Abstract
Recent emphasis has been placed on the role of epigenetic regulators and epigenetic marks as biomarkers for cancer diagnosis and prognosis, and as therapeutic targets for treatment. One such class of regulators is the protein arginine methyltransferase (PRMT) family. The present study examined available curated data regarding the expression and alteration of one of the least studied PRMT family members, PRMT8, in various types of cancer and cancer cell lines. Publicly available cancer data on PRMT8 expression were examined using the Human Protein Atlas and the Kaplan-Meier Plotter, and reverse transcription-polymerase chain reaction was used to screen a selection of human cell lines for variant-specific PRMT8 expression. High levels of PRMT8 expression in breast, ovarian and cervical cancer was observed. Additionally, in patients with breast and ovarian cancer, high PRMT8 expression was correlated with increased patient survival, whereas in gastric cancer, high PRMT8 expression was correlated with decreased patient survival. The present study also investigated the expression of PRMT8 variant 2, a novel transcript variant recently identified in our laboratory, in various cancer cell lines. Variant-specific expression of PRMT8 in numerous distinct cancer cell lines derived from different tissues, including the expression of the novel PRMT8 variant 2 in U87MG glioblastoma cells was demonstrated. The present study proposes the possibility of PRMT8 as a cancer biomarker, based on the high level of PRMT8 expression in various types of cancer, particularly in tissues that would not normally be expected to express PRMT8, and on the correlation of PRMT8 and patient lifespan in several cancer types. Variant-specific expression of PRMT8 in diverse cancer cell lines suggests the possibility of alternate PRMT8 isoforms to have diverse effects on cancer cell phenotypes.
Keywords: protein arginine methyltransferase 8, biomarker, breast cancer, ovarian cancer, gastric cancer
Introduction
Protein arginine methyltransferases (PRMTs) are one class of epigenetic regulator that have come under investigation recently for their potential roles as diagnostic biomarkers and therapeutic targets in various cancer types and subtypes. PRMT1, in particular, has been investigated for the correlation of its expression with various cancer types, its role in oncogenesis and its ability to act as a prognostic biomarker (1–6). For example, prostate tumor grade has been identified to correlate positively with histone 4 arginine 3 (H4R3) methylation, which is mediated by PRMT1 (7–9). In addition to PRMT1, PRMT4 (also termed coactivator-associated arginine methyltransferase 1) (10,11), PRMT5 (12) and PRMT6 (2) have also been studied with respect to their presence and activities in various cancer types. PRMT8, which was first described in 2005, has been the subject of limited study, despite the understanding that it contains a high level of structural homology (>80% sequence identity) to PRMT1, a well-studied PRMT family member with strong correlative and mechanistic ties to cancer (13).
Particular PRMT protein isoforms have been investigated in order to determine the specific roles of their protein products in cancer, suggesting the possibility for the use of PRMT transcript variants as cancer biomarkers (3,14–16). Our previous study (17) demonstrated the endogenous expression of a novel transcript variant of PRMT8 in long-lived, fibroblast growth factor 2 (FGF2)-treated primary human dermal fibroblasts cultured in low oxygen. This study also demonstrated that knockdown of PRMT8 was sufficient to halt proliferation and cause cell death in primary human dermal fibroblasts and U87MG glioblastoma cells, suggesting that PRMT8 expression is necessary for cellular viability and/or proliferation in healthy and neoplastic cells. Given the absence of studies investigating PRMT8 expression in association with cancer diagnosis or prognosis, the high homology of PRMT8 to the comparatively well-studied PRMT1, and the identification of a novel PRMT8 variant that brings to mind the isoform-specific effects of PRMT1 that are currently being teased out in various biological contexts, the present study set out to explore the potential for use of PRMT8 as a cancer biomarker, and to investigate the possibility of variant-specific expression and the effects of PRMT8 in distinct cancer subtypes.
Materials and methods
Bioinformatics analysis
The cancer tissue atlas of the Human Protein Atlas (HPA; proteinatlas.org) (18) was utilized to generate a list of cancer types detailing the expression level of PRMT8. PRMT8 staining was performed with the rabbit anti-PRMT8 primary antibody (cat. no. HPA039747; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) (18). HPA is a public database that curates histological images of 44 normal human tissues and the 20 most common types of cancer. In total, 216 cancer samples were used to generate profiles of various proteins using immunohistochemistry. Protein expression profiles were generated by staining samples from 44 normal human tissues, 20 different cancer types, 46 human cell lines and 6 patient-derived hematopoietic cell types. Pathologists annotated individual image files using internal annotation software by scoring the intensity of staining, percentage positivity and staining localization. The HPA was accessed in October 2015 using version 13.
