Abstract
The discovery in the 1980s that protein kinase C (PKC) is a receptor for the tumor-promoting phorbol esters fueled the dogma that PKC is an oncoprotein. Yet 30+ years of clinical trials for cancer using PKC inhibitors not only failed, but in some instances worsened patient outcome. The recent analysis of cancer-associated mutations, from diverse cancers and throughout the PKC family, revealed that PKC isozymes are generally inactivated in cancer, supporting a tumor suppressive function. In keeping with a bona fide tumor suppressive role, germ line causal loss of function (LOF) mutations in one isozyme have recently been identified in lymphoproliferative disorders. Thus, strategies in cancer treatment should focus on restoring, rather than inhibiting, PKC.
Keywords: PKC, phorbol esters, tumor suppressor, diacylglycerol, LOF
The PKC Family
PKC is a family of enzymes whose members play a pivotal role in cell signaling. PKC isozymes transduce the plethora of signals resulting from receptor-mediated hydrolysis of phospholipids, playing critical roles in diverse cellular functions, including controlling the balance between cell survival and cell death [1–4]. There are nine genes in mammals that encode PKC family members, with an even greater number of isozymes when one includes splice variants. Family members are classified into three subfamilies based on their cofactor dependence (see Text Box 1 and Figure I therein): conventional PKC isozymes (α, β, with two common splice variants, βI and βII, and γ) are activated by diacylglycerol and Ca2+, novel PKC isozymes (δ, ε, η and θ) are activated by diacylglycerol alone, and atypical PKC isozymes (ζ and ι) are regulated by protein:protein interactions. All isozymes are processed by a series of ordered phosphorylations and conformational transitions to yield a signaling-competent enzyme that is maintained in an autoinhibited conformation until binding of the correct second messengers (or protein scaffolds for atypical PKC isozymes [5]) [6, 7] (see Figure 1 for ‘life cycle’ of a conventional PKC). This processing depends on the kinase complex mTORC2 for most PKC isozymes (including all the conventional PKC isozymes) and PDK-1 for all PKC isozymes; in the absence of phosphorylation, diacylglycerol-sensitive PKC isozymes are in an open and proteolytically-labile conformation. Thus, mechanisms that prevent phosphorylation (such as depletion of PDK-1 or inhibition of mTORC2) prevent the accumulation of conventional and novel PKC isozymes in the cell. Following phosphorylation, PKC isozymes are activated by agonist binding to the diacylglycerol-sensing C1 domain and, in the case of conventional PKC isozymes, Ca2+ binding to the Ca2+-sensing C2 domain. This leads to conformational changes that break intramolecular contacts to ‘open’ PKC, resulting in release of an autoinhibitory pseudosubstrate segment from the substrate-binding cavity to permit substrate phosphorylation. The open conformation of PKC is sensitive to dephosphorylation and subsequent degradation (see Figure 1). Thus, the signaling output of PKC is critically dependent on 1] the rate of catalysis determined by the architecture of the kinase domain, 2] allosteric mechanisms that reversibly control the ‘closed’ (autoinhibited) and ‘open’ (active) conformation of the kinase, and 3] the steady-state levels of the enzyme. The complex nature of its regulation allows for multiple ways to either enhance or impair PKC activity.
Text Box.
The PKC Family
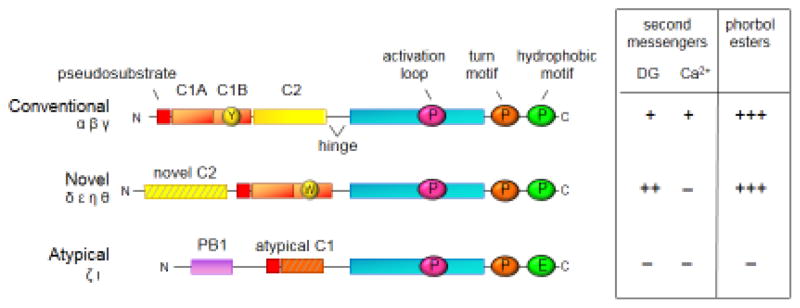
The nine PKC isozymes share a common architecture of an N-terminal regulatory moiety that constrains the catalytic activity of a C-terminal kinase domain (Figure I). All PKC isozymes have an autoinhibitory pseudosubstrate segment (red) that occupies the substrate-binding cavity in the inactive conformation. It is released from the substrate-binding cavity following engagement of various modules to appropriate second messengers or scaffolds, thus permitting substrate phosphorylation and downstream signaling. These modules are the C1 (orange), C2 (yellow), and PB1 (lavender) domains. The C1 domain serves as a diacylglycerol sensor in the conventional and novel PKC isozymes, but not in the atypical PKC isozymes because their C1 domain (striped) has a ring of basic residues surrounding the binding cleft precludes ligand binding. The stoichiometry of binding of diacylglycerol, or their functional analogues, the phorbol esters, is one mole ligand per mole PKC, revealing that only one of the tandem C1 domains binds ligand in the context of the full-length protein. A single amino acid difference from Tyr (Y in yellow circle) to Trp (W in yellow circle) in the C1B domain increases the affinity of novel PKC isozymes for diacylglycerol by two orders of magnitude compared to conventional PKC isozymes. This allows novel PKC isozymes to respond to agonist-evoked increases in diacylglycerol alone, without the need for increases in intracellular Ca2+. The C2 domain in conventional PKC isozymes serves as a Ca2+-sensor which targets these PKC isozymes to plasma membrane via a recognition motif for phosphatidylinositol-4,5-bisphosphate (PIP2), a lipid localized to plasma membrane. Although novel PKC isozymes have a C2 domain (striped), it lacks key Asp residues that coordinate Ca2+ and thus is not a Ca2+ sensor. The PB1 domain present in atypical PKC isozymes, which respond to neither Ca2+ nor diacyglycerol, mediates binding to protein scaffolds, interactions that promote release of the pseudosubstrate to allow localized signaling. The kinase domain (cyan) is regulated by three post-translational phosphorylations that ‘process’ PKC isozymes into a mature, autoinhibited, and stable species. PDK-1 catalyzes the phosphorylation on the activation loop (pink circle), a segment at the entrance to the active site, an event that triggers two tightly-coupled phosphorylations on the C-terminal tail at positions termed the turn motif (orange) and the hydrophobic motif (green). These C-terminal phosphorylations depend on the kinase activity of PKC and also on mTORC2; whether mTORC2 directly phosphorylates these sites, or whether it functions as a chaperone to allow newly-synthesized PKC to adopt a phosphorylatable conformation, remains to be determined. Note that atypical PKC isozymes differ in that their phosphorylation at the turn motif is co-translationally modified by ribosome-bound mTORC2 and that they have an acidic residue (Glu) at the hydrophobic motif. Cancer-associated LOF mutations have been identified throughout the PKC family and throughout the domain structure.
