Abstract
Clear cell renal cell carcinoma (ccRCC) is the most common type of neoplasm affecting the adult kidney. Previous studies on ccRCC have focused on microRNAs, a class of small non-coding RNAs that are important in cancer development and progression. The present study aimed to investigate the potential role of microRNA (miR)-30e-3p in ccRCC. The results revealed that overexpression of miR-30e-3p in the A498 and 786O ccRCC cell lines was able to inhibit cell invasion and migration. The expression level of Snail1, a potential target gene of miR-30e-3p, was inversely correlated with miR-30e-3p expression in ccRCC tissues and cell lines. Furthermore, Snail1 was revealed to be directly regulated by miR-30e-3p and had an important role in mediating the biological effects of miR-30e-3p in ccRCC. Restoration of Snail1 expression was able to reverse the inhibitory properties of miR-30e-3p. Therefore, the results of the current study suggest that miR-30e-3p exerts its anticancer functions through direct targeting of Snail1 in ccRCC cells, and may be a novel therapeutic agent for this form of cancer.
Keywords: microRNA-30e-3p, clear cell renal cell carcinoma, snail, migration, invasion
Introduction
Renal cell carcinoma (RCC) is the most common kidney malignancy with the highest mortality rate among types of urinary cancer (1) and its incidence has been rising in the last few decades (2). According to the World Health Organization classification of RCC, clear cell (cc)RCC is the most prevalent histological subtype, accounting for 70–80% of all RCC cases (3), and metastatic lesions are detected in ~30% of patients at the time of initial presentation due to the absence of symptoms (4). Although radical nephrectomy may effectively cure early and local ccRCC, the prognosis of metastatic ccRCC remains poor (5). Therefore, the identification of novel biomarkers, including microRNA (miRNA), for the prediction of early metastasis following nephrectomy and the development of targeted therapeutic regimens is required for the management of ccRCC (6).
Previous studies have revealed the involvement of miRNA in the pathogenesis and progression of various human malignancies, including ccRCC (7,8). The identification of specific miRNA expression profiles or signatures may facilitate early diagnosis and serve as a prognostic predictor for various forms of cancer (9). Dysregulation of miRNAs has been associated with the biological characteristics of ccRCC. Furthermore, miRNA-targeted treatment approaches have demonstrated potential efficacy in controlling the aggressive pathology of ccRCC, and miRNAs have been implicated in numerous steps during ccRCC occurrence and development, including proliferation, adhesion, invasion and metastasis (10–12).
The aim of the current study was to examine the role of miRNA (miR)-30e-3p on the invasion and migration of two ccRCC cell lines and investigate the potential underlying mechanisms. The results demonstrated that miR-30e-3p potentiated the invasive and migratory behavior of ccRCC cells through the inhibition of Snail1.
Materials and methods
Patients and tissue samples
Human primary ccRCC tissues were obtained from eight pairs of age and gender matched new-onset patients (six pairs of male and two pairs of female patients) who did not receive chemotherapy and underwent radical nephrectomy at The Department of Urology, Wenzhou Central Hospital (Wenzhou, China) between October 2013 and May 2014. All tissue samples were obtained following informed consent and were immediately stored in liquid nitrogen. The diagnoses were verified by pathologists at Wenzhou Central Hospital. The study protocol was approved by the Ethics Committee of Wenzhou Central Hospital.
RNA extraction and miRNA expression assay
miRNA was extracted using an mirVana™ miRNA Isolation kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR, Thermo Fisher Scientific, Inc.) was performed using 10 ng of total RNA and specific stem-loop real-time primers with a LightCycler® 480 system (Roche Molecular Diagnostics, Pleasanton, CA, USA) and a TaqMan® Universal PCR Master mix (Thermo Fisher Scientific, Inc.). All primers were obtained from TaqMan® miRNA assays (Thermo Fisher Scientific, Inc.). cDNA was subjected to denaturing (95°C, 10 sec), annealing (55°C, 15 sec), and extension (72°C, 30 sec) for 45 cycles. The relative expression of miR-30e-3p, normalized to U6 small nucleolar RNA, was quantified using the 2−ΔΔCq method (13). PCR amplification was performed using the following primers: U6 sense, 5′-GCTTCGGCAGCACATATACTAAAAT-3′, antisense 5′-CGCTTCACGAATTTGCGTGTCAT-3′;. miR-30e-3p sense, 5′-GGGCTTTCAGTCGGATGTTTACAGC-3′; antisense, 5′-CAGTGCGTGTCGTGGAGT-3′.
