Abstract
Multidrug resistance (MDR) is the primary barrier to the success of chemotherapy for lung adenocarcinoma. MicroRNA (miR)-134, which is downregulated in lung adenocarcinoma, influences cell proliferation, apoptosis and invasion of lung adenocarcinoma. However, the function of miR-134 in the MDR of lung adenocarcinoma remains unclear. In the present study, it was identified that miR-134 expression is significantly downregulated in A549/cisplatin MDR lung adenocarcinoma cells, as compared with A549 parental cells. miR-134 regulates the sensitivity of lung adenocarcinoma cells to certain anticancer drugs. Furthermore, it was demonstrated that forkhead box M1 and multidrug resistance-associated protein 1 are functional targets of miR-134. These data revealed an important role for miR-134 in the regulation of MDR in lung adenocarcinoma.
Keywords: microRNA-134, multidrug resistance, forkhead box M1, multidrug resistance-associated protein 1, lung adenocarcinoma
Introduction
Chemotherapy is one of the most effective treatments for advanced lung adenocarcinoma (1). However, cancerous cells frequently develop multidrug resistance (MDR) (2). MDR is the intrinsic or acquired simultaneous resistance to unrelated therapeutics, and is arguably one of the largest barriers to the successful chemotherapeutic treatment of cancer (1,2). Previous studies have demonstrated that multiple cellular processes, including DNA repair, cell apoptosis and proliferation, may be important in the development of MDR (3,4). However, the precise underlying mechanisms of MDR have not been fully elucidated.
MicroRNAs (miRNAs or miRs) are small, noncoding RNA molecules that negatively regulate a large number of protein-encoding genes via messenger (m)RNA degradation or translational silencing (5). They are involved in tumor proliferation, apoptosis, invasion and angiogenesis (6). Evidence has emerged for the role of miRNAs in modulating the drug sensitivity and resistance of cells, and numerous studies have demonstrated that the elevation or inhibition of miRNA expression may modulate MDR in cancer cells (7–10).
In the present study, it was observed that miR-134 is significantly downregulated in A549/cisplatin (CDDP) MDR lung adenocarcinoma cells, compared with parental A549 cells. It was also identified that the overexpression of miR-134 increased lung adenocarcinoma cell sensitivity to certain anticancer drugs. Furthermore, it was demonstrated that miR-134 may serve an essential role in the development of MDR in human lung adenocarcinoma cells by regulating the forkhead box M1 (FOXM1)/multidrug resistance-associated protein 1 (MRP1) signaling pathway.
Materials and methods
Cell culture and transfection
Human lung adenocarcinoma A549 cells were purchased from the American Type Culture Collection (Manassas, VA, USA), and the A549/CDDP cell line was established in the present study. The cells were maintained in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 100 µg/ml penicillin/streptomycin at 37°C with 5% CO2.
miR-134 mimics or inhibitors, their negative controls (NC and anti-NC, respectively) and FOXM1 small interfering (si)RNAs were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). Transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Tissue samples
Advanced lung adenocarcinoma tissue samples (males, 22; females, 8; age range, 40–62 years) were collected from Yinzhou People's Hospital (Ningbo, China) between January 2009 and March 2010. Informed consent was obtained from all the patients, and the Review Board of the Hospital Ethics Committee approved the study protocol. Prior to surgery, the patients were treated with 100 mg/m2 CDDP every 3 weeks for a maximum of 5 cycles. According to the ‘Response Evaluation Criteria in Solid Tumors’ of the World Health Organization (11), patients were divided into ‘sensitive’ (complete response or partial response) or ‘insensitive’ (stable disease or progressive disease) groups.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells or tissue samples using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA (1 µg) was reverse transcribed into complementary DNA in a final volume of 20 µl according to the protocol of the manufacturer of PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). The expression levels of miR-134 were evaluated using TaqMan® MicroRNA Assays (Applied Biosystems; Thermo Fisher Scientific, Inc.), and U6 small nuclear RNA levels were used for normalization. The primer sequences for miR-134 were as follows: Forward, 5′-ACACTCCAGCTGGGTGTGACTGGTTGAC-3′ and reverse,5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCCTCTG-3′. FOXM1 expression was evaluated by SYBR Green qPCR assay (Takara Biotechnology Co., Ltd.), and GAPDH was used as an endogenous control. The primer sequences for FOXM1 were as follows: Forward, 5′-GCTTGCCAGAGTCCTTTTTGC-3′ and reverse, 5′-CCACCTGAGTTCTCGTCAATGC-3′. qPCR (20-µl reaction volume) was performed on an ABI 7500 Real-Time PCR System using the following protocol: 95°C for 10 sec, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. Data analysis was performed using the 2−ΔΔCq method (12).