Figure I.
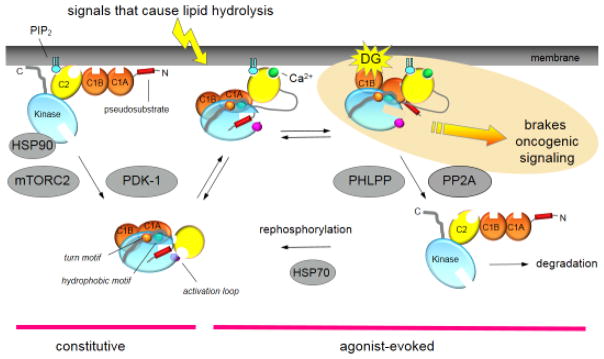
Figure 1.
Cartoon showing the multiple inputs regulating the function of a conventional PKC. Newly-synthesized PKC is processed by a series of ordered phosphorylations that depend on the binding of Hsp90 to a conserved PXXP motif in the kinase domain [74], the kinase complex mTORC2 [75], and PDK-1 (reviewed in [6]). Phosphorylation at three priming sites, the activation loop, the turn motif, and the hydrophobic motif, allow PKC to adopt an autonhibited conformation in which the Ca2+-sensing C2 domain (yellow) clamps the autoinhibitory pseudosubstrate segment (red) in the substrate-binding cavity of the kinase domain (cyan), and the diacyglycerol-sensing C1 domains (orange) become masked. Upon elevation of intracellular Ca2+, the C2 domain engages on the plasma membrane via Ca2+ coordination to anionic lipids and binding of plasma membrane-localized PIP2 to a surface on the domain. This positions PKC to bind its membrane-embedded ligand, diacylglycerol (DG), an event that releases the pseudosubstrate for maximal activation of PKC. This active PKC phosphorylates downstream substrates, such as K-RAS, to suppress oncogenic signaling. The open conformation of PKC is sensitive to dephosphorylation, with the first event being dephosphorylation of the hydrophoboic motif catalyzed by PHLPP; subsequent dephosphorylation by PP2A produces a fully dephosphorylated PKC that is shunted for degradation by a proteosomal pathway. However, binding of HSP70 to the dephosphorylated turn motif allows PKC to become rephosphorylated to sustain the signaling lifetime of the enzyme. Phorbol esters (not shown) bind with two-orders of magnitude higher affinity than diacylglycerol (highlighted in yellow) to the C1B domain and are not readily metabolized, trapping PKC in the open, active conformation and resulting in chronic loss, or down-regulation, of PKC. Novel PKC isozymes are regulated by similar mechanisms except their C2 domain does not function as a Ca2+ sensor. Atypical PKC isozymes are activated upon binding to specific protein scaffolds that tether the pseudosubstrate out of the substrate-binding cavity. Proteins indicated in grey are key regulators of the steady-state levels of PKC: HSP70, HSP90, mTORC2, and PDK-1 function to increase the steady-state levels of PKC by permitting processing phosphorylations, and PHLPP and PP2A decrease the steady-state levels of PKC by catalyzing the dephosphorylation of PKC. Targeting any of these proteins in cancer treatments will disrupt the balance of PKC signaling.