Cell culture and transfection
The A498 and 786O human ccRCC cell lines were obtained from The Cell Bank of the Type Culture Collection of The Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 50 mg/ml penicillin/streptomycin. All cell lines were maintained at 37°C in a humidified incubator containing 5% CO2. The mimics for control miRNA and miR-30e-3p were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The cells were transfected with microRNA mimics (50 nM) or SNAI1 recombinant plasmid (200 ng, designed and synthesized by Shanghai GenePharma Co., Ltd.) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were collected 24 h following transfection for further analyses.
Transwell invasion assay
Invasion assays were performed using 24-well Matrigel invasion chambers (8-µm pore size; Corning Incorporated, Corning, NY, USA). A total of 1×105 cells suspended in 200 µl serum-free RPMI-1640 medium were seeded into the upper chamber of the Transwell inserts, and 500 µl RPMI-1640 medium supplemented with 10% FBS was added to the lower part of the chamber. At 24 h following incubation, the cells on the lower side of the membrane were fixed in cold methanol and stained with 2% crystal violet for 20 min at room temperature (Sigma-Aldrich, Merck Millipore, Darmstadt, Germany). The cells under the microscopic light fields (Leica Microsystems, GmbH, Wetzlar, Germany) in each chamber were photographed and counted.
Scratch migration assay
To determine cell migration, 5×105 cells were seeded into 6-well plates and cultured until 90–95% confluency was reached. Prior to scratching, cells were starved for 24 h in medium with 1% FBS. Similar-sized wounds were introduced to monolayer cells using a sterile 200 µl pipette tip. The cells were subsequently washed three times with the PBS to remove cell debris, and the culture was continued in RPMI-1640 medium supplemented with 10% FBS at 37°C. The speed of wound closure was monitored and photographed at 24 h. Migration activity was calculated as the percentage of wound closure with the initial wound width defined as 100%.
miRNA target predictions and immunohistochemistry assay
TargetScan (http://www.targetscan.org/), and miRanda (http://microrna.sanger.ac.uk/sequences/index.shtml) algorithms were employed to search for putative protein coding gene targets of miR-30e-3p. Paraffin-embedded tissue sections (4-µm thick) were dewaxed and rehydrated in graded ethanol. Endogenous peroxidase activity was blocked by incubation with 0.3% hydrogen peroxide for 30 min at room temperature. Subsequently, slides were placed in an autoclave cooker filled with 1 mmol/l EDTA (pH 8.0), and antigen retrieval was performed by steam heating at 60°C for 10 min. The primary anti-Snail1 monoclonal antibody (#ab82846, Abcam, Cambridge, MA, USA) was diluted to 1:50 and incubated with the sections at 4°C in a humid chamber overnight. Following incubation with a mouse biotinylated secondary antibody horseradish peroxidase (HRP)-labeled goat anti-rabbit immunoglobulin (Ig) G (#A0208; dilution, 1:1,000; Beyotime Institute of Biotechnology, Shanghai, China), the antigen-antibody reaction was visualized with 3′3-diaminobenzidine serving as the chromogen (Sigma-Aldrich, Merck Millipore). Images were captured using a fluorescence microscope (IX-71; Olympus Corporation, Tokyo, Japan) with a camera. Digital images were recorded and analyzed using Image-Pro Plus 5.0 software (Media Cybernetics, Inc., Rockville, MD, USA).