Kaplan-Meier plots were used to assess survival differences at the gene expression level using the Kaplan-Meier Plotter (KM Plotter; kmplot.com) (19). KM Plotter is an integrative data analysis tool that curates gene expression data from Affymetrix microarrays from the Gene Expression Omnibus (National Center for Biotechnology Information, Bethesda, MD, USA), the European Genome-phenome Archive (European Bioinformatics Institute, Hinxton, UK) and The Cancer Genome Atlas (National Cancer Institute and National Human Genome Research Institute, Bethesda, MD, USA). KM Plotter is capable of assessing the effect of 70,632 genes on the survival of 4,142 breast, 2,437 lung, 1,648 ovarian and 1,065 gastric cancer patients. Gene expression and clinical data are managed on a PostgreSQL server and each database is updated twice yearly. Prognostic values for specific genes are split into two groups according to the quartile expression of a proposed biomarker. Patient cohorts are then compared using a Kaplan-Meier survival plot and P-values; hazard ratios with 95% confidence intervals are also calculated. PRMT8 was analyzed by selecting the median value of PRMT8 expression as the cut off for high and low PRMT8 groups. A univariate Cox regression analysis was performed to calculate hazard ratios and P-values. KM Plotter was accessed in September 2015 for all analyses.
Cell culture
The CRL-2073 human teratocarcinoma cell line was obtained from American Tissue Culture Collection (Manassas, VA, USA). The U-2OS human bone osteosarcoma cell line was a gift from the Billiar lab at the Worcester Polytechnic Institute (Worcester, MA, USA). The adult human Caco-2 colorectal adenocarcinoma cell line was a gift from the Weathers lab at the Worcester Polytechnic Institute. The adult human A172 and U87MG glioblastoma cell lines, and the MCF-10A, SK-BR-3 and MDA-MB-231 mammary tissue cell lines were gifts from the Jain lab at the Worcester Polytechnic Institute. Cells were cultured in medium consisting of Dulbecco's modified Eagle's medium (DMEM) Ham's F12 (50:50; MediaTech, Inc., Manassas, VA, USA) with 10% Fetal Clone III (HyClone; GE Healthcare Life Sciences, Logan, UT, USA). The DMEM [without L-Glutamine (L-Gln) or phenol red] was supplemented with 4 mM fresh L-Gln (MediaTech) prior to use. Cultures were performed in a 37°C incubator in a humidified environment of 5% CO2 and 19% O2. Human WA09 embryonic stem cells (WiCell, Madison, WI, USA) were cultured on mitomycin C-treated mouse embryonic fibroblasts seeded at 10,000 cells/cm2 onto 0.1% gelatin coated 6-well plates using 80% Knockout™ DMEM (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 20% Knockout™ serum replacement supplemented with 2.0 mM L-Gln, 0.055 mM 2-mercaptoethanol and 4 ng/ml FGF2, as recommended by the supplier. All cells were harvested on day 7.
Reverse transcription-polymerase chain reaction (RT-PCR)
RNA was isolated using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol and quantified by spectrophotometry (NanoDrop 2000; NanoDrop Technologies, Thermo Fisher Scientific, Inc.). In total, 1 µg total RNA was used to perform first strand cDNA synthesis using qScript™ cDNA SuperMix (Quanta Biosciences, Beverly, MA, USA). For PCR, 50 ng first-strand cDNA was used as a template for each reaction. PCR was performed using 12.5 µl GoTaq (Promega Corporation, Madison, WI, USA) and 0.2 mM each of forward and reverse primers (primer sequences in Table I). PCR cycling for PRMT8 was performed as follows: Initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 94°C for 15 sec, annealing at primer-specific annealing temperature for 30 sec and extension at 72°C for 1 min. Final extension was performed at 72°C for 10 min and samples were held at 4°C until use. PCR cycling for β-actin was performed as follows: Initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 15 sec, annealing at primer-specific annealing temperature for 15 sec and extension at 72°C for 15 sec. Final extension was performed at 72°C for 7 min and the samples held at 4°C until use. PCR cycling for PRMT8 variant 1: Initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at a primer-specific annealing temperature for 30 sec and extension at 72°C for 30 sec. Final extension was performed at 72°C for 10 min and the samples held at 4°C until use. PCR cycling for PRMT8 variant 2: Initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 15 sec, annealing at primer-specific annealing temperature for 15 sec and extension at 72°C for 15 sec. Final extension was performed at 72°C for 10 min and the samples held at 4°C until use. Amplification products were resolved on 2% agarose gels containing 0.5 µg/ml ethidium bromide in 1X Tris base, acetic acid and EDTA buffer, and images were captured using a Gel Doc XR System (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Table I.
