Abstract
Special AT-rich sequence-binding protein-1 (SATB1) is associated with cancer progression and poor clinical outcome. The present study aims to evaluate whether SATB1 affects the biological behaviors of prostate cancer (PCa), and furthermore, to elucidate whether this effect works through the epithelial-mesenchymal transition (EMT) pathway. Firstly, the expression of SATB1 was investigated in a series of PCa tissues as well as in a panel of PCa cell lines. Cell proliferation, migration and invasion were evaluated in SATB1 knockdown and overexpressed PCa cell lines by MTT and Transwell assays. The results showed that the expression of SATB1 was markedly upregulated in PCa tissues and all PCa cell lines (P<0.001). Ectopic expression of SATB1 promoted PCa cell proliferation and migration. Knockdown of SATB1 repressed the ability of cell proliferation and migration of PCa cells. In addition, inhibition of SATB1 could reverse the EMT processes through upregulation of E-cadherin and downregulation of vimentin. The present study provided evidence that SATB1 may act as a potential therapeutic target in PCa patients.
Keywords: SATB1, prostate cancer, metastasis, invasion, proliferation, epithelial-mesenchymal transition
Introduction
Prostate cancer (PCa) is one of the leading non-cutaneous malignancies, and the second most common cause of cancer-associated mortality in males (1). Epidemiological studies have suggested that multiple risk factors are involved in prostate carcinogenesis (2,3). A previous study determined that the high mortality rate of PCa led to an increased need to identify the factors affecting cancer progression and the need to understand the underlying molecular mechanisms. Although radical prostatectomy and radiation therapy are commonly used to improve the outcome of patients with PCa, the postoperative 5-year survival rate is low (4). There is currently no curative therapy for patients with metastatic PCa. Thus, the clinical treatment of PCa patients remains a challenge.
Special AT-rich sequence-binding protein-1 (SATB1) is a tissue specific nuclear protein that has emerged as a new regulator of oncogenic pathways (5). Previous studies demonstrated the potential involvement of SATB1 in various cancers (6,7). SATB1 participates in a variety of biologically important processes including cell proliferation, differentiation, apoptosis and the reprogramming of expression profiles. However, the expression and role of SATB1 in PCa remains unknown. The aim of the current study was to determine whether SATB1 affects the biological behaviors of PCa, and furthermore, to elucidate whether this effect works through the epithelial-mesenchymal transition (EMT) pathway.
Materials and methods
Human tissue samples
A total of 98 pairs of PCa tissue and matched normal prostate tissues were obtained from patients who had undergone surgical resection at Southern Medical University (Guangzhou, China) between December 2010 and November 2014. Consent was obtained from all patients, and the experimental protocols were approved by the local ethics committee of Southern Medical University. Clinical and pathological information were collected from a retrospective review of well-documented medical records, and the characteristics of patients were detailed.
Cell lines and culture conditions
The PC3, LNCaP, and 22Rv1 cell lines (American Type Culture Collection, Manassas, VA, USA) were cultured in RPMI-1640, supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The DU145 cell line was cultured in minimum essential Eagle's medium supplemented with 10% FBS and 2 mM L-glutamine with antibiotics. The human non-tumorigenic prostate epithelial cell line RWPE-1 was cultured in keratinocyte serum-free medium supplemented with 5 ng/ml human recombinant epidermal growth factor and 30 mg/ml bovine pituitary extract (Invitrogen; Thermo Fisher Scientific, Inc.). Cultures were maintained in a 5% CO2 humidified atmosphere at 37°C.