Western blot analysis
Cells were lysed on ice using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology). Protein concentration was quantified using a Bradford protein assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Protein (50 µg) was separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (Amersham; GE Healthcare Life Sciences, Chalfont, UK). The membranes were probed with human anti-Snail1 (#ab110490; dilution, 1:1,000; Abcam) and anti-GAPDH (#G8140; dilution, 1:1,000; United States Biological, Salem, MA, USA) primary antibodies overnight at 4°C, followed by incubation with the secondary antibody, HRP-labeled goat anti-rabbit IgG (#A0208; dilution, 1:1,000; Beyotime Institute of Biotechnology). The bands were detected with an enhanced chemiluminescence kit (GE Healthcare Life Sciences) and visualized with the ChemiDoc™ XRS system (Bio-Rad Laboratories, Inc.). GAPDH was used as a control.
Plasmid construction
To construct a luciferase reporter vector, the full-length wild-type 3′-untranslated region (3′-UTR) of Snail1, containing the putative miR-30e-3p binding site, was isolated from 786O cDNA using PCR amplification and cloned into the pGL3-luciferase reporter plasmid (Promega Corporation, Madison, WI, USA) downstream of the luciferase gene by Shanghai R&S Biotechnology Co., Ltd. (Shanghai, China). Point mutations in the putative miR-30e-3p binding seed region were created using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer's protocol.
To restore the expression levels of Snail1 protein, Snail1 without the 3′-UTR was amplified using a PCR Master mix kit (Thermo Fisher Scientific, Inc.). The thermocycling conditions consisted of 30 cycles at 98°C for 15 sec, 58°C at 90 sec and 72°C for 20 sec, with a final extension step at 72°C for 5 min. The primer sequences were as follows: Forward, 5′-ATGCCGCGCTCTTTCCTCGTC-3′; and reverse, 5′-TCAGCGGGGACATCCTGAGC-3′. The resulting PCR amplicons were cloned into a pGEM®-T vector system (Promega Corporation) according to the manufacturer's protocol. Briefly, pGEM®-T Vector and Rapid Ligation Buffer reactions were incubated for 1 h at room temperature. Subsequently, the ligated product was transformed into JM109 High Efficiency Competent Cells (Promega Corporation), coated onto LB solid medium, and stored at 37°C overnight. The positive clone was incubated in a LB liquid medium with agitation overnight. White colonies were selected and the insert fragments sequences were validated by DNA sequencing by Sangon Biotech Co., Ltd. (Shanghai, China).
Luciferase reporter assays
In total, 5×104c ells were seeded in triplicate in 24-well plates and grown to 70–80% confluency prior to transfection. A firefly luciferase reporter vector containing the Snail1 3′-UTR or its mutant 3′-UTR were co-transfected with miRNA mimics (50 nM) into the ccRCC cells using Lipofectamine® 2000 reagent, according to the manufacturer's protocol. Following a 48 h incubation at 37°C, all cells were harvested, lysed using the cell lysis solution provided in the kit (Promega Corporation) at 4°C and the luciferase activity was assayed using the Dual-Luciferase® Reporter assay kit (Promega Corporation) according to the manufacturer's protocol. The values were normalized against Renilla luciferase gene activity.