Western blot analysis
Cells were washed in PBS and lysed in radioimmunoprecipitation assay lysis buffer supplemented with protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). Total protein was quantified using a BCA Protein Assay kit (Beyotime Institute of Biotechnology, Nanjing, China). Proteins (30 µg) were separated by 8% SDS-PAGE and transferred to nitrocellulose membranes. Then, the membrane was blotted for 1 h at room temperature, with antibodies against FOXM1 (dilution, 1:500; catalog no. sc-376471; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), MRP1 (dilution, 1:500; catalog no. sc-58219; Santa Cruz Biotechnology, Inc.) and β-actin (dilution, 1:2,000; catalog no. sc-47778; Santa Cruz Biotechnology, Inc.). Upon washing three times, the blots were incubated with a horseradish peroxidase-conjugated secondary antibody (dilution, 1:2,000; catalog no. sc-2005; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Bands were visualized with a chemiluminescent detection system (Pierce ECL Western Blotting Substrate; Thermo Fisher Scientific, Inc.) and exposed in Molecular Imager® ChemiDoc™ XRS System (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell viability assay
The MTT assay was used to assess the 50% inhibition concentration (IC50) value for the drugs. Cells were seeded into 96-well plates at a concentration of 2×103 cells/well and incubated overnight under routine culture conditions. Then, cells were exposed to CDDP, vincristine (VCR) and 5-fluorouracil (5-FU) (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) at various concentrations ranging from 0 to 30 µg/ml for 48 h. Next, 10 µl of MTT solution was added to each well, and the plates were incubated for additional 3 h at 37°C. The absorbance of individual wells was read at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Statistical analysis
All data are expressed as the mean ± standard deviation. Statistical analyses were performed using the Student's t-test to differentiate the means of the various groups. The association between miR-134 and FOXM1 mRNA expression was explored by Pearson correlation. SPSS version 16.0 software (SPSS Inc., Chicago, IL, USA) was employed to analyze the data. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-134 is downregulated in MDR lung adenocarcinoma cells
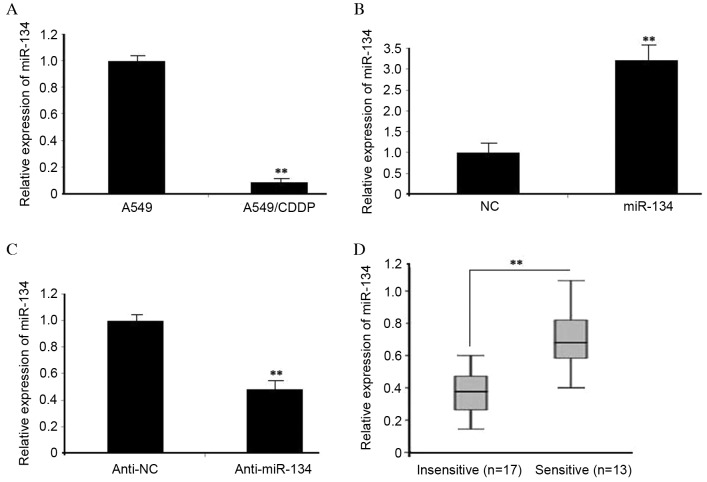
Based on the miRNA microarray data in our previous study (13), a total of 13 miRNAs were revealed to be differentially expressed (>2-fold change) in A549/CDDP cells, compared with A549 cells, among which, miR-134 was the most downregulated miRNA. The data were validated via RT-qPCR in A549/CDDP cells (Fig. 1A). The effect of miR-134 mimics was determined in A549/CDDP cells, which significantly increased miR-134 expression (P<0.01) (Fig. 1B). miR-134 inhibitors were observed to markedly suppress miR-134 expression (Fig. 1C). Next, RT-qPCR was used to compare the endogenous expression of miR-134 between the ‘insensitive’ and ‘sensitive’ groups, according to the patients’ response to CDDP. As presented in Fig. 1D, miR-134 expression was significantly downregulated in the ‘insensitive’ group tissues (n=17), compared with that in the ‘sensitive’ group (n=13) (P<0.01). The results suggest that miR-134 may be involved in the development of MDR in lung adenocarcinoma cells.