Immunohistochemistry (IHC)
Primary PCa tissues and adjacent normal tissues obtained following surgery were subjected to IHC analysis, according to the routine protocol. In brief, formalin-fixed and paraffin-embedded specimens were sectioned at 3–4 µm thickness for immunohistochemistry. The samples were deparaffinized with xylene, and rehydrated through a series of decreasing concentrations of ethanol to water. A high temperature antigen retrieval method was performed with citrate buffer solution (Maixin Bio, Fujian, China), and the slides were immersed in 100 µl of 3% hydrogen peroxide for 10 min at 37°C to block endogenous peroxidase activity. Subsequent to washing with PBS, the sections were incubated with 5% bovine serum albumin (BSA; Sigma-Aldrich, EMD Millipore, Billerica, MA, USA) for 30 min, followed by incubation with anti-SATB1 antibody (1:100, cat. no. ab92307, Abcam, Cambridge, UK) at 4°C overnight. Following washing with PBS, the sections were incubated with secondary antibody for 30 min at 37°C. The slides were stained with diaminobenzidine (as the color reagent) and hematoxylin (as a counterstain for nuclei). PBS was used as a negative control for the staining reactions. All slides were dehydrated in increasing concentrations of ethanol and xylene. Finally, all sections were mounted with neutral gum.
The IHC sections were scored by three pathologists independently to ensure inter-observer agreement. A total of five representative high power fields of fluorescence microscopy were selected. Each section was scored according to the intensity and percentage of positive cells (magnification, ×400). Staining intensity was scored as follows: 0, negative; 1, weakly positive; 2, moderately positive; and 3, strongly positive. The percentage of positive cells was also graded according to four categories: 1, 0–25% positive cells; 2, 26–50% positive cells; 3, 51–75% positive cells; and 4 >75% positive cells. Overall scores ≤3 were defined as negative; scores >3 were defined as positive.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from specimens or cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For RT-qPCR, RNA reverse transcribed to cDNA from 1 µg of total RNA was reverse transcribed in a final volume of 20 µl using random primers and a reverse transcription kit (Takara Biotechnology Co., Ltd., Dalian, China). According to the manufacturer's protocol, the reverse transcription was performed at 37°C for 15 min, and then at 85°C for 5 sec. RT-qPCR analysis was performed using a standard protocol from Power SYBR-Green (Takara Biotechnology Co., Ltd.). All protocols were performed according to the manufacturer's instructions. The 2−ΔΔCq values were normalized to those of GAPDH (8). Primers were synthesized by Shanghai Sangon Biological Engineering Technology Services (Shanghai, China). The primer sequences used for the studies are: SATB1 forward, 5′-GTGGAAGCCTTGGGAATCC-3′ and reverse, 5′-CTGACAGCTCTTCTTCTAGTT-3′ and GAPDH forward, 5′-TCGGAGTCAACGGATTTGGTCGT-3′ and reverse, 5′-TGCCATGGGTGGAATCATATTGGA-3′. The products of PCR were separated by 1.5% agarose gel electrophoresis, and the expression levels of the mRNA were detected, with GAPDH serving as the control. The RT-qPCR assays and data collection were performed using an ABI 7500 instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). Each sample was analyzed in triplicate.
Western blot analysis
Cells were lysed using the mammalian protein extraction reagent, radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Haimen, China), supplemented with a protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland) and PMSF (Roche Diagnostics). Proteins were separated by 10% SDS-PAGE (Sigma-Aldrich; EMD Millipore). Specific monoclonal antibodies against SATB1 (1:1,000, cat. no. ab92307, Abcam), vimentin (1:1,000, cat. no. ab92547, Abcam), E-cadherin (1:1,000, cat. no. ab11512, Abcam Inc.) and β-actin (1:5,000, cat. no. ab8227, Abcam) were applied for overnight incubation at 37°C. The secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2,000; cat. no. sc-2030, Santa Cruz Biotechnology, Inc., Dallas, TX, USA). An ECL chromogenic substrate was used to visualize the bands and the intensity of the bands was quantified by densitometry (Quantity One software; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell transfection
The SATB1 expression vector pcDNA3.1-SATB1 and SATB1-specific short hairpin (sh)RNA pGenesil2-SATB1 (the shRNA sequence was designed to target SATB1 as follows: 5′-GGAATGCTCTGAAGGACTTAC-3′) were constructed, and then transfected into 60%-confluent cells, respectively, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Following transfection, the transfected cells were allowed to recover for 24 h, and individual clones with stable transfection, screened with G418 (400 mg/ml) for two weeks, were then isolated. Thereafter, a single colony of stable cells was picked and maintained in culture medium (Gibco; Thermo Fisher Scientific, Inc.) containing 200 mg/ml G418 to further culture. All the subsequent experiments were performed using the stable cells from the single colony. Cells transfected with pcDNA3.1 or pGenesil2 control vector were used as negative controls.