RT-PCR primer sequences.
| Primer | Forward sequence (5–3′) | Reverse sequence (5–3′) | Amplicon (bp) |
|---|---|---|---|
| PRMT8 | GACTACGTCCACGCCCTGGTCACCTATTTTATT | GGTCTCGCACATTTTTGGCATTTGGCTTCATGG | 205 |
| PRMT8 v1 | AAGGAATCCGGAGCAGATGAGAAG | GGCATAGGAGTCGAAGTAATAATCTCTC | 458 |
| PRMT8 v2 | CTGTTTGAATGTGTGCCAGGTTG | GGCATAGGAGTCGAAGTAATAATCTCTC | 240 |
| β-actin | TCTGGCACCACACCTTCTACAA | CTTCTCCTTAATGTCACGCACG | 392 |
List of DNA sequences for RT-PCR primers used in Fig. 3. RT-PCR, reverse transcription-polymerase chain reaction; PRMT8, protein arginine methyltransferase 8; v, variant.
Results
PRMT8 is highly expressed in various types of cancer
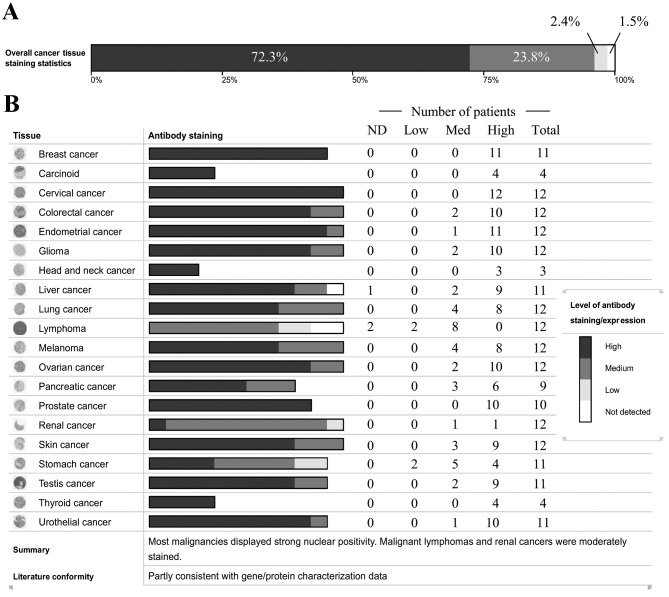
To explore the possibility of PRMT8 as a cancer biomarker, the cancer proteome, as curated by the HPA (18), was assessed for PRMT8 expression. Protein expression was measured by the HPA using immunohistochemical staining in patient-derived primary cancer tissue samples. The HPA analyzed 216 independent cancer samples from 20 of the most common types of cancer for 16,613 genes. PRMT8 expression was evaluated using a single primary antibody. Of all the types of human cancer tested, PRMT8 expression was detected in 98.5% of the tissue samples (Fig. 1A). PRMT8 is highly expressed in 72.3% of tissues, at moderate levels in 23.8% and at low-levels in 2.4% of patients analyzed. Fig. 1B depicts PRMT8 expression levels categorized by cancer type. From these data, PRMT8 is highly expressed in breast, glandular, cervical, head and neck, prostate and thyroid cancer. PRMT8 is expressed at moderate-high levels in colorectal, endometrial, brain, lung, ovarian, pancreatic, skin, testicular and urothelial cancer. PRMT8 is expressed at low-high levels in renal and stomach cancers. PRMT8 expression ranges from undetectable to high or from undetectable to moderate in liver and lymphatic cancer, respectively.
Figure 1.