Phorbol esters
The milky sap exuded by the Croton tiglium plant has been used over the millennia for various purposes ranging from poison arrows to cathartic treatments. Following the identification of phorbol esters as the active ingredient, models of skin carcinogenesis quickly established that phorbol esters are potent tumor promoters [8]. Painting sub-threshold amounts of a carcinogen such as DMBA on mouse skin had no effect, but if this was followed by closely spaced (twice a week) and repetitive applications of phorbol esters, papilomas developed (reviewed in [3]). The quest then began to find ‘the receptor’ for phorbol esters. This presented a challenge given the highly lipophilic nature of the compounds, which results in non-saturable binding to cell membranes. A conceptual break-through was made by Peter Blumberg, who synthesized phorbol esters with shorter acyl chains to decrease their lipophilicity [9]. These molecules now bound to cells in a saturable manner, allowing the identification of PKC as a high affinity receptor for phorbol esters [10]. While we now understand that phorbol esters bind the C1 domain, a module present in a variety of other signaling proteins [3], their most famous receptor is PKC. The tumor promoting properties of phorbol esters, coupled with the assumption that kinases are oncoproteins, set in motion the idea that PKC isozymes are oncoproteins. Thus began considerable efforts to target PKC in cancer with inhibitors [11]. However, these efforts were largely unsuccessful. Indeed, a meta analysis of five clinical trials for non-small cell lung carcinoma revealed that patient outcome worsened when PKC inhibitors were combined with chemotherapy compared to chemotherapy alone [12]. Why the disconnect?
Like the physiological agonist diacylglycerol, phorbol esters bind the C1 domain but, unlike diacylglycerol, they are not readily metabolized and result in constitutive activation of PKC. In its open and active conformation, PKC is sensitive to dephosphorylation (catalyzed by the PH domain leucine-rich repeat protein phosphatase (PHLPP) at the hydrophobic motif and PP2A phosphatases at the turn motif and activation loop) and subsequent degradation (Figure 1). Thus, while phorbol esters result in acute activation of PKC, this is followed by the chronic loss, or down-regulation, of PKC [13]. Indeed, in the era preceding genetic knockdown, overnight treatment with phorbol esters was a common and effective method to deplete cells of PKC. In the paradigm described above for carcinogen-induced tumor promotion, the daily application of phorbol esters would be expected to cause a loss of PKC. Supporting this, prolonged infusion with bryostatins, marine natural products that, like phorbol esters, also down-regulate PKC [14], resulted in a reduction in the levels of PKCα, PKCε, and PKCη in peripheral blood monocytes of patients with advanced metastatic cancers [15]. Although PKC is the best characterized target of phorbol esters, it should be noted that they have targets in addition to PKC [16] and, furthermore, also induce local inflammatory responses, functions that likely also contribute to their tumor-promoting properties. Nonetheless, one might ask if the tumor-promoting properties of phorbol esters result, in part, from the loss of PKC.
PKC in Cancer
So what actually happens in cancer? Analysis of approximately 50 of the now >1,000 mutations in PKC isozymes identified in diverse cancers has revealed that they are mostly loss-of-function (LOF, see Glossary) [17]. Specifically, two thirds of the cancer-associated mutations examined were inactivating and the remaining were inert. If one includes truncating mutations or indels (insertions or deletions), an even greater fraction is LOF, as truncating mutations or indels for each isozyme have been frequently reported in the 126 cancer genome studies in cBioPortal [18]. These are important observations in light of experiments showing haploinsufficiency for at least one isozyme, PKCβ, in a colon cancer cell line. DLD1 colon cancer cells harbor a LOF mutation on one allele (A509T) of PKCβ; its correction to wild-type by genome editing suppressed anchorage-independent growth, a hallmark of cancer, however deletion of the mutant allele to generate a hemizygous cell line resulted in intermediate suppression of anchorage-independent growth. This provides compelling evidence that indels or truncation mutations in PKC isozymes that lead to loss of one functional copy of a specific PKC isozyme will enhance tumor growth [17]. Importantly, in this analysis of cancer-associated mutations, not a single gain-of-function (GOF) mutation was identified. Inactivation occurred by diverse mechanisms, such as impeding second messenger binding, preventing processing phosphorylations, or inhibiting catalysis. Additionally, there are numerous mutations in PKC family members that can be predicted with high confidence to be LOF, since they occur in highly conserved motifs required for catalytic activity, such as the hallmark amino acid segments HRD, DFG, or APE [19]. A sampling of these mutations is listed in Table 1. And the recently developed software KinView, an interactive integrative visualization that annotates cancer-associated protein kinase mutations, has identified a number of additional LOF mutations in the PKC family, of which one in PKCβII was validated experimentally [20]. Neomorphic mutations in PKC may also redirect PKC away from its relevant substrates, not only subverting its normal function but also engaging novel signaling pathways. In this regard, mutations in PKCγ that alter substrate specificity have been identified in lung cancer samples [21]. Thus, cancer-associated mutations across the PKC family are generally LOF.
Table 1.
A sampling of mutations that occur in critical regions of the PKC kinase domain (VAIK, HRD, DFG, or APE motifs [19]) from various cancers. These are distinct from the characterized LOF mutations recently reported [17]. Data were extracted from the cBioPortal [18].