Cell viability measurement
The viability of the cells was analyzed by Thiazolyl blue (MTT; Sigma-Aldrich, EMD Millipore). Following transfection, cells were seeded on a 96-well plate at a density of 104 cells/well. Following 24, 48 and 72 h of incubation at 37°C, the culture medium was removed, and 5 mg/ml MTT was added to each well. The plates were subsequently incubated at 37°C for 4 h. The culture medium was replaced with 150 µl dimethyl sulfoxide/well, and the absorbance (A) at a wavelength of 490 nm was measured using 2104 EnVision® Multilabel Reader (cat. no. 2104–0010; PerkinElmer, Inc., Waltham, MA, USA).
Transwell invasion assay
Cell invasion assay was carried out using Transwell chambers with inserts of 8-mm pore size (Corning, Corning Inc., Corning, NY, USA). Briefly, cells suspended in serum-free media were plated into the upper chamber for invasion assay following transfection, and media supplemented with 10% FBS was placed into the lower chamber. After 24 h, the cells that had invaded through the membrane to the lower surface were fixed with methanol, stained with Crystal violet for 10 min, and counted.
Statistical analysis
The data are presented as the mean ± standard error of the mean. In total, three replicate wells were examined per assay, and each experiment was performed in triplicate. Statistical comparisons among groups were determined by Student's t-test. All data were analyzed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). All statistical tests were two-tailed and P<0.05 was considered to indicate a statistically significant difference.
Results
SATB1 is upregulated in human PCa tissues and cell lines
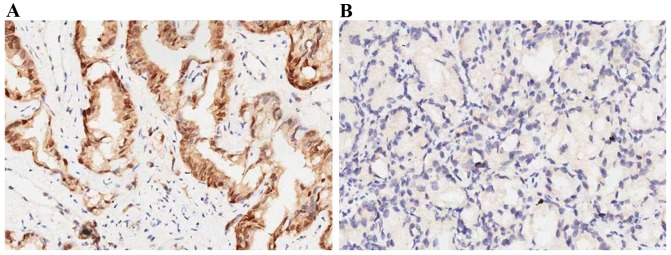
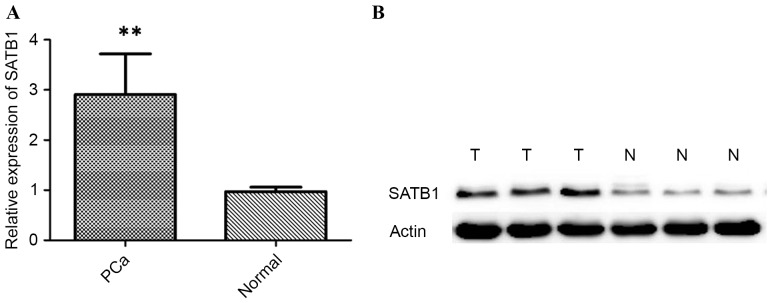
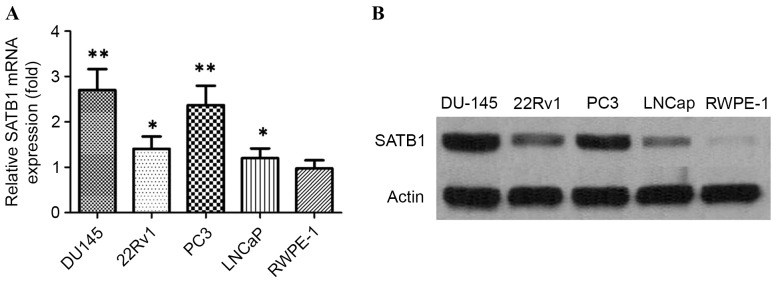
First, the expression levels of SATB1 were examined in 98 pairs of PCa tissue samples and normal prostate tissue samples were matched by performing immunohistochemistry, RT-qPCR and western blot analysis. The expression of SATB1 was revealed to be significantly upregulated in the cancerous tissues compared with the corresponding adjacent non-tumorous tissues (Fig. 1). Results showed that SATB1 protein