Abstract
Cells have evolved certain precautions to preserve their genomic content during mitosis and avoid potentially oncogenic errors. Aside from the well-established DNA damage and spindle assembly checkpoints, recent observations have identified an additional mitotic fail safe, referred to as the mitotic surveillance pathway. This pathway triggers a cell cycle arrest to block the growth of potentially unfit daughter cells, and is activated by both prolonged mitosis and centrosome loss. Recent genome-wide screens surprisingly revealed that 53BP1 and USP28 act upstream of p53 to mediate signaling through the mitotic surveillance pathway. Here, we review advances in our understanding of this fail-safe, and discuss how 53BP1 and USP28 adopt non-canonical roles to function in this pathway.
Keywords: Centrosome, mitosis, cell cycle arrest, checkpoint, surveillance
Evidence for a centrosome sensor
Genome integrity relies on the accurate segregation of chromosomes by the microtubule-based mitotic spindle. As the major microtubule organizing centers of animal cells, centrosomes guide the formation of the bipolar mitotic spindle. Concordant with this role, centrosome duplication is tightly controlled to ensure the presence of exactly two centrosomes in mitosis (Box 1). When this process errs, the assembly of too many or too few centrosomes leads to abnormal spindle formation that can promote chromosome missegregation. Recent studies have shown that the gain or loss of centrosomes activates a cell cycle arrest in daughter cells, even if mitosis is completed normally [1–3]. These observations suggest that cells can directly or indirectly sense an abnormal centrosome number to prevent further growth and guard against mitotic errors.
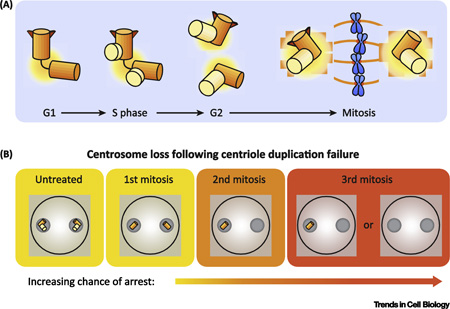
Text Box 1. Centrosome duplication cycle.
Centrosome duplication is a tightly regulated process. Cells begin G1 phase with a single centrosome, containing a pair of centrioles (Box Figure 1.1). At the start of S phase, centriole duplication begins with the assembly of a single cartwheel structure on the wall of each the two mother centrioles. This process requires Plk4 activity, the upstream “master regulator” kinase which promotes the recruitment of SAS6, a protein that oligomerizes into a cartwheel structure that provides a scaffold for assembly of the daughter centriole. Daughter centrioles begin assembly at the start S phase and are completed in G2 phase. Thus in G2 phase, the cell contains two centrosomes, each comprised of a pair of centrioles. The two centrosomes separate at the beginning of mitosis and form the two poles of the bipolar mitotic spindle upon which chromosomes are segregated. When the cell divides each cell inherits a single centrosome, allowing the cycle to start anew.
Box Figure 1A. Schematic of the centrosome duplication cycle.
Upon inhibition of Plk4, the centriole duplication process is blocked. The centriole contents of the cell are then diluted with each subsequent division, as illustrated in Box Figure 1.2.
Box Figure 1B. Dynamics of centriole loss following Plk4 inhibition.
Early clues to the existence of such a pathway arose from the observation that ablation of centrosomes in single cells led to daughter cells that arrest in G1 of the cell cycle [4, 5]. More recently, the manipulation of components required for centrosome duplication has provided further evidence for the requirement of centrosomes for continued cell growth. Genetic inactivation of the centriole protein SAS4 in the mouse embryo or in the developing mouse brain resulted in centrosome loss, delayed spindle assembly, and widespread apoptosis [6, 7]. Furthermore, disrupting centrosome duplication in human RPE1 cells by downregulation or inhibition of the kinase Plk4 resulted in cells progressing through 3 or 4 cell cycles before undergoing an irreversible arrest with 0 or 1 centrosomes (Box 1) [1, 2].
Importantly, the requirement for centrosomes for continued cell proliferation was alleviated by deletion of p53. However, centrosome loss did not trigger a cell cycle arrest through any known p53-dependent mechanisms, including DNA damage, Hippo signaling, mitotic errors and oxidative stress. Taken together, these data suggested that cells activate p53 in response to centrosome loss in a manner unique from previously defined pathways, leading to the proposal that a distinct “centrosome surveillance pathway” exists to curb the growth of cells that fail centrosome duplication [1, 2]. In this review, we highlight recent work to understand the molecular basis of this centrosome sensor, discussing the components involved, and the potential mechanism(s) by which the sensor is activated.