| Isozyme | Sample | Cancer | Mutation | Motif |
|---|---|---|---|---|
| PRKCA | TCGA-AA-A010-01.1581 | Colorectal | D481E | DFG |
| PRKCA | TCGA-EE-A2GC-06.3524 | Melanoma | A506V | APE |
| PRKCA | TCGA-FG-8191-01.186 | Low Grade Glioma | A506T | APE |
| PRKCB | TCGA-GF-A6C8-06.3531 | Melanoma | D484N | DFG |
| PRKCB | HCC1500_BREAST | Breast | A509T | APE |
| PRKCB | HCT15_LARGE_INTESTINE | Colorectal | A509T | APE |
| PRKCB | TCGA-BH-A0HB-01.1857 | Breast | A509V | APE |
| PRKCG | TCGA-BR-7716-01.2771 | Stomach | D498N | DFG |
| PRKCE | TCGA-F1-A448-01.2751 | Stomach | K437T | VAIK |
| PRKCH | TCGA-BR-8687-01.2786 | Stomach | F498V | DFG |
| PRKCI | TCGA-IB-7651-01.691 | Pancreatic | D378N | HRD |
| PRKCI | KYSE150_OESOPHAGUS | Esophageal | D396E | DFG |
| PRKCI | TCGA-05-4424-01.1371 | Lung Adenocarcinoma | D396E | DFG |
| PRKCI | TCGA-78-7155-01.1374 | Lung Adenocarcinoma | E423D | APE |
| PRKCZ | TCGA-E2-A15E-01.676 | Breast | E421K | APE |
Sampling of high confidence predicted LOF mutations in PKC isozymes
mutated residue in bold
The majority of PKC LOF mutations are heterozygous, leaving one WT allele, raising interesting mechanistic questions as to how these PKC LOF mutants are promoting tumorigenesis. One possibility is that PKC LOF mutants have unique functions, supported by observations that these mutants can suppress signaling by multiple PKC isoforms, discussed below; thus, they are functioning in a ‘super’ dominant negative manner by impacting signaling of other gene products. An evaluation of classical tumor suppressors demonstrates that often when tumor suppressors, such as p53, have one allele mutated and maintain a functional allele that the somatic variants are neomorphic (in this case the p53 mutants are now considered to be GOF mutants) [22–24]. In the case of PKC, it is possible that the mutant protein could act as a scaffold to sequester signalling components of other pathways, perhaps subverting them from their physiological function or altering their activity. A second possibility is that the WT allele is not lost in order to maintain a threshold of PKC signaling that is required to promote tumorigenesis, as it is well documented that complete loss or hyperactivation of signaling pathways can be detrimental to the process of tumorigenesis [25].
The importance of LOF mutations in PKC isozymes in promoting oncogenic signaling was demonstrated by correction of a mutant allele of PKCβ in a colon cancer-derived cell line: this not only suppressed anchorage-independent growth, noted above, but effectively suppressed tumor growth in a xenograft murine model [17]. This cell line also harbored an oncogenic mutation in K-RAS, underscoring the dominating tumor suppressive role of PKC, even in the context of one of the most potent oncogenes. Potentially contributing to the strong survival advantage conferred by LOF mutations, many of the inactivating mutations characterized for PKC are dominant negative towards the global signaling output of other PKC isozymes. This dominant negative effect on the activity of other PKC isozymes may result from the mutant PKC interfering with the phosphorylation of other PKC isozymes, because their phosphorylation requires common titratable components (see Figure 1). This was first reported by Parker and coworkers who showed that expression of a PKC isozyme that could not be processed by phosphorylation impeded the accumulation of other PKC isozymes [26]; one potential candidate underlying the mechanism for the dominant negative effect is PDK-1, required for the priming phosphorylations of all PKC isozymes and reported to be present at 10 nM in HeLa cells, considerably below the sum concentration of all the PKC isozymes (>100 nM) [27]. In the case of LOF mutations in cancer, one mutant allele of PKCβ was shown to suppress the steady-state levels of another PKC isozyme, PKCα, a suppression that was reversed upon correction of the mutant allele to wild-type [17]. But even truncation mutations or indels likely confer a survival advantage to cancer cells: as noted above, deletion of a mutant PKCβ allele revealed haploinsufficiency. Thus, the steady-state levels of PKC tune the suppression of survival signaling pathways, suggesting that PKC levels may serve as a predictor for patient survival, as discussed below. The consensus emerging now is that PKC acts as the brakes to oncogenic function, potentially by inactivating oncogenes and stabilizing tumor suppressors.
Bioinformatic and Clinical Insights into the Tumor Suppressive Function of PKC Isozymes
Analysis of genes most frequently co-mutated with PKC isozymes identified a significant number of oncogenes [17]. K-RASis a case in point. Its phosphorylation on Ser181 by PKC, leading to its release from the plasma membrane and relocalization to mitochondria where it was shown to promote apoptosis, was reported a decade ago [28]. Although phosphorylation on Ser181 was later reported to be necessary for tumor growth [29], a recent study by McCormick and colleagues supports a tumor suppressive function of this PKC-mediated phosphorylation, consistent with the earlier report. Strikingly, these authors showed that oral administration to mice of a weak PKC activator, prostratin (a weak phorbol ester [30]), repressed tumor formation in orthotopic models of human pancreatic cancer [31]. In addition to removing K-RAS from the membrane described originally [28], cell-based studies revealed that the PKC-mediated phosphorylation of K-RAS inhibited the interaction of K-RAS with calmodulin, an interaction that results in reduced activity of CaM kinase and thus enhanced malignancy of pancreatic cells. The success of prostratin-treatment in the animal model likely reflected activation by the drug being sufficiently weak to prevent the degradation of PKC, yet sufficient to enhance PKC activity for phosphorylation of K-RAS and subsequent suppression of oncogenic function. The co-occurrence of mutations in oncogenes such as K-Ras in tumors with LOF mutations in PKC support a model in which the oncogene is kept in check by the strong tumor suppressive function of relevant PKC isozymes. In support of this, studies following the discovery of PKC identified an abundance of substrates that control growth factor and survival signaling. For example, an inhibitory phosphorylation of the EGF receptor by PKC was reported by Hunter and colleagues over 30 years ago [32]. Thus, survival signaling following mutation of an oncogene may only be strongly favored when the braking action of PKC is lost. Such a mechanism is reminiscent of the two-step model for tumor promotion, wherein a subthreshold amount of a carcinogen might induce an oncogenic mutation, but this does not cause a tumor unless PKC is down-regulated with repetitive applications of phorbol esters.