was predominantly located in the nucleus. Positive staining of SATB1 was observed in 77/98 (78.57%) PCa cases, and in 18/98 (18.36%) adjacent non-neoplastic specimens, with statistical significance (P<0.001). The results of RT-qPCR and western blotting revealed that tumor tissues showed increased levels of SATB1 mRNA and protein compared with adjacent non-tumor tissues (P<0.01; Fig. 2). Additionally, the expression of SATB1 was examined in a panel of PCa cell lines, as well as an immortalized non-tumorigenic human prostate epithelial cell line RWPE-1. mRNA and protein levels of SATB1 were greatly elevated in all human prostate cancer cell lines in comparison to RWPE-1 cells (P<0.001), but expression levels of SATB1 varied among PCa cell lines (Fig. 3A and B).
Figure 1.
(A) Positive expression of SATB1 in the PCa tissues. (B) Absent expression of SATB1 in the adjacent normal prostate tissues (magnification, ×200). SATB1, special AT-rich sequence binding protein-1.
Figure 2.
(A) Reverse transcription-quantitative polymerase chain reaction demonstrating expression level of SATB1 mRNA in PCa tissues, and adjacent normal prostate tissues. **P<0.01. (B) Western blot analysis demonstrating expression level of SATB1 protein in PCa tissues, and adjacent normal prostate tissues. SATB1, special AT-rich sequence binding protein-1; PCa, prostate cancer.
Figure 3.
(A) Reverse transcription-quantitative polymerase chain reaction demonstrating expression level of SATB1 mRNA in PCa cell lines and RWPE-1 cells. **P<0.01, *P<0.05. (B) Western blots showing the expression of SATB1 protein in PCa cell lines and RWPE-1 cells. PCa, prostate cancer; SATB1, special AT-rich sequence binding protein-1.
SATB1 promotes PCa cell proliferation
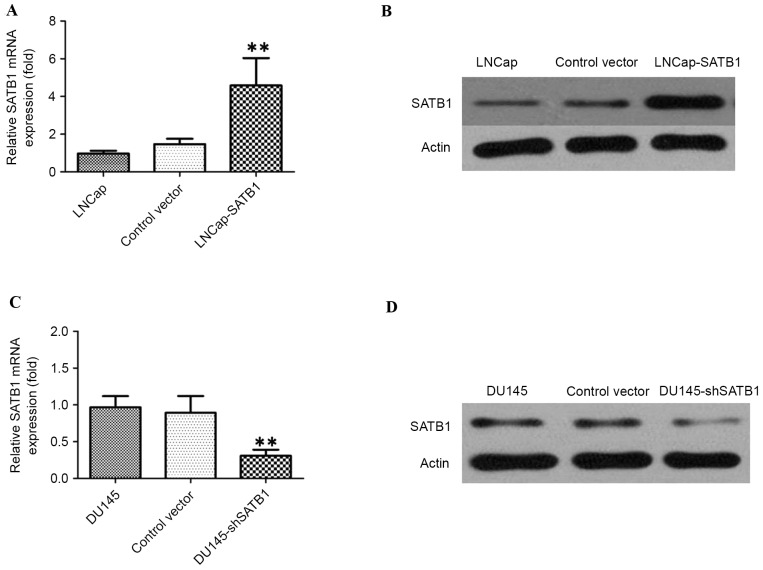
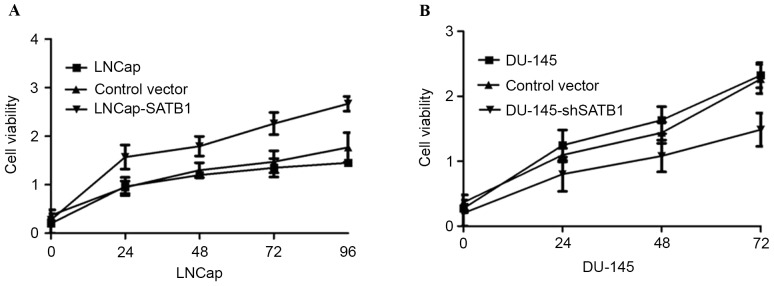
In order to test the oncogenic activity of SATB1 in PCa, LNCaP cells were used to establish SATB1-overexpressing cell lines and DU145 cells were used to establish SATB1-silencing cell lines by viral transduction. The levels of SATB1 in the resultant cell lines with forced SATB1 expression (designated as LNCap-SATB1) and silenced SATB1 expression (designated as