Figure 1.
miR-134 was decreased in MDR lung adenocarcinoma cells. (A) The expression levels of miR-134 in A549 and A549/CDDP cells were examined by RT-qPCR. U6 was used as a control. (B) A549/CDDP cells were transfected with miR-134 mimics, while (C) A549 cells were transfected with miR-134 inhibitors. The expression of miR-134 was detected by RT-qPCR at 48 h following transfection. (D) Relative expression levels of miR-134 were detected in ‘insensitive’ (n=17) and ‘sensitive’ (n=13) lung adenocarcinoma tissues via RT-qPCR. Data were obtained from three independent experiments (**P<0.01). miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CDDP, cisplatin; NC, negative control.
miR-134 regulates the sensitivity of lung adenocarcinoma cells to anticancer drugs
To investigate whether miR-134 had a direct role in the development of MDR in lung adenocarcinoma, A549 or A549/CDDP cells were transfected with miR-134 inhibitors or mimics. MTT assay revealed that A549 cells transfected with miR-134 inhibitors exhibited markedly decreased sensitivity to CDDP, VCR and 5-FU, as indicated by the IC50 value (P=0.025) (Fig. 2A). By contrast, the increased miR-134 expression levels in A549/CDDP cells resulted in an enhanced sensitivity to CDDP, VCR and 5-FU (Fig. 2B). The data indicated that the modulation of miR-134 expression was able to alter the sensitivity of lung adenocarcinoma cells to specific chemotherapeutic agents.
Figure 2.
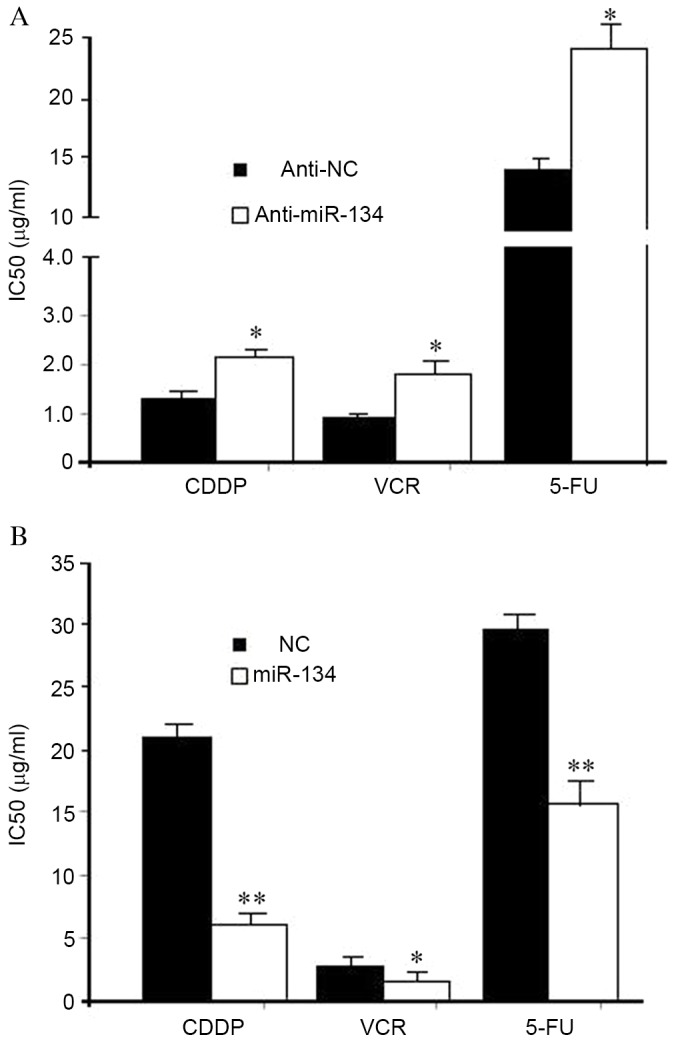