Clinical data provide strong support for a tumor suppressive role of PKC. Low levels of PKCα expression have recently been shown to predict poor outcome in T-cell Acute Lymphoblastic Leukemia (T-ALL) [33]. Similarly, low levels of PKCβII protein predict poor outcome in colorectal cancer [34]. And PKCβI, PKCβII, and PKCδ levels have been shown to be lower in high-grade and late-stage bladder cancer compared with normal, low-grade, or early-stage tissue [35–37]. Because the steady-state levels of PKC dictate its signaling output, reduced protein levels would result in reduced activity, supported by gene editing studies discussed above that reveal haploinsufficiency of at least one isozyme.
In hindsight, it is not surprising that cancer-associated mutations in PKC are LOF. In fact, the first cancer-associated mutation in a PKC isozyme, D294G in the C2 domain of PKCα, was identified by Joubert and colleagues over 20 years ago, in three types of cancers, and caused a clear loss of function [38]. Indeed, numerous cell biological studies suggested a tumor suppressive role of PKCα. For example, studies from Black and coworkers in the 1990s established a role for PKCα in suppressing cell growth [39, 40]. Levels of PKC protein are down-regulated in 60% of human colorectal cancers [41]. Moreover, genetically engineered mice deleted in PKCα developed colon tumors [42]. Despite evidence supporting a tumor suppressive role of PKC, other studies supporting an oncogenic role, coupled with the tumor promoting function of phorbol esters, created the dogma that PKC isozymes are oncoproteins [43].
Cementing the Role of PKC as a Tumor Suppressor
A hallmark of bona fide tumor-suppressing enzymes is the presence of germline mutations leading to the development of human proliferative disorders [44]. LOF mutations in classical tumor suppressors such as the lipid phosphatase PTEN and the kinase LKB1 (also known as STK11) are associated with early onset of human disorders with phenotypes associated with cancer [45, 46]. For example, LOF mutations in the kinase LKB1 lead to the development of Peutz-Jeghers syndrome [45], associated with the development of colonic hamartomas. Therefore, it is expected that LOF mutations in PKC isozymes would be associated with the development of human proliferative disorders that are associated with the development of cancer. This is indeed the case: LOF mutations in PKCδ are causal in juvenile systemic lupus erythematosus (JSLE) [47] and autoimmune lymphoproliferative syndrome and lead to increased proliferation and resistance to apoptosis in immune cells [48–50]. JSLE patients often develop B cell lymphomas [51]. Of 4 germline LOF PKCδ mutations identified to date in JSLE and autoimmune lymphoproliferative syndrome, somatic mutations at one of the sites (Arg614) have also been identified in three different colorectal tumors (cBioPortal; [18]). The frequent observation of somatic LOF mutations in PKC isozymes combined with the identification of germline causal mutations in lymphoproliferative disorders leaves little doubt about the importance of the tumor suppressive roles of PKC isozymes in cancer.
Exceptions to the Reversed Paradigm?
Although analysis of cancer-associated mutations in PKC are consistent with PKC isozymes generally serving as tumor suppressors, there may be cancer-specific and isozyme-specific contexts where that is not the case. Adult T Cell Leukemia (ATLL) may be one context where one isozyme, PKCβ, may have an oncogenic function. Results of whole genome and exome sequencing of ATLL patients have revealed that 33% of patients harbor mutations in PKCβ, with a hotspot at Asp427 [52]. Overexpression studies reveal that mutation of this residue to Asn, one common change at this site, results in enhanced activation of PKCβ, as assessed by accelerated phorbol ester-dependent membrane translocation and enhanced NF-κB transcription. These activating effects are, however, so great that it raises the question as to whether this enhanced open conformation of PKC may promote the down-regulation of the mutant protein. Whether ATLL patients with activating mutations in PKCβ maintain normal steady-state levels of this isozyme, or have reduced levels, remains to be determined.
Although no GOF mutations in cancer have been identified to date in PKCδ, this isozyme has roles both in survival and apoptotic pathways [3, 53]. For example, PKCδ promotes tumor progression in pancreatic cancer [54] and PKCδ-deficient mice have an increased incidence of lung tumors [55]. Another novel PKC, PKCε, may also have oncogenic roles, although no GOF mutations have been reported for this isozyme either. Transgenic mice overexpressing PKCε in the prostate develop preneoplastic lesions [43], and, conversely, genetic ablation of this isozyme in a transgenic mouse model of prostate adenocacinoma inhibits prostate cancer development and metastasis [56]. PKCε expression is frequently elevated not only in prostate tumors, but also those of breast and other cancers [43]. Some studies support the possibility that breast cancer may be one context-specific cancer where PKC may function as an oncoprotein: Reyland and colleagues have shown that elevated PKCδ mRNA levels negatively correlate with prognosis in Erb2-positive breast cancer, with mouse models suggesting that it is required for ErbB2-driven mammary gland tumorigenesis [57]. An earlier study also reported elevated PKCδ in breast cancer, but in this case in estrogen receptor-positive breast cancer [58]. Yet, evaluation of the mutational status of conventional and novel PKC isozymes in breast cancer suggests that PKC isozymes will be tumor suppressors in this cancer as well. Notably, there are several truncation and frameshift mutations observed in the genes of the conventional and novel PKC isozymes in human primary breast tumors. In addition, a previously characterized LOF mutation in PKCβ, A509V [17], is also observed in an invasive breast carcinoma. Taken together, these data point to a tumor suppressive role of PKC in breast cancer. Establishing whether PKC isozymes may play oncogenic roles in specific contexts in specific cancers awaits functional characterization of mutations in PKC isozymes in these cancers.