Statistical analysis
SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. Data were presented as the mean ± standard deviation. The statistical significance between groups was analyzed using the Student's t-test or Pearson's χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-30e-3p suppresses ccRCC cell invasion and migration
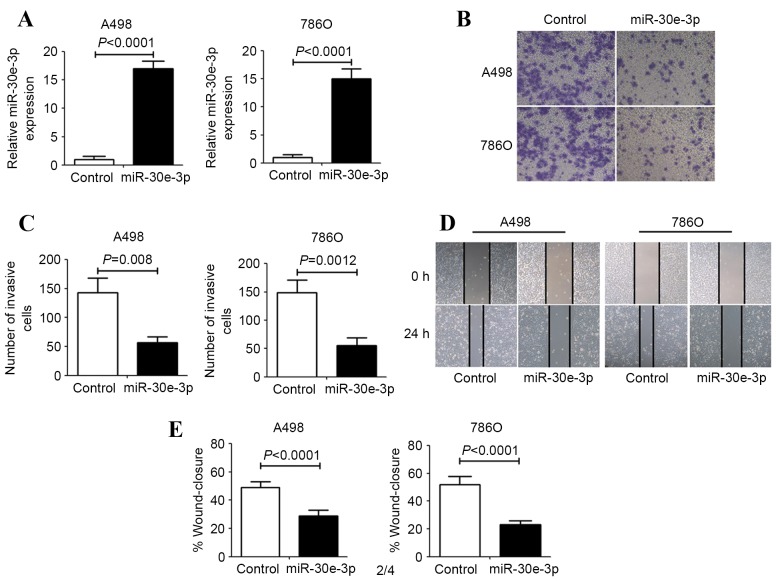
To elucidate the role of miR-30e-3p in ccRCC cells, miR-30e-3p was overexpressed in the A498 and 786O human ccRCC cell lines using miR-30e-3p mimics. As assessed by RT-qPCR, the expression of miR-30e-3p was significantly upregulated in ccRCC cells following transfection with miR-30e-3p mimics (P<0.0001; Fig. 1A). The effect of miR-30e-3p on the invasion and migration of human ccRCC cells was evaluated using Transwell invasion and scratch migration assays. Transwell invasion assays revealed that the number of invasive ccRCC cells was significantly decreased in the group transfected with miR-30e-3p mimics, compared with the control group (P<0.01; Fig. 1B and C). Concordant with this result, the scratch migration assays also demonstrated that miR-30e-3p mimics were able to significantly inhibit ccRCC cell migration (P<0.01; Fig. 1D and E).
Figure 1.
The effect of miR-30e-3p on ccRCC cell invasion and migration. A498 and 786O cells were transfected with control or miR-30e-3p mimics for 48 h. (A) Reverse transcription-quantitative polymerase chain reaction analysis was performed to examine the levels of miR-30e-3p expression. (B) Representative images of Transwell migration assays after 24 h of incubation (magnification, ×100). (C) Quantification of the number of invasive cells in the Transwell assay was performed by counting the number of cells in five high-power fields in each chamber. (D) Representative images were taken at 0 and 24 h to assess the cell migration into the wound gap (magnification, 40x). (E) The percentage of wound closure was calculated by measuring the distance migrated in five high-power fields. Data are expressed as the mean ± standard deviation (n=5). miR, microRNA; ccRCC, clear cell renal cell carcinoma.
miR-30e-3p negatively regulates Snail1 expression levels by directly targeting its 3′-UTR
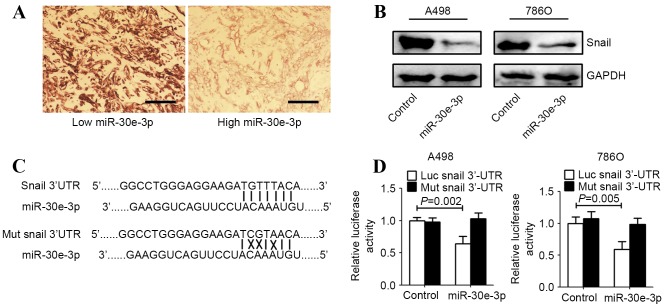
To investigate the underlying mechanisms by which miR-30e-3p influences cellular invasion and migration, TargetScan (http://www.targetscan.org/), and miRanda (http://microrna.sanger.ac.uk/sequences/index.shtml) algorithms were employed to search for putative protein coding gene targets of miR-30e-3p. Snail1 was identified by TargetScan and miRanda as a potential target of miR-30e-3p. The correlation between Snail1 and miR-30e-3p expression levels was evaluated in tissue samples from patients with ccRCC. The Snail1 levels in tumors with high levels of miR-30e-3p expression were markedly lower, compared with those in tumors with low levels of miR-30e-3p expression (Fig. 2A). Western blot analysis demonstrated that Snail1 expression was markedly downregulated in ccRCC cell lines following transfection with miR-30e-3p (Fig. 2B). To identify whether Snail1 is a direct target of miR-30e-3p in ccRCC cells, the potential seed sequence for miR-30e-3p in the 3′-UTR region of Snail1 was analyzed and the wild type and mutant Snail1 3′-UTR fragments were cloned into a luciferase reporter gene system (Fig. 2C). Wild-type or mutant Snail1 3′-UTR constructs were co-transfected with miR-30e-3p or control mimics into A498 and 786O cells. Luciferase activity was subsequently evaluated. Overexpression of miR-30e-3p in the two ccRCC cell lines resulted in significantly reduced luciferase activity for the wild-type constructs, whereas no alteration in luciferase activity was detected with the mutant Snail1 3′-UTR luciferase reporter plasmid (P<0.05; Fig. 2D).