Histological PRMT8 expression in cancer. (A) Overall PRMT8 expression intensity in all cancer types tested, represented as percentages of total cancer cases. Dark grey represents high PRMT8 expression (72.3% of cases). Grey represents medium PRMT8 expression (23.8% of cases). Light grey represents low PRMT expression (2.4% of cases). White represents PRMT8 expression at undetectable levels (1.5% of cases). (B) PRMT8 expression intensity in specified types of cancer represented by number of patients with varying PRMT8 expression. Cancer type is listed on the far left, histological antibody staining correlated with PRMT8 expression level is represented by the shade of the greyscale bars, and specific patient numbers with varying PRMT8 expression levels are listed on the right. Dark grey represents high PRMT8 expression, grey represents medium PRMT8 expression, light grey represents low PRMT8 expression, and white represents PRMT8 expression at undetectable levels. Data obtained from the Human Protein Atlas. PRMT8, protein arginine methyltransferase 8; ND, non-detectable level of protein.
Expression of PRMT8 correlates with patient survival in several cancer types
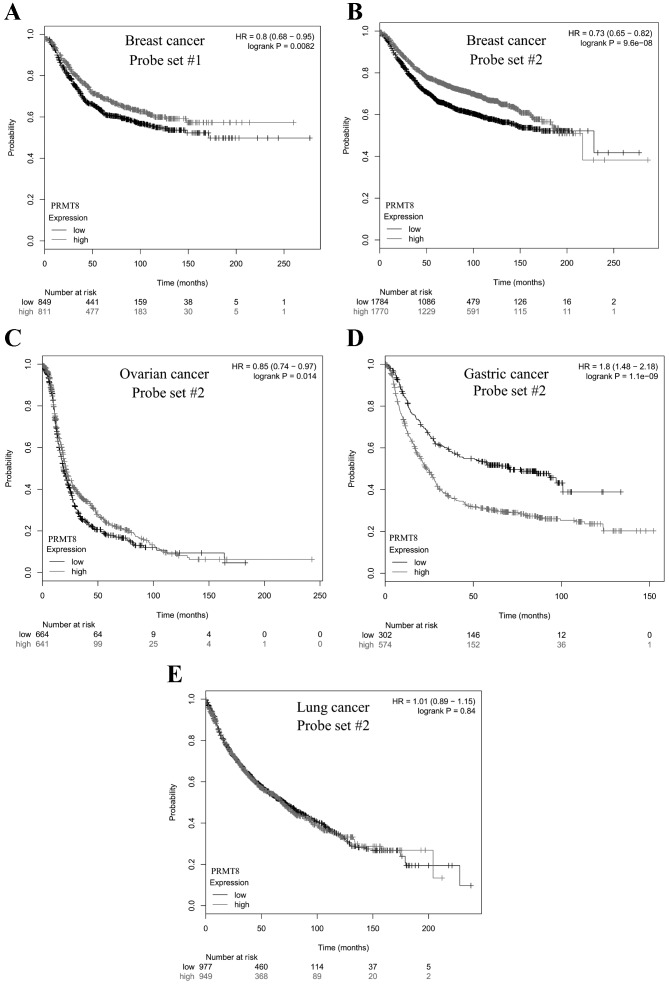
Ideally, a good biomarker correlates with a measurable outcome, a certain metric that can be used to characterize the disease state, such as patient survival. KM Plotter was used to analyze microarray data from 10,188 cancer samples (19) for the purpose of correlating PRMT8 expression with patient survival. A total of 2 probe sets for PRMT8 were included in the microarray data curated by the KM Plotter. Probe sets 1 and 2 were used to test PRMT8 expression levels in 1,660 and 3,554 patients with breast cancer, respectively. Patients with high levels of PRMT8 expression were revealed to survive significantly longer, compared with patients with low PRMT8 expression (P=8.2×10-3 and P=9.6×10-8, respectively; Fig. 2A and B). This was also the case when probe set 2 was used to measure PRMT8 expression in patients with ovarian cancer, where high levels of PRMT8 expression correlated significantly with increased patient survival (n=1,305; P=0.014; Fig. 2C). Conversely, when probe set 2 was used to measure PRMT8 expression in patients with gastric cancer, high levels of PRMT8 expression were significantly correlated with decreased patient survival (n=876; P=1.1×10−9; Fig. 2D). When probe set 2 was used to measure PRMT8 expression in patients with non-small-cell lung cancer, no significant correlation was identified between patient survival and PRMT8 expression levels (n=1,926; P=0.84; Fig. 2E).