Atypical PKC isozymes should be considered as their own family as they are not regulated by the canonical diacylglycerol pathways. Rather, these enzymes have particularly low catalytic activity and their interaction on protein scaffolds tethers them in an open and active conformation near their protein substrates [5, 59, 60]. While LOF mutations have been identified in PKCζ [17, 61] and PKCι [62], the PRKCI gene is part of the 3q amplicon and considerable evidence supports a role for PKCι as an oncoprotein [63]. Notably, Fields and coworkers have identified a clear oncogenic role for PKCι in lung squamous cell carcinomas (LSCC) [64, 65]. They identified SOX2, a master transcriptional regulator of stemness, as a direct substrate of PKCι and showed that phosphorylation of SOX2 is required for the expression of hedgehog acetyl transferase, events that are necessary to maintain growth in soft agar. Lastly, Glioblastoma may also be a cancer in which atypical PKC isozymes function as oncoproteins: Ghosh and coworkers showed that high atypical PKC immunoreactivity, detecting primarily PKCι, correlated with poor disease prognosis in patients with glioblastoma; furthermore, an atypical PKC inhibitor reduced tumor growth in a mouse model of glioblastoma [66]. Whether GOF mutations in cancer occur for the atypical PKC isozymes remains to be determined.
Balancing the Signaling Output of PKC
Although an abundance of LOF mutations in PKC isozymes have been identified in cancer, GOF mutations in PKC do occur in disease. However, rather than being associated with survival-promoting pathophysiologies, they occur in degenerative diseases. In addition, these mutations enhance the agonist-dependent activation of PKC (for example by loosening autoinhibitory constraints), rather than causing constitutive activation. This is not surprising as mutations that lock PKC in the active conformation would be expected to have the paradoxical effect of being LOF because they would destabilize the enzyme and shunt it to degradation. In particular, germline mutations that enhance the activity of PKC have been identified in neurodegenerative diseases: activity-enhancing mutations in PKCγ cause spinocerebellar ataxia [67] and activity-enhancing variants in PKCα cosegregate with affected individuals in families with Alzheimer’s Disease [68]. Additionally, a polymorphism in PKCη that enhances its activity is associated with increased risk for cerebral infarction (stroke) [69], arthritis [70], and severe gastric atrophy [71]. This underscores the importance of precisely controlling the signaling output of PKC to control the balance between survival and degeneration: too little activity promotes a survival advantage whereas as too much activity promotes degeneration. In cancer, loss of PKC function confers a survival advantage by removing the brakes to oncogenic signaling. And in degenerative disease, enhanced PKC function promotes cell death.
Concluding Remarks
Targeting tumor suppressors in cancer is a major challenge. In the case of PKC, it would involve finding small molecules to stabilize the active conformation of the enzyme without promoting its down-regulation (see Outstanding Questions). In this regard, the ability of oral administration of the weak PKC activator, prostratin, to reduce tumor volume in mouse xenograft studies of pancreatic cancer lends promise to this therapeutic avenue [31]. Alternatively, modulating the activity of enzymes that control the steady-state levels of PKC in the cell might be another mechanism to promote PKC signaling. It should be noted that the dominant role of PKC isozymes in suppressing survival signaling suggests caution in targeting proteins that also regulate PKC steady-state levels. Notably, mTOR inhibitors [72] and Hsp90 inhibitors [73], currently in use in the clinic, prevent processing of PKC [74, 75]. Thus, these drugs will have the detrimental effect of removing the tumor suppressive function of PKC. Innovative approaches to restore PKC activity, coupled to chemotherapeutics targeted towards the primary oncogenic drivers, will likely increase the efficacy of cancer therapies. For example, K-RAS driven cancers that harbour mutations in PKC are likely to respond well to therapies that enhance or stablize PKC [31]. With the advent of personalized medicine, patients with LOF mutations in PKC isozymes or low levels of PKC expression in specific cancers will be particularly good candidates for approaches to restore PKC activity.
Outstanding Questions Box.
Will germline LOF mutations in PKCs be associated with other hereditary diseases associated with increased proliferative or cell survival phenotypes? Along these lines are individuals with heterozygous germline LOF mutations predisposed to developing cancer at an earlier age?
Are there contexts when PKCs are oncogenes? Truncating events are correlated and supportive of a tumor suppressive role of PKCs in cancer, however truncating events are rare for the atypical isoform, PKCζ. Does this suggest that PKCζ, may be an outlier and have an oncogenic role in cancer?