Identifying components of the centrosome surveillance pathway
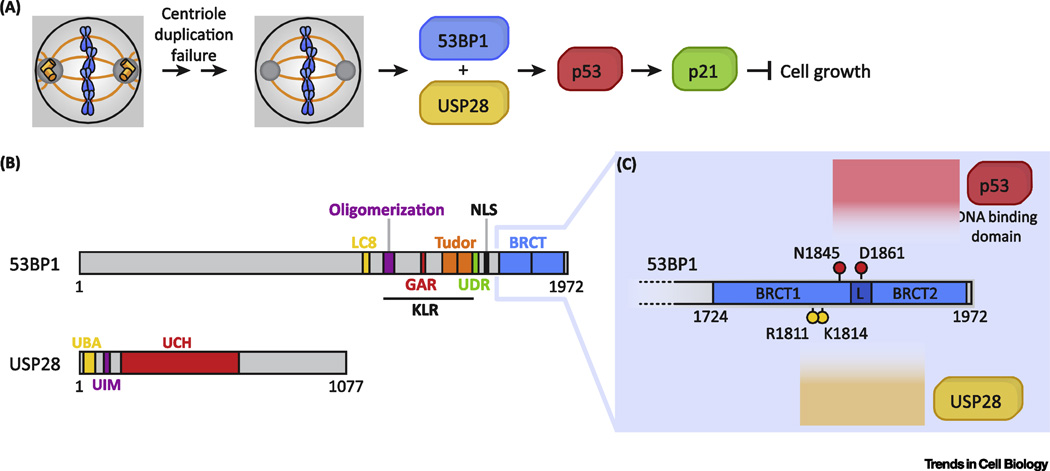
A centrosome surveillance pathway would be expected to consist of components which, when disrupted, would halt signaling and allow cells to proliferate despite centrosome loss. Recent studies have exploited this logic and used genome-wide CRISPR/Cas9 knockout screening technology [8] to identify components of this pathway [9–11]. The screens were designed to enrich for sgRNAs that allow the continued growth of cells that lose centrosomes after Plk4 inhibition. All the screens observed an enrichment of sgRNAs targeting p53, 53BP1 and USP28, while two screens also identified sgRNAs targeting TRIM37 (Figure 1A). Loss of 53BP1, USP28, and TRIM37 all prevented p53 stabilization and G1 arrest following centrosome loss. Importantly, knockout of the essential centriole component SAS6 also triggered a 53BP1, USP28, and p53-dependent G1 arrest, demonstrating that the cell cycle arrest was likely to be caused by centrosome loss and not by the loss of Plk4 activity per se [10].
Figure 1. Centrosome surveillance components.
A) A schematic of known components involved in cell cycle arrest following centrosome loss.
B) Diagram of the domain structure of 53BP1. The N-terminal region of 53BP1 is a disordered region containing multiple ATM phosphorylation sites, while the C-terminal half contains the following domains: LC8 (dynein light chain 8) binding, oligomerization, GAR (glycine-and-arginine rich), tandem Tudor (recognizes methylated residues), UDR (ubiquitination-dependent recruitment), NLS (nuclear localization signal), tandem BRCT. USP28 contains the domains: UBA (ubiquitin-associated), UIM (ubiquitin-interaction motif), UCH (ubiquitin carboxyhydroxylase).
C) Diagram of important residues in the 53BP1 tandem BRCT domains for mediating interactions with the DNA-binding domain of p53, and with USP28. Modified from [20].