Figure 2.
miR-30e-3p directly targeted Snail1 in ccRCC cells. A498 and 786O cells were transfected with control or miR-30e-3p mimics for 48 h. (A) Snail1 protein expression was detected in ccRCC tissue using immunohistochemistry. Scale bar=100 µm. (B) Western blot analysis revealed that upregulation of miR-30e-3p markedly decreased Snail protein expression in ccRCC cells. GAPDH was used as an internal control. (C) The full-length 3′-UTR sequences of wild-type and mutant Snail1 were cloned into the pGL3-promoter luciferase vector. The putative binding sites between miR-30e-3p and the 3′-UTR sequence of Snail1 are presented (solid lines indicate matching base pairs and crosses indicate non-matching base pairs). (D) A498 and 786O cells were co-transfected with a luciferase reporter plasmid carrying the full-length 3′-UTR sequences of wild-type or mutant Snail1 and control or miR-30e-3p mimics. A dual-luciferase reporter system analysis was performed. Data are expressed as the mean ± standard deviation (n=5). 3′-UTR, 3′-untranslated region; miR, microRNA; ccRCC, clear cell renal cell carcinoma; luc, luciferase; mut, mutant.
Reintroduction of Snail1 rescues miR-30e-3p-induced effects on ccRCC
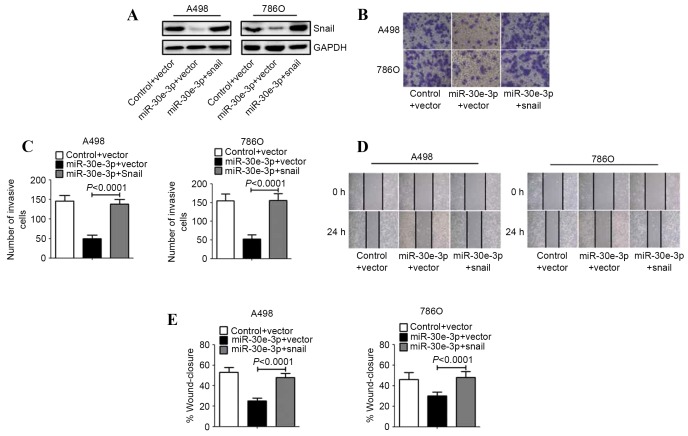
To determine whether the suppression of ccRCC cell invasion and migration triggered by miR-30e-3p was mediated by Snail1, A498 and 786O cells were transfected with a Snail1 construct lacking a 3′-UTR, or an empty vector. Western blot analysis revealed increased expression of Snail protein in the ccRCC cells transfected with the Snail1 construct, compared with those transfected with the empty vector (Fig. 3A). Transwell invasion assays revealed that restored expression of Snail1 abrogated the miR-30e-3p mediated suppression of the invasion of ccRCC cells (Fig. 3B and C). Similarly, the overexpression of Snail1 resulted in the restoration of ccRCC cell migration (Fig. 3D and E).
Figure 3.
Overexpression of Snail1 reverses the inhibitory effect of miR-30e-3p. A498 and 786O cells were co-transfected with control or miR-30e-3p mimics and the empty vector or Snail1 recombinant plasmid for 48 h. (A) The expression of Snail1 protein was analyzed by western blot analysis. GAPDH was used as an internal control. (B) Transwell assays were performed to assess the invasiveness of the cells (magnification, ×100). (C) Representative micrographs of the invasion of A498 and 786O cells in Transwells. (D) Representative images of the scratch migration assay in A498 and 786O cell lines at 0 and 24 h (magnification, ×40). (E) The percentage of the distance migrated by ccRCC cells was quantified. Data are expressed as the mean ± standard deviation (n=5). miR, microRNA.