DU145-shSATB1) were verified by RT-qPCR and western blotting (Fig. 4). Therefore, it was investigated whether SATB1 would affect human PCa cell proliferation. MTT assay showed that the LNCap-SATB1 cell line had a significant increase in cell proliferation compared with the control. By contrast, DU145-shSATB1 cell line had lower rates of cell proliferation (Fig. 5A and B).
Figure 4.
(A) RT-qPCR demonstrating that expression level of SATB1 mRNA in LNCap-SATB1 cells was significantly increased compared with control cells. **P<0.01. (B) Western blot analysis demonstrating that the expression of SATB1 protein in LNCap-SATB1 cells was significantly increased compared with control cells. (C) RT-qPCR demonstrating that expression level of SATB1 mRNA in DU145-shSATB1 cells was significantly decreased compared with control cells. (D) Western blots analysis demonstrating that the expression of SATB1 protein in DU145-shSATB1 cells was significantly decreased compared with control cells. RT-qPCR, reverse transcription quantitative polymerase chain reaction; SATB1, special AT-rich sequence binding protein; sh, short hairpin RNA.
Figure 5.
(A) Overexpression of SATB1 promotes cell growth of LNCaP cells. (B) Silencing of SATB1 inhibits the cell growth of DU-145 cells. SATB1, special AT-rich sequence binding protein; sh, short hairpin RNA.
SATB1 regulates PCa cell migration and invasion
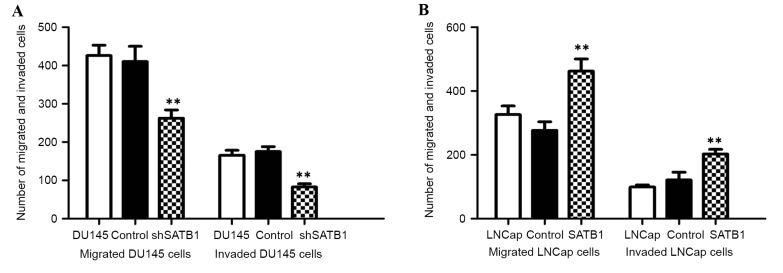
Migration and invasion assay were performed to investigate the effects of SATB1 on the migratory and invasive behaviors of PCa cells in vitro by Transwell assay. The results demonstrated that the number of DU145-shSATB1 cells that transited the artificial basement membrane or/and Matrigel matrix was significantly lower than control cells, while the migration and invasion capability was significantly increased in LNCaP-SATB1 cells (P<0.01). However, no significant difference was observed between the pcDNA3.1 empty vector-transfected group and the control group (Fig. 6A and B). These data confirmed that ectopic expression of SATB1 in LNCaP cells by the pcDNA3.1-SATB1 vector could promote their capability of migration and invasion in PCa in vitro.