53BP1, USP28, and p53 have all been reported to act in DNA damage response pathways. Therefore, one interpretation is that these components are being activated by DNA damage resulting from centrosome loss. Indeed, 53BP1 is a large scaffolding protein that plays a well-established role in DNA double-strand break repair [12, 13], while the deubiquitinase USP28 is a binding partner of 53BP1 that is reported to have a minor role in DNA repair and DNA damage signaling [14, 15] (Box 2, Figure 1B). However, several lines of evidence argue against the simple explanation that DNA damage is responsible for triggering the centrosome surveillance pathway. First, no increase in DNA damage was observed in cells that lost centrosomes [1, 2, 10]. Second, if centrosome loss activated DNA damage signaling, other components of the DNA damage signaling pathway would be expected to emerge from the genome-wide screens - for example, knockout of core components of the DNA damage response such as ATM, ATR, Chk1, and Chk2 would also produce viable knockouts and disrupt DNA damage signaling. However, while knockout of Chk2 enabled cells to escape from DNA damage induced arrest, it was unable to alleviate growth arrest following centrosome loss [10], suggesting that the DNA damage signaling pathway and the centrosome surveillance pathway are genetically separable. Finally, disrupting 53BP1 recruitment to DNA damage foci, either by mutation of its recruitment domain [9] or by knockout of upstream recruitment factor RNF168 [10], resulted in cells that still arrested following centrosome loss, showing that the role of 53BP1 in the centrosome surveillance pathway does not require its recruitment to sites of DNA damage. This hypothesis is further supported by biochemical work showing that both p53 and USP28 interact with the tandem-BRCT domains of 53BP1 [15–17], which are dispensable for the DNA damage response role of 53BP1 [18], but are required for the centrosome surveillance pathway [9]. Taken together, these data suggest a new role for 53BP1 in the centrosome surveillance pathway that is independent of its canonical role in DNA damage signaling.
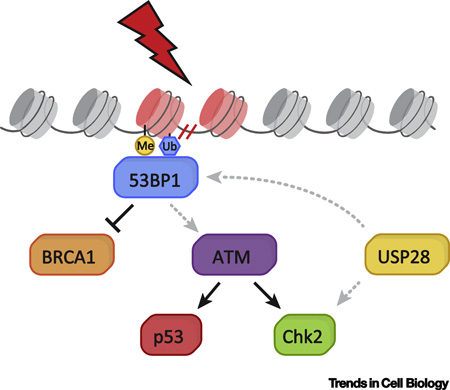
Text Box 2. The role of 53BP1 and USP28 in the DNA damage response.
53BP1 is best known for its role in the DNA damage response. It is recruited to sites of DNA double-strand breaks (DSBs), where it acts to amplify ATM signaling and scaffold the recruitment of DSB-responsive factors (Box Figure 2). 53BP1 functions in G1 to promote non-homologous end-joining (NHEJ) of DSBs and suppress homology-directed repair (HDR) (reviewed in [13] and [12]). Following DNA damage, 53BP1 is recruited to methylated and ubiquitylated histones through its Tudor and UDR domains, respectively. The unstructured N-terminal half of 53BP1 contains 28 S/TQ sites that are phosphorylated by ATM following DNA damage, and are required for its recruitment of NHEJ-promoting factors. The functions of the remaining domains of 53BP1 are less well-understood. The oligomerization domain of 53BP1 contributes to chromatin binding, but its role in NHEJ remains unclear. The BRCT repeats of 53BP1 do not play a major role in the DNA damage response, though they have been shown to support DSB repair in G1 heterochromatin [33]. Finally, the role of the glycine-arginine rich (GAR) domain, dynein light chain 8 (LC8) binding motif, and kinetochore localization region (KLR) remain poorly understood.
In contrast to 53BP1, little is known about how USP28 functions. This E3 ligase has been shown to be recruited to DSBs through the BRCT repeats of 53BP1 [15], and has been shown to stabilize the DNA damage effector Chk2 [14] (Box Figure 2). However, both of these roles have only a minor influence on DNA damage repair.
Box Figure 2. Canonical role of 53BP1 and USP28 in the DNA damage response.
53BP1 and USP28 in centrosome surveillance
If not acting through their canonical role in DNA damage response, how then do 53BP1 and USP28 function in the centrosome surveillance pathway? Previous work has shown that both p53 and USP28 directly interact with 53BP1 through its BRCT domains [14, 19]. The most direct model that can be drawn is that, once an triggered by an upstream stimulus, 53BP1 acts as a scaffold to recruit both USP28 and p53 in close proximity, thus allowing USP28 to deubiquitinate p53 and modify its activity. Consistent with this model, 53BP1 rescue experiments found that the BRCT repeats were required for 53BP1 signaling in the centrosome surveillance pathway [9]. Furthermore, USP28 and p53 bind the BRCT repeats at different interfaces, and it has been shown that 53BP1 and USP28 share a co-regulatory role in supporting p53 functions [20] (Figure 1B–C). While 53BP1, USP28, and p53 have not yet been demonstrated to form a ternary complex, USP28 was able to deubiquitinate p53 in vitro [9]. Taken together, a body of evidence supports a model in which a 53BP1-USP28 complex modulates p53 activity in response to centrosome loss.