Discussion
The identification and validation of novel biomarkers for ccRCC is a priority not only for the prediction of patient prognosis and use in the clinical follow-up for ccRCC, but also for defining novel therapeutic strategies. Despite advances in cancer therapy, there remain limitations to the management of this disease, and patients with metastatic ccRCC have a high mortality risk (2). A number of previous studies have identified that miRNAs may serve as biomarkers for cancer risk stratification, outcome prediction and the classification of histological subtypes (14,15). Therefore, identification of miRNAs and their targets involved in tumor pathology may provide insight into the prognosis and aid the development of therapies for patients with ccRCC.
Snail1 was revealed to be a potential target gene for miR-30e-3p in the current study, which demonstrated that miR-30e-3p may negatively regulate Snail1 expression by directly targeting the 3′-UTR of Snail1 mRNA in ccRCC cells. Snail1 is a zinc finger-containing transcription factor that was initially identified in Drosophila, and is required for processes underlying embryogenesis, including mesoderm and neural crest formation and central nervous system development (16,17). Snail1 functions through its zinc-finger motif and the E-cadherin promoter E-box region (sequence CAGGTG) on the main chain in order to suppress E-cadherin expression (18). The downregulation of E-cadherin expression directly results in an epithelial-mesenchymal transition, alterations in cell morphology, loss of polarity, structural instability and the destruction of the connections between epithelial cells, leading to tumor cell metastasis and invasion from the original site of the tumor (19). Previous studies have provided evidence that the levels of Snail1 expression are a potential independent prognostic biomarker for disease recurrence and poor survival in patients with localized ccRCC following nephrectomy (20,21). In addition, the small interfering RNA-mediated downregulation of Snail1 expression in ccRCC cell lines results in increased expression of E-cadherin, together with the inhibition of the cell invasion through Matrigel in vitro (22). These data imply that Snail1 overexpression may be an important event in ccRCC metastasis.
In the current study, computational prediction with TargetScan and miRanda algorithm software revealed that an evolutionarily conserved region in the 3′-UTR of Snail1 possessed a complementary matching region with the seed sequence of miR-30e-3p. Subsequently, Snail1 was demonstrated to be a potential direct target gene for miR-30e-3p, and miR-30e-3p negatively regulated Snail1 expression by directly targeting the 3′-UTR of Snail mRNA in ccRCC cells. In addition, the present study demonstrated that the inhibitory effect of miR-30e-3p on the invasion and migration of ccRCC cells was reversed by overexpression of Snail1 protein, suggesting that miR-30e-3p may inhibit ccRCC metastasis by suppressing Snail expression.
The results of the present study revealed a potential underlying molecular mechanism by which miR-30e-3p may reduce the invasion and migration of human ccRCC cells through suppression of Snail1. Therefore, Snail1 may be a potential therapeutic target for patients with ccRCC.