miR-134 regulates the sensitivity of lung adenocarcinoma cells to anticancer drugs. (A) Knockdown of miR-134 increased the IC50 values of A549 cells to CDDP, VCR and 5-FU. (B) Ectopic expression of miR-134 sensitized A549/CDDP cells to CDDP, VCR and 5-FU. Data were obtained in three independent experiments. *P<0.05, **P<0.01. miR, microRNA; CDDP, cisplatin; VCR, vincristine; MDR, multidrug resistance; 5-FU, 5-fluorouracil; NC, negative control; IC50, 50% inhibitory concentration.
Inhibition of FOXM1 sensitizes A549/CDDP cells to anticancer drugs
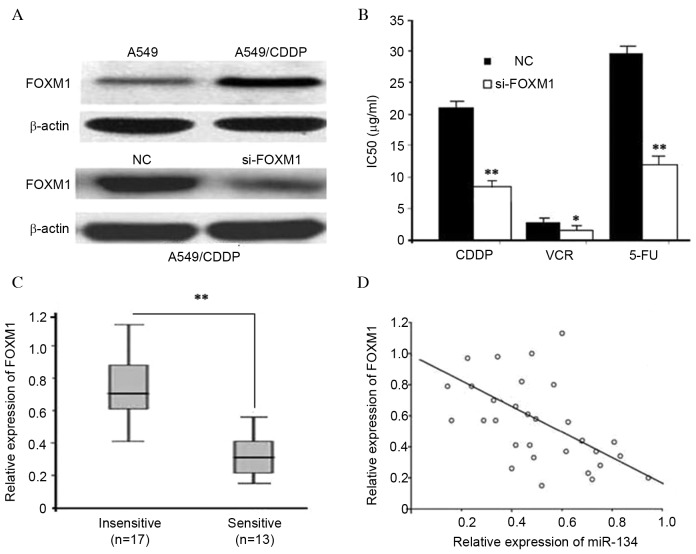
It was previously reported that miR-134 downregulates FOXM1 expression by directly targeting its 3′ untranslated region (UTR) in lung adenocarcinoma cells (14). To explore the functions of FOXM1 in MDR, western blotting was used to detect FOXM1 expression. As presented in Fig. 3A, A549/CDDP cells exhibited significantly higher FOXM1 expression levels, as compared with A549 cells (P<0.01). MTT assays revealed that FOXM1 siRNA was able to enhance cell sensitivity to CDDP, VCR and 5-FU, which recapitulated the effects of miR-134 (Figs. 2B and 3B). RT-qPCR was then used to compare the endogenous expression levels of FOXM1 between the ‘insensitive’ and ‘sensitive’ groups. As presented in Fig. 3C, FOXM1 was significantly upregulated in the ‘insensitive’ group tissues (n=17), compared with the ‘sensitive’ group tissues (n=13). Furthermore, the inverse correlation between miR-134 and FOXM1 mRNA expression was verified by linear regression analysis (r=−0.651; P<0.01; Fig 3D). The results suggest that FOXM1 targeting may be one underlying mechanism by which miR-134 regulates MDR in human lung adenocarcinoma cells.