Figure 6.
(A) Inhibition of SATB1 suppresses the ability of migration and invasion of DU-145 cells. **P<0.01 vs. control. (B) Ectopic expression of SATB1 promotes the ability of migration and invasion of LNCaP cells. SATB1, special AT-rich sequence binding protein; sh, short hairpin RNA.
Expression of SATB1 regulates EMT
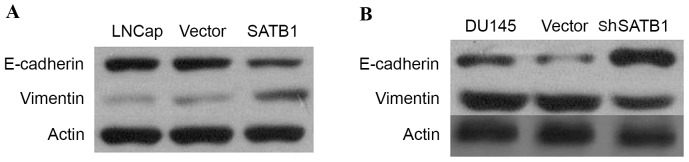
To study whether SATB1 could regulate EMT, western blotting was performed to demonstrate expression of EMT-relevant markers. The results indicated that E-cadherin expression was significantly increased in DU145-shSATB1 cells, and expression of vimentin was greatly reduced in DU145-shSATB1 cells, indicating that SATB1 is involved in the EMT process in PCa (P<0.01; Fig. 7).
Figure 7.
(A) Western blotting demonstrating that E-cadherin expression was significantly decreased in LNCap-SATB1 cells, and expression of vimentin was greatly upregulated in LNCap-SATB1 cells. (B) Western blotting demonstrating that E-cadherin expression was significantly increased in DU145-shSATB1 cells, and expression of vimentin was greatly reduced in DU145-shSATB1 cells. SATB1, special AT-rich sequence binding protein; sh, short hairpin RNA.
Discussion
PCa is one of the most common types of cancer in elderly men, as well as the second leading cause of cancer-associated mortality in males. There is a great need for identification of factors affecting PCa progression, as well as an understanding of their molecular events. Over the past number of decades, growing evidence has indicated that SATB1 may serve a critical role in tumor initiation and progression (9). Importantly, several studies reported that SIRT1 overexpression correlated positively with more aggressive capability of numerous tumors (10,11). However, a number of studies have detected the expression and function of SATB1 in PCa. The present study reinforces the theory that SATB1 is an important promoter for PCa progression, and notably, SATB1 was identified as a pivotal modulator of the molecular and functional characteristics of EMT in PCa.
SATB1, as a regulator in T cell differentiation, has been verified to serve a critical role in metastatic breast cancer (12). SATB1 can act as a tumor suppressor gene or oncogene in the initiation and development of tumors. In the present study, SATB1 expression was upregulated in clinical PCa tissues compared with the corresponding non-tumor tissues. The present study demonstrated that SATB1 expression was upregulated in human PCa cell lines compared with normal prostate epithelial cell. Gain and loss of function experiments were then performed for two PCa cell lines, LNCaP and DU145, to determine the possibility of SATB1 as a therapeutic target of PCa. The shRNA technique was employed to knockdown the SATB1 expression in DU145 cells in which SATB1 expression is relatively high; and SATB1 levels were overexpressed by viral transduction in LNCaP cells in which SATB1 expression is relatively low. The results revealed that the depletion of SATB1 expression had an anti-tumorigenic effect in DU145 cells, overexpressed SATB1 in LNCaP cells could result in increased cell proliferation. Consequently, SATB1 appeared to be a critical factor for the proliferation of PCa cancer cells.
Metastasis is the main cause of mortality in patients with cancer (13,14). Previous studies demonstrated that SATB1 serves an important role in the process of invasion and migration (15,16). Consistent with this, the findings presented in the present study demonstrate that knockdown of endogenous SATB1 in PCa cell lines greatly reduces cell migration and invasion capacity in vitro, suggesting that SATB1 has an important role in PCa invasiveness. EMT is an early event in which cancer cells switch to an aggressive phenotype (17). In the present study, knockdown of SATB1 reverses the EMT process through upregulation of E-cadherin in PCa cells, suggesting that SATB1 promotes PCa aggressiveness through EMT.