Will neomorphic mutations occur in PKC isozymes wherein mutations result in loss of signaling in canonical PKC signaling pathways but new gains in function arise that regulate unique downstream signaling pathways? Some mutations in substrate binding regions of PKC isoforms suggest this will be the case.
Can therapies be developed to enhance PKC signaling output without promoting down-regulation? Recent studies using prostatin indicate this may be possible. Furthermore, structural information and molecular modeling should aid in development of more efficacious and isozyme specific PKC agonists that could, for example, enhance signaling by PKC by decreasing intramolecular contacts that mediate autoinhibition.
How important are the dominant negative effects of PKC mutants and how does this impact the signaling from other PKC isozymes and AKT which is also regulated by some of the common titratable elements that regulate PKC, such as PDK-1 and mTORC2? What mechanisms are underpinning the observed dominant negative effects?
Trends Box.
Functional characterization of somatic mutations in the PKC family of kinases reveals that a majority of mutations are LOF and no mutations are GOF, reversing the paradigm that PKCs are oncogenes
Germline LOF mutations in PKCδ are associated with cancer-like syndromes including lymphoproliferative disorders and JSLE, further supporting the role of PKC isozymes as tumor suppressors
LOF mutant PKC isoforms act in a dominant negative manner to suppress PKC signaling
These studies contribute to a larger theme in the field that many kinases will have tumor suppressive roles and inactivating mutations in kinases will be prevalent in cancer
Acknowledgments
We thank Andrew Hudson for advice on bioinformatics analysis. This work was supported by NIH GM 43154 (A. C. N.), National Cancer Institute and Cancer Research UK (J. B.).
None of the authors have any competing interests.
Abbreviations
- GOF
gain-of-function
- JSLE
juvenile systemic lupus erythematosus
- LOF
loss-of-function
- PKC
protein kinase C Key words
Glossary
- Down-regulation
Decreased steady-state levels of a protein that can result from decreased transcription, destabilizing mutations, or altered post-translational modification leading to increased protein degradation. With reference to PKC, down-regulation signifies the loss of protein following prolonged treatment with phorbol esters.
- Gain-of-function (GOF) mutation
A mutation that results in increased function of a protein or the gain of a new molecular activity. These mutations are generally missense mutations that eliminate mechanisms of inhibition resulting in constitutive activation of the protein.
- Germline mutation
Inherited mutation that occurs in germ cells and is present in a majority of cells in the human body.
- Loss-of-function (LOF) mutation
A mutation that results in decreased protein expression or compromised protein function. LOF mutations can be missense mutations that result in a change in a single amino acid but are more commonly indels (insertions or deletions) or nonsense mutations that introduce a stop codon early in the gene.
- Neomorphic mutation
A mutation that leads to loss of the normal function of a protein and gain of a novel molecular function that will promote phenotypes associated with cancer.
- Oncogene
A gene that harbors frequent gain-of-function mutations or has increased expression in cancer leading to acquisition of phenotypes associated with cancer, such as increased proliferation or survival.
- Oncoprotein
The protein product of an oncogene.
- Somatic mutation
a mutation acquired in a cell from damage to DNA that is then passed on through cell division.
- Tumor suppressor
A gene that harbors frequent loss-of-function mutations or deletions in cancer; it generally acts to suppress proliferation and growth or promote cell death under certain physiological conditions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosse C, et al. PKC and the control of localized signal dynamics. Nat Rev Mol Cell Biol. 2010;11(2):103–12. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- 2.Newton AC. Protein kinase C: poised to signal. Am J Physiol Endocrinol Metab. 2010;298(3):E395–402. doi: 10.1152/ajpendo.00477.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7(4):281–94. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 4.Battaini F, Mochly-Rosen D. Happy birthday protein kinase C: past, present and future of a superfamily. Pharmacol Res. 2007;55(6):461–6. doi: 10.1016/j.phrs.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tobias IS, Newton AC. Protein Scaffolds Control Localized Protein Kinase Czeta Activity. J Biol Chem. 2016;291(26):13809–22. doi: 10.1074/jbc.M116.729483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antal CE, Newton AC. Tuning the signalling output of protein kinase C. Biochem Soc Trans. 2014;42(6):1477–83. doi: 10.1042/BST20140172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antal CE, et al. Intramolecular C2 Domain-Mediated Autoinhibition of Protein Kinase C betaII. Cell Rep. 2015;12(8):1252–60. doi: 10.1016/j.celrep.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumberg PM, et al. Phorbol ester receptors and the in vitro effects of tumor promoters. Ann N Y Acad Sci. 1983;407:303–15. doi: 10.1111/j.1749-6632.1983.tb47836.x. [DOI] [PubMed] [Google Scholar]
- 9.Driedger PE, Blumberg PM. Specific Binding of Phorbol Ester Tumor Promoters. Proc Natl Acad Sci U S A. 1980;77:567–571. doi: 10.1073/pnas.77.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castagna M, et al. Direct Activation of Calcium-activated, Phospholipid-dependent Protein Kinase by Tumor-promoting Phorbol Esters. J Biol Chem. 1982;257:7847–7851. [PubMed] [Google Scholar]