Figure 2.
Kaplan-Meier survival plot of cancer patients with varying PRMT8 expression. Differences in PRMT8 expression in breast cancer patients were determined with Affymetrix (A) probe set #1 or (B-E) #2 using a microarray, and patient survival was plotted over time. The gray curve consists of all patients with high PRMT8 expression. The black curve consists of all patients with low PRMT8 expression. (A) In 1,660 patients with breast cancer analyzed with probe set #1, there was a significant positive correlation between high PRMT8 expression and patient survival (P=8.2e-03). (B) In 3,554 patients with breast cancer analyzed with probe set #2, there was a significant positive correlation between high PRMT8 expression and patient survival (P=9.6e-08). (C) In 1,305 patients with ovarian cancer analyzed with probe set #2, there was a significant positive correlation between high PRMT8 expression and patient survival (P=0.014). (D) In 876 patients with gastric cancer, there was a significant negative correlation between high PRMT8 expression and patient survival (P=1.1e-09). (E) In 1,926 patients with non-small cell lung cancer, there was no significant correlation between high PRMT8 expression and patient survival (P=0.84). Data obtained from Kaplan-Meier Plotter (kmplot.com) (19). PRMT8, protein arginine methyltransferase 8; HR, hazard ratio.
PRMT8 variant 2 is expressed in the U87MG tumorigenic glioblastoma cell line
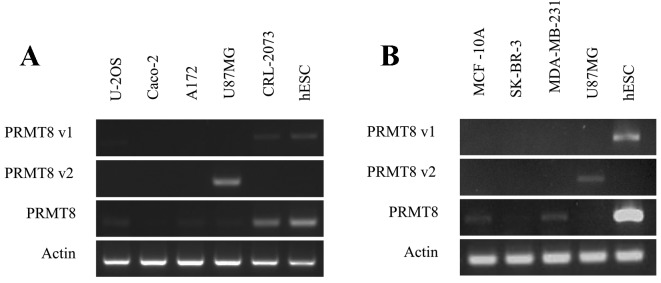
While data from the KM Plotter is useful for determining whether PRMT8 is a potentially useful biomarker for specific types of cancer, the effect of variant-specific expression on patient survival cannot be determined from these datasets. To gain insight into the variant-specific expression and function that PRMT8 may have in various types of cancer, the present study evaluated individual cell lines for specific PRMT8 variants using RT-PCR. This preliminary assay revealed that the U-2OS osteosarcoma, the A172 and U87MG glioblastoma, the CRL-2073 teratocarcinoma line and the three MCF-10A, SK-BR-3, and MDA-MB-231 mammary gland cell lines, all express PRMT8 at varying levels (Fig. 3A and B). Certain cell lines, including SK-BR-3, expressed PRMT8 at barely detectable levels under the conditions tested. The Caco-2 colorectal adenocarcinoma line does not express detectable levels of PRMT8 (Fig. 3A). Of the cell lines tested, and under the conditions used, only the U87MG glioblastoma cell line expressed detectable levels of PRMT8 variant 2 (Fig. 3A). The U-2OS osteosarcoma and the CRL-2073 teratocarcinoma cell line express detectable levels of PRMT8 variant 1, and the A172 glioblastoma cell line and the aforementioned three mammary gland lines do note express detectable levels of PRMT8 variant 1 or PRMT8 variant 2 (Fig. 3B). In Fig. 3A and B, WA09 human embryonic stem cells (hESCs) were used as a positive control for PRMT8 expression, as well as a positive control for PRMT8 variant 1 expression. U87MG cells were used in Fig. 3B as a positive control for PRMT8 variant 2 expression.
Figure 3.
Variant-specific PRMT8 expression in various cancer cell lines. (A) PRMT8 expression was measured by RT-PCR in the human CRL-2073 teratocarcinoma, human U-2OS bone osteosarcoma, adult human Caco-2 colorectal adenocarcinoma and adult human A172 and U87MG glioblastoma cell lines. hESCs were used as a positive control for PRMT8 expression. β-actin was used as a loading control. (B) PRMT8 expression was measured by RT-PCR in (non-tumorigenic) human MCF-10A mammary gland epithelial cells, human SK-BR-3 mammary gland adenocarcinoma cells, and human MDA-MB-231 mammary gland adenocarcinoma cell lines. hESCs were used as a positive control for PRMT8 and PRMT8 variant 1 and, based on the data from 3A, U87MG was used as a positive control for PRMT8 variant 2. PRMT8, protein arginine methyltransferase 8; RT-PCR, reverse transcription-polymerase chain reaction; hESCs, human embryonic stem cells.
