination of TP53 mutation status via DNA sequencing, p53 protein expression by western blotting or immunohistochemistry and PLAC1 expression by qPCR may serve in many cases to predict patient prognosis and inform treatment. We do not suggest that our simple TP53-PLAC1 axis is all that is involved by any means, especially in light of the more comprehensive in vitro PLAC1P1 transcription model (18). However, taking the basic data presented here, we propose that assessment of both TP53 and PLAC1 status could potentially be beneficial to women diagnosed with serous ovarian cancer by identifying those patients with TP53 mutated tumors and/ or p53 protein is absent. In these women the disease is likely to be more aggressive and their survival reduced. Knowing this, more targeted/adjuvant therapies may be called for from the outset.
Figure 5.
A graphic representation of p53 protein binding blocking RXRα and LXRβ transcription factor binding at the PLAC1P1 promoter (above) and the inability of mutant p53 protein to bind and displace RXRα and LXRβ transcription factor binding (below). Note that p53 null tumors will have the same inability to repress PLAC1P1 transcription.
Acknowledgments
This study was supported in part by NIH R01CA99908 and R01CA184101 to Kimberly K. Leslie, NIH K12HD000849 to Henry D. Reyes, the University of Iowa Carver College of Medicine Department of Obstetrics and Gynecology Research Development Fund, and DOD Collaborative Undergraduate HBCU Student Summer training program Awards W81XWH-12-1-0117 and W81XWH-13-1-0178 to Nonye Ibik. We also acknowledge the Department of Obstetrics and Gynecology Women's Health Tissue Repository and Gynecologic Malignancy Repository (Dr Donna Santillan, Director), the continued invaluable assistance of the University of Iowa Institute of Human Genetics Genomics Facility (in particular Garry Hauser and Mary Boes) and Dr Kristina Thiel for her editorial expertise and advice.
References
- 1.Cocchia M, Huber R, Pantano S, Chen EY, Ma P, Forabosco A, Ko MS, Schlessinger D. PLAC1 an Xq26 gene with placenta-specific expression. Genomics. 2000;68:305–312. doi: 10.1006/geno.2000.6302. [DOI] [PubMed] [Google Scholar]
- 2.Massabbal E, Parveen S, Weisoly DL, Nelson DM, Smith SD, Fant M. PLAC1 expression increases during trophoblast differentiation: Evidence for regulatory interactions with the fibroblast growth factor-7 (FGF-7) axis. Mol Reprod Dev. 2005;71:299–304. doi: 10.1002/mrd.20272. [DOI] [PubMed] [Google Scholar]
- 3.Fant M, Barerra-Saldana H, Dubinsky W, Poindexter B, Bick R. The PLAC1 protein localizes to membranous compartments in the apical region of the syncytiotrophoblast. Mol Reprod Dev. 2007;74:922–929. doi: 10.1002/mrd.20673. [DOI] [PubMed] [Google Scholar]
- 4.Devor EJ. Placenta-specific protein 1 is conserved throughout the Placentalia under purifying selection. Sci World J. 2014;2014:537356. doi: 10.1155/2014/537356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackman SM, Kong X, Fant ME. Plac1 (placenta-specific 1) is essential for normal placental and embryonic development. Mol Reprod Dev. 2012;79:564–572. doi: 10.1002/mrd.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang W-L, Yang Q, Zhang H, Lin H-Y, Zhou Z, Lu X, Zhu C, Xue LQ, Wang H. Role of placenta-specific protein 1 in trophoblast invasion and migration. Reproduction. 2014;148:343–352. doi: 10.1530/REP-14-0052. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Pang X-W, Liu F-F, Dong X-Y, Wang H-C, Wang S, Zhang Y, Chen WF. PLAC1/CP1 gene expression and autologous humoral immunity in gastric cancer patients. Beijing Da Xue Xue Bao. 2006;38:124–127. In Chinese. [PubMed] [Google Scholar]
- 8.Koslowski M, Sahin U, Mitnacht-Kraus R, Seitz G, Huber C, Türeci O. A placenta-specific gene ectopically activated in many human cancers is essentially involved in malignant cell processes. Cancer Res. 2007;67:9528–9534. doi: 10.1158/0008-5472.CAN-07-1350. [DOI] [PubMed] [Google Scholar]
- 9.Silva WA, Jr, Gnjatic S, Ritter E, Chua R, Cohen T, Hsu M, Jungbluth AA, Altorki NK, Chen YT, Old LJ, et al. PLAC1 a trophoblast-specific cell surface protein, is expressed in a range of human tumors and elicits spontaneous antibody responses. Cancer Immun. 2007;7:18. [PMC free article] [PubMed] [Google Scholar]
- 10.Dong X-Y, Peng J-R, Ye Y-J, Chen H-S, Zhang L-J, Pang XW, Li Y, Zhang Y, Wang S, Fant ME, et al. Plac1 is a tumor-specific antigen capable of eliciting spontaneous antibody responses in human cancer patients. Int J Cancer. 2008;122:2038–2043. doi: 10.1002/ijc.23341. [DOI] [PubMed] [Google Scholar]