While little is known about the upstream mechanism of ‘sensing’ centrosome loss, it is likely that the known pathway components 53BP1, USP28, and p53 play an indirect role in monitoring centrosome number, as none of the components show a clear localization to the centrosome [10]. One plausible interpretation is that centrosome loss results in changes in microtubule nucleation that could trigger cell cycle arrest. During mitosis, microtubules of the mitotic spindle are captured by the kinetochores, a complex protein structure that forms at the centromeres of chromosomes and serves to link chromatin to the mitotic spindle [21]. The kinetochore has a layered ultrastructure, consisting of the inner and outer plates, and beyond that, the fibrous corona. One intriguing observation is that at mitosis onset, 53BP1 relocalizes from DNA damage foci to the corona region of kinetochores [22] – a behavior with no known function. Specifically, 53BP1 is recruited to kinetochores during chromosome condensation, and depleted following microtubule attachment [22]. 53BP1 colocalizes with other corona proteins such as CENP-E and components of the spindle assembly checkpoint (SAC); however, unlike SAC components and other corona proteins, 53BP1 does not persist at kinetochores if the SAC is reactivated by disrupting spindle assembly with microtubule poisons [9]. This unique localization pattern suggests that 53BP1 could act at the kinetochore to respond to perturbations in microtubule nucleation and trigger activation of the centrosome surveillance pathway. Testing the role of 53BP1 kinetochore localization is an important area of future study.
Centrosome amplification arrests the cell cycle through a distinct pathway(s)
It is important to note that as with centrosome loss, the production of too many centrosomes has also been shown to activate a p53-dependent cell cycle arrest [3]. This raises the question of whether the 53BP1-USP28-p53 signaling axis is activated by both centrosome loss and gain. While the idea of a universal centrosome sensor is attractive, knockout of 53BP1 or USP28 did not rescue the cell cycle arrest caused by supernumerary centrosomes [10]. Similarly, while LATS2 signaling was shown to relieve an arrest caused by tetraploidization (a condition associated with excess centrosomes), LATS2 was not required for centrosome loss surveillance [10, 23]. Taken together, while both centrosome loss and centrosome over-amplification stabilize p53 to arrest cell growth, the upstream signaling components do not appear to be shared. Thus, there is unlikely to be a single, unified “centrosome counting mechanism"; instead, each condition is likely to be indirectly detected through “symptoms” associated with either loss or gain of centrosomes.
A mitotic clock
A key defect observed in cells lacking centrosomes is that they are slower to assemble spindles, and thus spend longer in mitosis [2, 9–11]. An earlier pioneering study demonstrated that a prolonged mitosis that surpasses a threshold duration (1.5 hours in RPE1 cells) is sufficient to trigger a p53-dependent cell cycle arrest in daughter cells, despite the completion of an otherwise normal division [24]. Analysis of 53BP1−/− and USP28−/− cells that lack centrosomes showed that most divisions surpassed the threshold of the mitotic timer. Despite the extended mitotic duration, acentrosomal 53BP1−/− and USP28−/− populations continue to proliferate, suggesting that the mitotic timer was no longer functioning [10]. Indeed, closer examination of the mitotic timer in these knockout backgrounds confirmed that 53BP1 and USP28 are essential not only for the centrosome surveillance pathway, but also for mitotic timer functionality [9–11]. This observation raised the possibility that centrosome loss triggers a cell cycle arrest by causing a mitotic delay.