Figure 3.
Inhibition of FOXM1 sensitizes A549/CDDP cells to anticancer drugs. (A) Differential FOXM1 expression in A549 vs. A549/CDDP cells, as determined by western blot analysis. (B) FOXM1 silencing sensitized A549/CDDP cells to CDDP, VCR and 5-FU (*P<0.05, **P<0.01). (C) Relative expression levels of FOXM1 mRNA were detected in ‘insensitive’ (n=17) and ‘sensitive’ (n=13) lung adenocarcinoma tissues via RT-qPCR (**P<0.01). (D) The expression levels of miR-134 and FOXM1 mRNA were inversely correlated among all the tissue samples (n=30), as indicated by two-tailed Pearson's correlation analysis (r=−0.651; P<0.01). miR, microRNA; CDDP, cisplatin; VCR, vincristine; MDR, multidrug resistance; 5-FU, 5-fluorouracil; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; FOXM1, forkhead box M1; si, small interfering; IC50, 50% inhibitory concentration; mRNA, messenger RNA.
miR-134 regulates MRP1 expression
MRP1 has been reported to be overexpressed in drug-resistant lung adenocarcinoma cells (15–17). Therefore, it was investigated in the current study whether miR-134 is able to regulate MRP1 expression. The results revealed that MRP1 protein expression levels were markedly decreased in miR-134 mimic-transfected cells (P<0.01) (Fig. 4A). By contrast, there was a small increase in the protein expression levels of MRP1 following transfection with miR-134 inhibitors (Fig. 4A). As MRP1was not the putative target of miR-134, as determined by TargetScanHuman 7.1 (http://www.targetscan.org/vert_71/), it was investigated whether FOXM1 is able to regulate MRP1 expression. As presented in Fig. 4B, FOXM1 siRNA significantly downregulated MRP1 expression, compared with NC siRNA (P<0.01), which resembled the inhibitory effects of miR-134. The results demonstrated that miR-134 may prevent MDR in human lung adenocarcinoma cells, partly by regulating the FOXM1/MRP1 signaling pathway.
Figure 4.
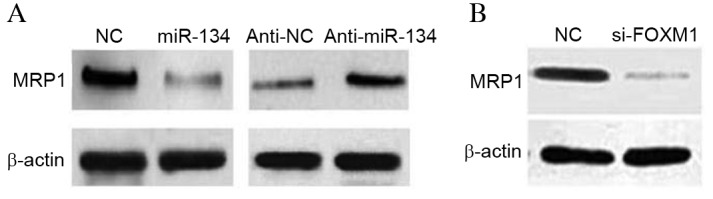
miR-134 regulates MRP1 expression. (A) A549/cisplatin cells were transfected with miR-134 mimics or miR-134 inhibitors. Western blotting was used to detect MRP1 expression. (B) Knockdown of FOXM1 decreased MRP1 expression levels. β-actin was used as an internal control. miR, microRNA; FOXM1, forkhead box M1; NC, negative control; si, small interfering; MRP1, multidrug resistance-associated protein 1.
Discussion
In order to improve the current understanding of the biological mechanisms of chemoresistance in lung adenocarcinoma cells, and to investigate potential reversion approaches, the A549/CDDP MDR A549 cell line was established in the current study. Based on previously obtained miRNA microarray data (13), a series of miRNAs have been hypothesized to be associated with chemoresistance. The present study focused on miR-134, which was determined to be the most downregulated miRNA in A549/CDDP cells (13).