- 11.Liu F-F, Dong X-Y, Pang X-W, Xing Q, Wang H-C, Zhang HG, Li Y, Yin YH, Fant M, Ye YJ, et al. The specific immune response to tumor antigen CP1 and its correlation with improved survival in colon cancer patients. Gastroenterology. 2008;134:998–1006. doi: 10.1053/j.gastro.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 12.Tchabo NE, Mhawech-Fauceglia P, Caballero OL, Villella J, Beck AF, Miliotto AJ, Liao J, Andrews C, Lele S, Old LJ, et al. Expression and serum immunoreactivity of developmentally restricted differentiation antigens in epithelial ovarian cancer. Cancer Immun. 2009;9:6. [PMC free article] [PubMed] [Google Scholar]
- 13.Devor EJ, Leslie KK. The oncoplacental gene placenta-specific protein 1 is highly expressed in endometrial tumors and cell lines. Obstet Gynecol Int. 2013;2013:807849. doi: 10.1155/2013/807849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devor EJ, Reyes HD, Santillan DA, Santillan MK, Onukwugha C, Goodheart MJ, Leslie KK. Placenta-specific protein 1: A potential key to many oncofetal-placental OB/GYN research questions. Obstet Gynecol Int. 2014;2014:678984. doi: 10.1155/2014/678984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghods R, Ghahremani MH, Madjd Z, Asgari M, Abolhasani M, Tavasoli S, Mahmoudi AR, Darzi M, Pasalar P, Jeddi-Tehrani M, et al. High placenta-specific 1/low prostate-specific antigen expression pattern in high-grade prostate adenocarcinoma. Cancer Immunol Immunother. 2014;63:1319–1327. doi: 10.1007/s00262-014-1594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F-F, Shen D, Kang X, Zhang C, Song Q. New tumor antigen PLAC1/CP1, a potentially useful prognostic marker and immunotherapy target for gastric adenocarcinoma. J Clin Pathol. 2015;68:913–916. doi: 10.1136/jclinpath-2015-202978. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Moradin A, Schlessinger D, Nagaraja R. RXRα and LXR activate two promoters in placenta- and tumor-specific expression of PLAC1. Placenta. 2011;32:877–884. doi: 10.1016/j.placenta.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Schlessinger D, Nagaraja R. T antigen transformation reveals Tp53/RB-dependent route to PLAC1 transcription activation in primary fibroblasts. Oncogenesis. 2013;2:e67. doi: 10.1038/oncsis.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santillan MK, Leslie KK, Hamilton WS, Boese BJ, Ahuja M, Hunter SK, Santillan DA. 'Collection of a lifetime: A practical approach to developing a longitudinal collection of women's healthcare biological samples'. Eur J Obstet Gynecol Reprod Biol. 2014;179:94–99. doi: 10.1016/j.ejogrb.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 22.Joerger AC, Fersht AR. Structure-function-rescue: The diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Xiao Z, Ren EC. Redefining the p53 response element. Proc Natl Acad Sci USA. 2009;106:14373–14378. doi: 10.1073/pnas.0903284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devor EJ. Placenta-specific protein 1 (PLAC1) is a unique onco-fetal-placental protein and an underappreciated therapeutic target in cancer. Integr Cancer Sci Therap. 2016;3:479–483. doi: 10.15761/ICST.1000192. [DOI] [Google Scholar]
- 25.Brachova P, Thiel KW, Leslie KK. The consequence of oncomorphic TP53 mutations in ovarian cancer. Int J Mol Sci. 2013;14:19257–19275. doi: 10.3390/ijms140919257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blagosklonny MV. p53 from complexity to simplicity: Mutant p53 stabilization, gain-of-function, and dominant-negative effect. FASEB J. 2000;14:1901–1907. doi: 10.1096/fj.99-1078rev. [DOI] [PubMed] [Google Scholar]
- 27.Lane DP. Cancer p53 guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 28.Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Kraus WL, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brachova P, Mueting SR, Carlson MJ, Goodheart MJ, Button AM, Mott SL, Dai D, Thiel KW, Devor EJ, Leslie KK. TP53 oncomorphic mutations predict resistance to platinum- and taxane-based standard chemotherapy in patients diagnosed with advanced serous ovarian carcinoma. Int J Oncol. 2015;46:607–618. doi: 10.3892/ijo.2014.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, Huarte M. To repress or not to repress: This is the guardian's question. Trends Cell Biol. 2011;21:344–353. doi: 10.1016/j.tcb.2011.04.002. [DOI] [PubMed] [Google Scholar]