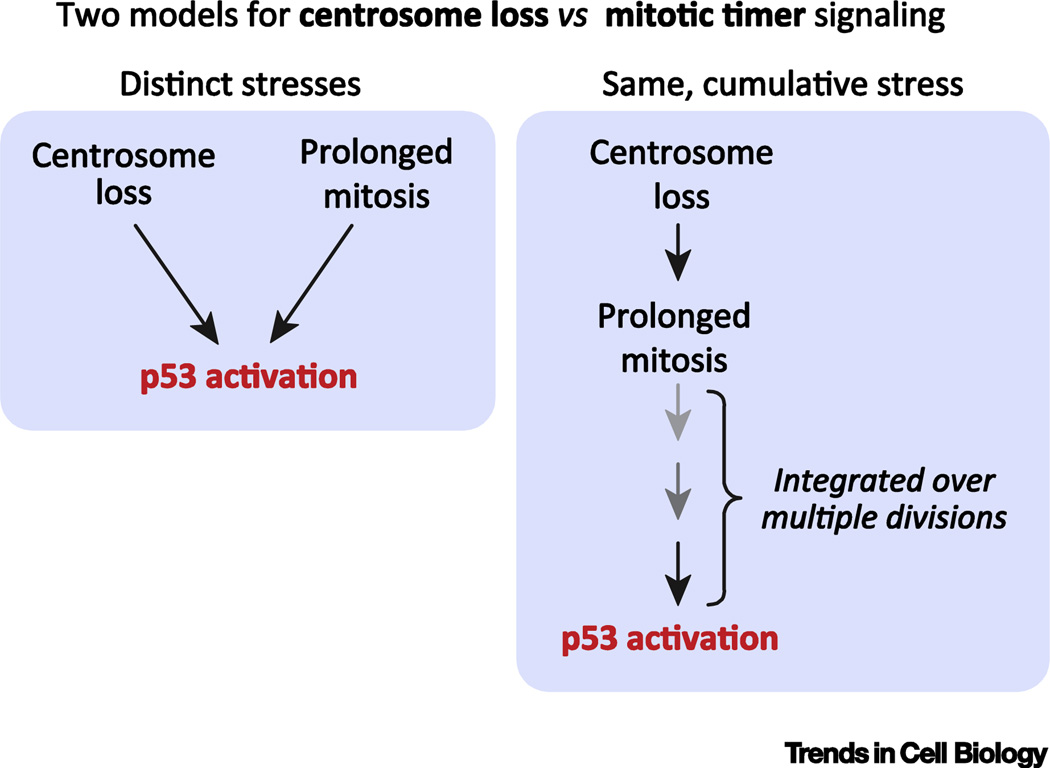
While activation of the mitotic timer is a promising premise for why cells arrest following centrosome loss, experiments to test this hypothesis have thus far proved inconclusive. Careful tracking of cell lineages has shown that cells spend progressively more time in mitosis as centrosomes are depleted. Nevertheless, many cells that arrest after centrosome loss had divided below the mitotic threshold window of 1.5 hours [1, 2]. This suggests that an increased duration of a single division is unable to account for the penetrance of the arrest that occurred following centrosome loss. It is possible, however, that cells “integrate” mitotic stress over several divisions (Figure 2). For example, two sequential, moderately prolonged mitoses that do not exceed the mitotic timer threshold may act to arrest cells by the same mechanism as a single prolonged division that exceeds 1.5 hours in duration. Support for such a “memory” model comes from earlier work investigating how cells respond to a prolonged mitosis [25]. This showed that transient treatment with a p38 MAP kinase inhibitor was sufficient to allow the proliferation of daughters arising from a prolonged (>1.5 hour) mitosis. These daughters divide with normal timing, but the resulting granddaughter cells arrest, despite a previous mitosis of normal duration. This suggests that the “granddaughter” cells can “recall” the stress from a prolonged mitosis more than one full cell cycle earlier. Testing whether cells can integrate mitotic stress over multiple cell cycles requires long-term cell lineage analysis. An alternative possibility, however, is that rather than being coupled, centrosome loss and prolonged mitosis are two distinct stresses that that feed into a common 53BP1-USP28-p53 signaling pathway.
Figure 2. Mitotic timer.
A) A schematic illustrating the two models for how the centrosome surveillance and mitotic timer stimuli activate the 53BP1-USP28-p53 signaling axis. Modified from [11].
The role of TRIM37 as a bypass of the centrosome surveillance response
The E3 ligase TRIM37 was also identified in the genome-wide CRISPR screens for increased growth following centrosome loss, and is an intriguing mechanistic outlier, distinct from the 53BP1-USP28-p53 axis [9, 11]. Knockout of TRIM37 prevents p53 stabilization and allows cells to escape arrest following centrosome loss – but unlike 53BP1 and USP28, its depletion does not disrupt mitotic timer function [11]. Studies of TRIM37−/− cells suggest it relieves centrosome surveillance through a distinct mode of action from the 53BP1-USP28 module. TRIM37 depletion enables the formation of foci that contain an array of centrosome components. These foci persist after loss of centrosomes, and nocodazole washout assays show that, like centrosomes, they are able to nucleate microtubule growth [11]. TRIM37−/− cells do not show a dramatic increase in mitotic duration like 53BP1−/− and USP28−/− cells, and instead can efficiently form the mitotic spindle in the absence of centrosomes. This suggests that the centrosome-like foci in TRIM37−/− cells may serve as surrogate MTOCs that increase the speed of spindle assembly to “bypass” the arrest triggered by centrosome loss.