The downregulation of miR-134 is a frequent occurrence in various types of cancer, suggesting that miR-134 may be important in tumorigenesis and tumor progression (18–20). Sun et al (18) indicated that miR-134 served a pivotal role in non-small cell lung cancer through inhibiting cell proliferation, migration and invasion, and promoting apoptosis by targeting the oncogenic cyclin D1 gene. Gao et al (19) reported that miR-134 suppresses endometrial cancer stem cells by targeting protein O-glucosyltransferase 1. Our previous study suggested that miR-134 inhibited the epithelial-mesenchymal transition by targeting FOXM1 in lung adenocarcinoma cells (14). In the current study, it was demonstrated that ectopic miR-134 expression was able to reverse MDR in lung adenocarcinoma cells via targeting the expression of FOXM1. To the best of our knowledge, the present study is the first to elucidate the association between miR-134 expression and MDR in human lung adenocarcinoma.
FOXM1 belongs to a family of FOX transcription factors (21). The overexpression of FOXM1 has been identified in a variety of aggressive human carcinomas, including lung cancer (22,23). Other studies have revealed that FOXM1 and its downstream DNA damage repair targets [breast cancer (BRCA) 1, BRCA2 and X-ray repair cross-complementing protein 1], increased CDDP resistance in various types of cancer cells (24,25). The results of the present study suggest that FOXM1 was upregulated in A549/CDDP cells. Thus, knocking down FOXM1 may reverse MDR in lung adenocarcinoma cells.
The alteration of MRP1 expression has previously been associated with certain lung diseases, and this protein may be pivotal in protecting the lungs via its ability to efflux an array of drugs to sub-lethal levels (26,27). Previous studies have compiled numerous reports on the key role of MRP1 in MDR (15–17,27). In the present study, it was demonstrated that MRP1 is negatively regulated by miR-134, and that FOXM1 siRNA significantly downregulated MRP1 expression in lung adenocarcinoma cells. The data indicate that the miR-134/FOXM1/MRP1 signaling pathway may be important in regulating MDR in lung adenocarcinoma cells.
In conclusion, miR-134 is a novel miRNA that is able to regulate MDR in lung adenocarcinoma. By regulating the expression of FOXM1 and MRP1, miR-134 overexpression promotes drug-induced apoptosis in MDR lung adenocarcinoma cells. The miR-134/FOXM1/MRP1 signaling pathway provides novel insight into the mechanisms underlying drug resistance, and the restoration of miR-134 expression may be a potential therapeutic strategy for the treatment of MDR in lung adenocarcinoma in the future.
Acknowledgements
The present study was supported by the Zhejiang Provincial Natural Science Foundation of China (grant no. LY14H160002) and the Yinzhou Science and Technology Bureau (grant no. 2013-107).
References
- 1.Szákacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 3.Li K, Chen B, Xu L, Feng J, Xia G, Cheng J, Wang J, Gao F, Wang X. Reversal of multidrug resistance by cisplatin-loaded magnetic Fe3O4 nanoparticles in A549/DDP lung cancer cells in vitro and in vivo. Int J Nanomedicine. 2013;8:1867–1877. doi: 10.2147/IJN.S43752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuss S, Polcari D, Geissler M, Brassard D, Mauzeroll J. Assessment of multidrug resistance on cell coculture patterns using scanning electrochemical microscopy. Proc Natl Acad Sci USA. 2013;110:9249–9254. doi: 10.1073/pnas.1214809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu K, Liang X, Cui D, Wu Y, Shi W, Liu J. miR-1915 inhibits Bcl-2 to modulate multidrug resistance by increasing drug-sensitivity in human colorectal carcinoma cells. Mol Carcinog. 2013;52:70–78. doi: 10.1002/mc.21832. [DOI] [PubMed] [Google Scholar]
- 6.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 7.Xu K, Liang X, Shen K, Cui D, Zheng Y, Xu J, Fan Z, Qiu Y, Li Q, Ni L, Liu J. miR-297 modulates multidrug resistance in human colorectal carcinoma by down-regulating MRP-2. Biochem J. 2012;446:291–300. doi: 10.1042/BJ20120386. [DOI] [PubMed] [Google Scholar]