Discussion
Correlations of PRMT expression with oncogenic disease states, and the activity of PRMT family members in cancer cell phenotypes, have been previously studied, generally and in isoform-specific manners (3,14–16). This is particularly true of PRMT1, the PRMT family member most homologous to PRMT8 (13), the expression of which is amplified in numerous breast cancer tissues and has been demonstrated to correlate with patient age, tumor grade and menopausal status (2,3). In lung and bladder cancer, PRMT1 and 6 demonstrated elevated expression and regulated cancer cell growth, as knockdown leads to decreased proliferation in each type of cancer cell line (2). PRMT4 was revealed to be highly expressed in prostatic intraepithelial neoplasia and in prostatic adenocarcinoma, compared with benign prostate tissue (10), as well as in colorectal cancer, where PRMT4 was identified to regulate p53 and nuclear factor-κB target gene transcription in colorectal adenocarcinoma cells (11).
The present study reports that PRMT8 is expressed at high levels in numerous types of cancer, which is notable given that, in mature organisms, PRMT8 tends to be localized to brain tissue (13), suggesting a role for PRMT8 dysregulation in oncogenesis or the maintenance of cancer cell phenotypes. This appears particularly provocative given the increasing importance attributed to epigenetics in cancer and cancer therapies (20,21), and in light of our recent study demonstrating that the knockdown of PRMT8 is sufficient to kill U87MG glioblastoma cells in vitro (17). The HPA curation of the cancer proteome demonstrated that PRMT8 is moderately-highly expressed in various cancerous tissues, including those of non-neural origin that would not be otherwise expected to express high levels of PRMT8, comprising breast, glandular, prostate, and thyroid tissues.
Additionally, the present study reports that high PRMT8 expression correlates significantly with patient survival in several different types of cancer, demonstrating a positive association in patients with breast and ovarian cancer, a negative correlation in patients with gastric cancer and no significant correlation in patients with non-small cell lung cancer. These varied tissue-specific effects may be due to expression of different PRMT8 transcript variants, as the probe set data were not variant-specific, preventing uncoupling of the effects of specific PRMT8 transcript variants on patient survival. The present study believes that this possibility merits future investigation into the use of variant-specific PRMT8 expression as a cancer biomarker; much in the same way that variant-specific expression of PRMT1 has been explored (3,14–16). It is worth noting that, even in the absence of available data to uncouple variant-specific PRMT8 expression and correlation with patient survival, the potential for total PRMT8 expression to be used as a prognostic biomarker and for consideration as a possible therapeutic target stands on its own. This is particularly true in gastric cancer, where the negative correlation between PRMT8 expression and patient survival time is considerable: Half of patients with low levels of PRMT8 expression had succumbed to the disease by ~8 years post-analysis; whereas half of patients with high levels of PRMT8 were deceased by ~2 years post-analysis.
The novel transcript variant of PRMT8 that was recently identified (17), PRMT8 variant 2, is unique in that its protein product lacks a portion of the N-terminus of the full-length PRMT8 isoform, which normally harbors a hydrophobic myristoylation motif that confers localization to the plasma membrane (13). As the protein product of PRMT8 variant 2 lacks the glycine residue near the N-terminus that is able to be myristoylated, the protein product of PRMT8 variant 2 instead localizes to the nucleus (Hernandez et al, unpublished data). This finding is consistent with a previous study (22), which demonstrated that, while the full-length isoform of PRMT8 localizes to the plasma membrane, two truncated isoforms translated from in-frame methionine codons and lacking the N-terminal glycine residue that may be myristoylated, demonstrated strong patterns of nuclear localization. Nuclear localization, rather than anchorage of the protein into a membrane, appears more fitting for a protein with suspected epigenetic activity, particularly given the demonstrated ability of full-length PRMT8 to bind histone H4 and nucleosome assembly protein 3 in vitro (13). That the protein product of PRMT8 variant 2 exhibits nuclear localization suggests that the expression of PRMT8 variant 2 in various types of cancer, including the demonstrated expression in U87MG glioblastoma cells, may contribute to the development of the cancer cell phenotype through epigenetic mechanisms due to its arginine methyltransferase activity. This hypothesis is supported by a recent report that PRMT8 positively regulates p53 expression due to etopside-induced DNA damage in U-2OS osteosarcoma cells (23). Additional investigation into the role of PRMT8 variants and their protein products in various cancer cells may reveal information about cancer cell phenotypes, and may propose other uses for PRMT8 as a cancer biomarker or even as a therapeutic target.