In conclusion, SATB1 was reported to be upregulated in PCa tissues and cell lines. SATB1 also promoted cell proliferation, migration and increased invasive capability. In addition, inhibition of SATB1 could reverse EMT through up-regulation of epithelial markers E-cadherin in an established human PCa cell line. SATB1 was demonstrated to serve a crucial role in the progression of PCa, and may function as a therapeutic target for PCa patients.
References
- 1.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rebbeck TR. Prostate cancer genetics: Variation by race, ethnicity, and geography. Semin Radiat Oncol. 2017;27:3–10. doi: 10.1016/j.semradonc.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu N, Chen HJ, Chen SH, Xue XY, Chen H, Zheng QS, Wei Y, Li XD, Huang JB, Cai H, Sun XL. Upregulation of Talin-1 expression associates with advanced pathological features and predicts lymph node metastases and biochemical recurrence of prostate cancer. Medicine (Baltimore) 2016;95:e4326. doi: 10.1097/MD.0000000000004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, Eisenberger MA, Higano C, Bubley GJ, Dreicer R, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the prostate cancer clinical trials working group. J Clin Oncol. 2008;26:1148–1159. doi: 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 6.Cheng C, Lu X, Wang G, Zheng L, Shu X, Zhu S, Liu K, Wu K, Tong Q. Expression of SATB1 and heparanase in gastric cancer and its relationship to clinicopathologic features. APMIS. 2010;118:855–863. doi: 10.1111/j.1600-0463.2010.02673.x. [DOI] [PubMed] [Google Scholar]
- 7.Baba H, Ishikawa T, Mogushi K, Ishiguro M, Uetake H, Tanaka H, Sugihara K. Identification of SATB1 as a specific biomarker for lymph node metastasis in colorectal cancer. Anticancer Res. 2016;36:4069–4076. [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Patani N, Jiang W, Mansel R, Newbold R, Mokbel K. The mRNA expression of SATB1 and SATB2 in human breast cancer. Cancer Cell Int. 2009;9:18. doi: 10.1186/1475-2867-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Tian X, Ji H, Guan X, Xu W, Dong B, Zhao M, Wei M, Ye C, Sun Y, et al. Expression of SATB1 promotes the growth and metastasis of colorectal cancer. PLoS One. 2014;9:e100413. doi: 10.1371/journal.pone.0100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu X, Cheng C, Zhu S, Yang Y, Zheng L, Wang G, Shu X, Wu K, Liu K, Tong Q. SATB1 is an independent prognostic marker for gastric cancer in a Chinese population. Oncol Rep. 2010;24:981–987. doi: 10.3892/or.2010.981. [DOI] [PubMed] [Google Scholar]
- 12.Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H, He J, Han P, Tian D. Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int. 2012;32:1064–1078. doi: 10.1111/j.1478-3231.2012.02815.x. [DOI] [PubMed] [Google Scholar]
- 13.Kohwi-Shigematsu T, Poterlowicz K, Ordinario E, Han HJ, Botchkarev VA, Kohwi Y. Genome organizing function of SATB1 in tumor progression. Semin Cancer Biol. 2013;23:72–79. doi: 10.1016/j.semcancer.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 15.Chu SH, Ma YB, Feng DF, Zhang H, Zhu ZA, Li ZQ, Jiang PC. Upregulation of SATB1 is associated with the development and progression of glioma. J Transl Med. 2012;10:149. doi: 10.1186/1479-5876-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mir R, Pradhan SJ, Galande S. Chromatin organizer SATB1 as a novel molecular target for cancer therapy. Curr Drug Targets. 2012;13:1603–1615. doi: 10.2174/138945012803530008. [DOI] [PubMed] [Google Scholar]
- 17.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]