Acknowledgements
This study was supported by a scientific fund from The Technology Bureau of Wenzhou City (grant no. Y20140287).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Cairns P. Renal cell carcinoma. Cancer Biomark. 2010;9:461–473. doi: 10.3233/CBM-2011-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Beltran A, Carrasco JC, Cheng L, Scarpelli M, Kirkali Z, Montironi R. 2009 update on the classification of renal epithelial tumors in adults. Int J Urol. 2009;16:432–443. doi: 10.1111/j.1442-2042.2009.02302.x. [DOI] [PubMed] [Google Scholar]
- 4.Cindolo L, Patard JJ, Chiodini P, Schips L, Ficarra V, Tostain J, de La Taille A, Altieri V, Lobel B, Zigeuner RE, et al. Comparison of predictive accuracy of four prognostic models for nonmetastatic renal cell carcinoma after nephrectomy: A multicenter european study. Cancer. 2005;104:1362–1371. doi: 10.1002/cncr.21331. [DOI] [PubMed] [Google Scholar]
- 5.Patil S, Ishill N, Deluca J, Motzer RJ. Stage migration and increasing proportion of favorable-prognosis metastatic renal cell carcinoma patients: Implications for clinical trial design and interpretation. Cancer. 2010;116:347–354. doi: 10.1002/cncr.24713. [DOI] [PubMed] [Google Scholar]
- 6.Mangolini A, Bonon A, Volinia S, Lanza G, Gambari R, Pinton P, Russo GR, Del Senno L, Dell'Atti L, Aguiari G. Differential expression of microrna501-5p affects the aggressiveness of clear cell renal carcinoma. FEBS Open Bio. 2014;4:952–965. doi: 10.1016/j.fob.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Ruan A, Wang X, Han W, Wang R, Lou N, Ruan H, Qiu B, Yang H, Zhang X. Mir-129-3p, as a diagnostic and prognostic biomarker for renal cell carcinoma, attenuates cell migration and invasion via downregulating multiple metastasis-related genes. J Cancer Res Clin Oncol. 2014;140:1295–1304. doi: 10.1007/s00432-014-1690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maher ER. Genomics and epigenomics of renal cell carcinoma. Semin Cancer Biol. 2013;23:10–17. doi: 10.1016/j.semcancer.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Butz H, Szabó PM, Nofech-Mozes R, Rotondo F, Kovacs K, Mirham L, Girgis H, Boles D, Patocs A, Yousef GM. Integrative bioinformatics analysis reveals new prognostic biomarkers of clear cell renal cell carcinoma. Clini Chem. 2014;60:1314–1326. doi: 10.1373/clinchem.2014.225854. [DOI] [PubMed] [Google Scholar]
- 10.Su Z, Chen D, Li Y, Zhang E, Yu Z, Chen T, Jiang Z, Ni L, Yang S, Gui Y, et al. Microrna-184 functions as tumor suppressor in renal cell carcinoma. Exp Ther Med. 2015;9:961–966. doi: 10.3892/etm.2015.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, Shen D, Li H, Zhang Y, Lv X, Huang Q, Gao Y, Li X, Gu L, Xiu S, et al. Microrna-185 inhibits cell proliferation and induces cell apoptosis by targeting vegfa directly in von hippel-lindau-inactivated clear cell renal cell carcinoma. Urol Oncol. 2015;33:169. e1–11. doi: 10.1016/j.urolonc.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Ma Y, Yu D, Zhao J, Ma P. Mir-377 functions as a tumor suppressor in human clear cell renal cell carcinoma by targeting ets1. Biomed Pharmacother. 2015;70:64–71. doi: 10.1016/j.biopha.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Di Leva G, Croce CM. MiRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrique R, Luis AS, Jerónimo C. The epigenetics of renal cell tumors: From biology to biomarkers. Front Genet. 2012;3:94. doi: 10.3389/fgene.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: Slowly going into the fast lane of development and cancer. Gen. 2000;257:1–12. doi: 10.1016/S0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 17.Ohkubo T, Ozawa M. The transcription factor snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci. 2004;117:1675–1685. doi: 10.1242/jcs.01004. [DOI] [PubMed] [Google Scholar]
- 18.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, De Herreros A García. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 19.Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Höfler H, Becker KF. Differential expression of the epithelial-mesenchymal transition regulators snail, sip1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Liu Y, Liu H, Zhang W, An H, Xu J. Snail predicts recurrence and survival of patients with localized clear cell renal cell carcinoma after surgical resection. Urol Oncol. 2015;33(69):e1–e10. doi: 10.1016/j.urolonc.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Cai J. Roles of transcriptional factor snail and adhesion factor E-cadherin in clear cell renal cell carcinoma. Exp Ther Med. 2013;6:1489–1493. doi: 10.3892/etm.2013.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikami S, Katsube K, Oya M, Ishida M, Kosaka T, Mizuno R, Mukai M, Okada Y. Expression of snail and slug in renal cell carcinoma: E-cadherin repressor snail is associated with cancer invasion and prognosis. Lab Invest. 2011;91:1443–1458. doi: 10.1038/labinvest.2011.111. [DOI] [PubMed] [Google Scholar]