Acknowledgements
The authors would like to thank Kris Billiar, Anjana Jain and Pam Weathers at Worcester Polytechnic Institute for their generous gifts of U-2OS; A172, U87MG, MCF-10A, SK-BR-3 and MDA-MB-231; and Caco-2 cells, respectively. The present study was supported by the National Institutes of Health award (grant no. R01GM85456) and the National Science Foundation Integrative Graduate Education and Research Traineeship Training Program (grant no. DGE 1144804).
References
- 1.Cheung N, Chan L, Thompson A, Cleary ML, So C. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 2.Yoshimatsu M, Toyokawa G, Hayami S, Unoki M, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int J Cancer. 2011;128:562–573. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 3.Mathioudaki K, Scorilas A, Ardavanis A, Lymberi P, Tsiambas E, Devetzi M, Apostolaki A, Talieri M. Clinical evaluation of PRMT1 gene expression in breast cancer. Tumor Biol. 2011;32:575–582. doi: 10.1007/s13277-010-0153-2. [DOI] [PubMed] [Google Scholar]
- 4.Mathioudaki K, Papadokostopoulou A, Scorilas A, Xynopoulos D, Agnanti N, Talieri M. The PRMT1 gene expression pattern in colon cancer. Br J Cancer. 2008;99:2094–2099. doi: 10.1038/sj.bjc.6604807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papadokostopoulou A, Mathioudaki K, Scorilas A, Xynopoulos D, Ardavanis A, Kouroumalis E, Talieri M. Colon cancer and protein arginine methyltransferase 1 gene expression. Anticancer Res. 2009;29:1361–1366. [PubMed] [Google Scholar]
- 6.Li B, Liu L, Li X, Wu L. miR-503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem Biophys Res Commun. 2015;464:982–987. doi: 10.1016/j.bbrc.2015.06.169. [DOI] [PubMed] [Google Scholar]
- 7.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 9.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, Allis CD. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/S0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 10.Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, Zhang S, Li L, Pan CX, Hu Z, et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83–89. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- 11.Kim YR, Lee BK, Park RY, Nguyen NT, Bae JA, Kwon DD, Jung C. Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer. 2010;10:197. doi: 10.1186/1471-2407-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 14.Goulet I, Gauvin G, Boisvenue S, Côté J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J Biol Chem. 2007;282:33009–33021. doi: 10.1074/jbc.M704349200. [DOI] [PubMed] [Google Scholar]
- 15.Zhong J, Cao RX, Zu XY, Hong T, Yang J, Liu L, Xiao XH, Ding WJ, Zhao Q, Liu JH, Wen GB. Identification and characterization of novel spliced variants of PRMT2 in breast carcinoma. FEBS J. 2012;279:316–335. doi: 10.1111/j.1742-4658.2011.08426.x. [DOI] [PubMed] [Google Scholar]
- 16.Scorilas A, Black MH, Talieri M, Diamandis EP. Genomic organization, physical mapping, and expression analysis of the human protein arginine methyltransferase 1 gene. Biochem Biophys Res Commun. 2000;278:349–359. doi: 10.1006/bbrc.2000.3807. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez S, Dominko T. Novel protein arginine methyltransferase 8 isoform is essential for cell proliferation. J Cell Biochem. 2016;117:2056–2066. doi: 10.1002/jcb.25508. [DOI] [PubMed] [Google Scholar]
- 18.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 19.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 20.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kousaka A, Mori Y, Koyama Y, Taneda T, Miyata S, Tohyama M. The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience. 2009;163:1146–1157. doi: 10.1016/j.neuroscience.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 23.Sammons MA, Zhu J, Berger SL. A chromatin-focused siRNA screen for regulators of p53-dependent transcription. G3 (Bethesda) 2016;6:2671–2678. doi: 10.1534/g3.116.031534. [DOI] [PMC free article] [PubMed] [Google Scholar]