In summary, TRIM37 plays an intriguing role in the regulation of centrosomal components, independent of the 53BP1-USP28 signaling module. The shortening of mitotic duration in TRIM37−/− cells supports a model in which cumulative prolonged mitoses trigger the centrosome surveillance pathway. Nevertheless, it remains possible that centrosome loss is instead detected through the loss of some output of centrosome activity, which is compensated for by the centrosome-like foci generated in TRIM37−/− cells.
Concluding remarks: A new mode of mitotic surveillance
The “centrosome surveillance” pathway was originally identified based on its requirement to arrest cells following centrosome loss. However, more recent work has demonstrated that centrosome surveillance pathway components are also required to respond to prolonged mitosis. We therefore propose that in the future, a more appropriate descriptive name for the 53BP1-USP28-p53 signaling mechanism will be the “mitotic surveillance pathway”. This name respects the fact that current evidence suggests the pathway is triggered by stresses that perturb mitosis. It will be interesting to test if additional stresses encountered during mitosis are also able to activate this pathway.
A major unanswered question is why has the mitotic surveillance pathway evolved? One possibility is that this pathway acts as a guardian of genome integrity, given that a mitotic delay or centrosome loss both increase the probability of mitotic errors. Indeed, while cells with a compromised surveillance pathway continue to proliferate after centrosome loss, they generate an acentrosomal population that displays inefficient spindle assembly and increased rates of chromosome missegregation and cytokinesis failure [1, 2, 11]. An increased mitotic duration could thus act as a ‘quality control sensor’ to identify and eliminate cells that delay in mitosis due to persistent spindle defects.
However, while 53BP1-knockout mice are tumor-prone and display high rates of aneuploidy in tumor cells [26, 27], USP28-knockout mice display no tumor phenotype [28, 29]. It is therefore currently unclear if the evolutionary pressure to maintain the mitotic surveillance pathway rests on the basis of preserving mitotic integrity. One possibility is that rather than maintaining proper centrosome number for mitotic spindle assembly, the mitotic surveillance pathway helps to maintain correct centrosome number to ensure each cell contains a centrosome for assembly of the primary cilium, an important cellular signaling hub. An additional interesting possibility is that the mitotic surveillance pathway acts to prevent the proliferation of differentiated cells that have inactivated centrosome-mediated microtubule nucleation [1]. Understanding the physiological relevance of the mitotic surveillance pathway remains an exciting area of future research.
While the organismal role of the mitotic surveillance pathway remains to be elucidated, there are likely to be cell and tissue type differences in signaling through this pathway. One example where the mitotic surveillance pathway must be inactive is in early mouse embryos, which proliferate in the absence of centrosomes until the 64-cell stage [30], before becoming sensitive to centrosome loss later in development [6]. Additionally, it is clear that the mitotic surveillance pathway is not present in flies, which lack clear 53BP1 and USP28 homologues [31, 32]. Thus, while the spindle assembly checkpoint serves to guard against chromosome segregation errors and is conserved from yeast to humans, the mitotic surveillance pathway may be restricted to vertebrates.
Understanding the mechanistic basis of the mitotic surveillance pathway will be an exciting area of future research. Important future directions include dissecting the interactions between 53BP1 and USP28 that activate downstream p53-p21 signaling, as well as screening for the upstream components that transduce the signal (see Outstanding Questions). Indeed, the genome-wide knockout screens for cells that can proliferate following centrosome loss has so far only yielded hits that also inactivate or circumvent the mitotic timer. This raises the question of whether screens can be designed to identify proteins uniquely required for mitotic timer or centrosome surveillance pathways. Targeted future screens could therefore reveal information about the upstream network(s) that signal to the 53BP1-USP28 module.
Outstanding questions Box.
How is the loss of centrosomes detected and signaled to the 53BP1-USP28-p53 axis?
Does centrosome loss trigger the 53BP1-USP28-p53 axis through the same mechanism as a single prolonged mitosis?
Is the localization of 53BP1 to the kinetochores important for sensing the perturbations that activate the surveillance pathway?