- 8.An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P, Nie Y, Zhao Q. miR-23b-23p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015;6:e1766. doi: 10.1038/cddis.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Yang J, Xu G, Wang W, Liu C, Yang H, Yu Z, Lei Q, Xiao L, Xiong J, et al. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2015;6:3147–3164. doi: 10.18632/oncotarget.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ottman R, Nguyen C, Lorch R, Chakrabarti R. MicroRNA expressions associated with progression of prostate cancer cells to antiandrogen therapy resistance. Mol Cancer. 2014;13:1. doi: 10.1186/1476-4598-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumors: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Wang Y, Song Y, Fu Z, Yu W. miR-27a regulates cisplatin resistance and metastasis by targeting RKIP in human lung adenocarcinoma cells. Mol Cancer. 2014;13:193. doi: 10.1186/1476-4598-13-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Wang Y, Luo J, Fu Z, Ying J, Yu Y, Yu W. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 n non-small cell lung cancer cells. FEBS Lett. 2012;586:3721–3725. doi: 10.1016/j.febslet.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 15.Young LC, Campling BG, Cole SP, Deeley RG, Gerlach JH. Multidrug resistance proteins MRP3, MRP1 and MRP2 in lung cancer: Correlation of protein levels with drug response and messenger RNA levels. Clin Cancer Res. 2001;7:1798–1804. [PubMed] [Google Scholar]
- 16.Doubre H, Césari D, Mairovitz A, Bénac C, Chantot-Bastaraud S, Dagnon K, Antoine M, Danel C, Bernaudin JF, Fleury-Feith J. Multidrug resistance-associated protein (MRP1) is overexpressed in DNA aneuploid carcinomatous cells in non-small cell lung cancer (NSCLC) Int J Cancer. 2005;113:568–574. doi: 10.1002/ijc.20617. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Li ZN, Du YJ, Li XQ, Bao QL, Chen P. Expression of MRP1, BCRP, LRP and ERCC1 in advanced non-small-cell lung cancer: Correlation with response to chemotherapy and survival. Clin Lung Cancer. 2009;10:414–421. doi: 10.3816/CLC.2009.n.078. [DOI] [PubMed] [Google Scholar]
- 18.Sun CC, Li SJ, Li DJ. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget. 2016;7:35960–35978. doi: 10.18632/oncotarget.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS Lett. 2015;589:207–214. doi: 10.1016/j.febslet.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Kim J, Mueller AC, Dey B, Yang Y, Lee DH, Hachmann J, Finderle S, Park DM, Christensen J, et al. Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B and glioblastoma. Cell Death Diff. 2014;21:720–734. doi: 10.1038/cdd.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Ahmad A, Li Y, Banerjee S, Kong D, Sarkar FH. Forkhead box M1 transcription factor: A novel target for cancer therapy. Cancer Treat Rev. 2010;36:151–156. doi: 10.1016/j.ctrv.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, Huang S. Aberrant FOXM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–6219. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 23.Chiu WT, Huang YF, Tsai HY, Chen CC, Chang CH, Huang SC, Hsu KF, Chou CY. FOXM1 confers to epithelial-mesenchymal transition, stemness and chemoresistance in epithelial ovarian carcinoma cells. Oncotarget. 2015;6:2349–2365. doi: 10.18632/oncotarget.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–1016. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, Myatt SS, Lam EW. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodge G, Holmes M, Jersmann H, Jersmann H, Reynolds PN, Hodge S. The drug efflux pump Pgp1 in pro-inflammatory lymphocytes is a target for novel treatment strategies in COPD. Respir Res. 2013;14:63. doi: 10.1186/1465-9921-14-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu JF, Pokharel D, Bebawy M. MRP1 and its role in anticancer drug resistance. Drug Metab Rev. 2015;47:406–419. doi: 10.3109/03602532.2015.1105253. [DOI] [PubMed] [Google Scholar]