How do 53BP1 and USP28 act to regulate p53 activity?
What is the physiological relevance of centrosome surveillance? In what cell/tissue types is the pathway active?
How do cells trigger a p53-dependent arrest in response to centrosome amplification?
TRENDS.
Cells have developed quality control mechanisms to protect genome integrity in mitosis.
Cells can trigger a cell cycle arrest in response to a delayed mitosis or centrosome loss.
This response is p53-dependent, but independent of known p53-activating signaling pathways suggesting the existence of a novel “mitotic surveillance pathway”.
Genome-wide screens reveal that 53BP1 and USP28 activate p53 in this surveillance response.
The 53BP1-USP28-p53 axis may serve as a form of mitotic quality control by preventing the growth of cells that have an increased chance of making mitotic errors.
Acknowledgments
This work was supported by a research grant from the National Institutes of Health GM 114119 (to A.J.H.) and the NSF GRFP (to B.G.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Wong YL, et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambrus BG, et al. p53 protects against genome instability following centriole duplication failure. The Journal of Cell Biology. 2015;210:63–77. doi: 10.1083/jcb.201502089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland AJ, et al. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes & Development. 2012;26:2684–2689. doi: 10.1101/gad.207027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinchcliffe EH, et al. Requirement of a Centrosomal Activity for Cell Cycle Progression Through G1 into S Phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- 5.Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proceedings of the National Academy of Sciences. 2014;111:E1491–E1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Insolera R, et al. Cortical neurogenesis in the absence of centrioles. Nature Neuroscience. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong CS, et al. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife. 2016;5:e16270. doi: 10.7554/eLife.16270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambrus BG, et al. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. The Journal of Cell Biology. 2016;214:143–153. doi: 10.1083/jcb.201604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meitinger F, et al. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. The Journal of Cell Biology. 2016;214:155–166. doi: 10.1083/jcb.201604081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends in Cell Biology. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 14.Zhang D, et al. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 15.Knobel PA, et al. USP28 is recruited to sites of DNA damage by the tandem BRCT domains of 53BP1 but plays a minor role in double-strand break metabolism. Molecular and Cellular Biology. 2014;34:2062–2074. doi: 10.1128/MCB.00197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joo WS, et al. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes & Development. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derbyshire DJ, et al. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J. 2002;21:3863–3872. doi: 10.1093/emboj/cdf383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward I, et al. The tandem BRCT domain of 53BP1 is not required for its repair function. Journal of Biological Chemistry. 2006;281:38472–38477. doi: 10.1074/jbc.M607577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwabuchi K, et al. Two cellular proteins that bind to wild-type but not mutant p53. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:6098–6102. doi: 10.1073/pnas.91.13.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuella-Martin R, et al. 53BP1 Integrates DNA Repair and p53-Dependent Cell Fate Decisions via Distinct Mechanisms. Molecular Cell. 2016 doi: 10.1016/j.molcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 22.Jullien D, et al. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. Journal of Cell Science. 2002;115:71–79. doi: 10.1242/jcs.115.1.71. [DOI] [PubMed] [Google Scholar]
- 23.Ganem NJ, et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uetake Y, Sluder G. Prolonged Prometaphase Blocks Daughter Cell Proliferation Despite Normal Completion of Mitosis. Curr. Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward IM, et al. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Molecular and Cellular Biology. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward IM, et al. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Molecular and Cellular Biology. 2005;25:10079–10086. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schülein-Völk C, et al. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. CellReports. 2014;9:1099–1109. doi: 10.1016/j.celrep.2014.09.057. [DOI] [PubMed] [Google Scholar]
- 29.Diefenbacher ME, et al. The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. The Journal of Clinical Investigation. 2014;124:3407–3418. doi: 10.1172/JCI73733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szollosi D, et al. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. Journal of Cell Science. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- 31.Li L, et al. The Drosophila ubiquitin-specific protease Puffyeye regulates dMyc-mediated growth. Development. 2013;140:4776–4787. doi: 10.1242/dev.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sreesankar E, et al. Drosophila Rif1 is an essential gene and controls late developmental events by direct interaction with PP1-87B. Sci Rep. 2015;5:10679. doi: 10.1038/srep10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldock RA, et al. ATM Localization and Heterochromatin Repair Depend on Direct Interaction of the 53BP1-BRCT2 Domain with γH2AX. Cell Reports. 2015 doi: 10.1016/j.celrep.2015.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]