- 11.Mackay HJ, Twelves CJ. Targeting the protein kinase C family: are we there yet? Nat Rev Cancer. 2007;7(7):554–62. doi: 10.1038/nrc2168. [DOI] [PubMed] [Google Scholar]
- 12.Zhang LL, et al. The protein kinase C (PKC) inhibitors combined with chemotherapy in the treatment of advanced non-small cell lung cancer: meta-analysis of randomized controlled trials. Clin Transl Oncol. 2015;17(5):371–7. doi: 10.1007/s12094-014-1241-3. [DOI] [PubMed] [Google Scholar]
- 13.Jaken S, et al. Characterization of phorbol ester receptors and their down-modulation in GH4C1 rat pituitary cells. Cancer Res. 1981;41(6):2175–81. [PubMed] [Google Scholar]
- 14.de Vries DJ, et al. Demonstration of sub-nanomolar affinity of bryostatin 1 for the phorbol ester receptor in rat brain. Biochem Pharmacol. 1988;37(21):4069–73. doi: 10.1016/0006-2952(88)90097-4. [DOI] [PubMed] [Google Scholar]
- 15.Marshall JL, et al. Phase I study of prolonged infusion Bryostatin-1 in patients with advanced malignancies. Cancer Biol Ther. 2002;1(4):409–16. doi: 10.4161/cbt.1.4.17. [DOI] [PubMed] [Google Scholar]
- 16.Kazanietz MG. Eyes wide shut: protein kinase C isozymes are not the only receptors for the phorbol ester tumor promoters. Mol Carcinog. 2000;28(1):5–11. doi: 10.1002/(sici)1098-2744(200005)28:1<5::aid-mc2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 17.Antal CE, et al. Cancer-Associated Protein Kinase C Mutations Reveal Kinase’s Role as Tumor Suppressor. Cell. 2015;160(3):489–502. doi: 10.1016/j.cell.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meharena HS, et al. Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS Biol. 2013;11(10):e1001680. doi: 10.1371/journal.pbio.1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McSkimming DI, et al. KinView: a visual comparative sequence analysis tool for integrated kinome research. Mol Biosyst. 2016;12(12):3651–3665. doi: 10.1039/c6mb00466k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creixell P, et al. Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell. 2015;163(1):202–17. doi: 10.1016/j.cell.2015.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 24.Dittmer D, et al. Gain of function mutations in p53. Nat Genet. 1993;4(1):42–6. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature. 2015;521(7552):357–61. doi: 10.1038/nature14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Paramio P, et al. The broad specificity of dominant inhibitory protein kinase C mutants infers a common step in phosphorylation. Biochem J. 1998;333( Pt 3):631–6. doi: 10.1042/bj3330631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hein MY, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163(3):712–23. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 28.Bivona TG, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21(4):481–93. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Barcelo C, et al. Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Res. 2014;74(4):1190–9. doi: 10.1158/0008-5472.CAN-13-1750. [DOI] [PubMed] [Google Scholar]
- 30.Szallasi Z, Blumberg PM. Prostratin, a nonpromoting phorbol ester, inhibits induction by phorbol 12-myristate 13-acetate of ornithine decarboxylase, edema, and hyperplasia in CD-1 mouse skin. Cancer Res. 1991;51(19):5355–60. [PubMed] [Google Scholar]
- 31.Wang MT, et al. K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell. 2015;163(5):1237–51. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 32.Hunter T, et al. Protein Kinase C Phosphorylation of the EGF Receptor at a Threonine Residue Close to the Cytoplasmic Face of the Plasma Membrane. Nature. 1984;311:480–483. doi: 10.1038/311480a0. [DOI] [PubMed] [Google Scholar]
- 33.Milani G, et al. Low PKCalpha expression within the MRD-HR stratum defines a new subgroup of childhood T-ALL with very poor outcome. Oncotarget. 2014;5(14):5234–45. doi: 10.18632/oncotarget.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dowling CM, et al. Protein kinase C beta II suppresses colorectal cancer by regulating IGF-1 mediated cell survival. Oncotarget. 2016;7(15):20919–33. doi: 10.18632/oncotarget.8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koren R, et al. Protein kinase C (PKC) isoenzymes immunohistochemistry in lymph node revealing solution-fixed, paraffin-embedded bladder tumors. Appl Immunohistochem Mol Morphol. 2000;8(2):166–71. doi: 10.1097/00129039-200006000-00013. [DOI] [PubMed] [Google Scholar]
- 36.Langzam L, et al. Patterns of protein kinase C isoenzyme expression in transitional cell carcinoma of bladder. Relation to degree of malignancy. Am J Clin Pathol. 2001;116(3):377–85. doi: 10.1309/1VKK-HWH7-YVJN-7UF7. [DOI] [PubMed] [Google Scholar]
- 37.Varga A, et al. Tumor grade-dependent alterations in the protein kinase C isoform pattern in urinary bladder carcinomas. Eur Urol. 2004;46(4):462–5. doi: 10.1016/j.eururo.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Prevostel C, et al. The natural protein kinase C alpha mutant is present in human thyroid neoplasms. Oncogene. 1995;11(4):669–74. [PubMed] [Google Scholar]
- 39.Saxon ML, et al. Activation of protein kinase C isozymes is associated with post-mitotic events in intestinal epithelial cells in situ. J Cell Biol. 1994;126(3):747–63. doi: 10.1083/jcb.126.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frey MR, et al. Protein kinase C isozyme-mediated cell cycle arrest involves induction of p21(waf1/cip1) and p27(kip1) and hypophosphorylation of the retinoblastoma protein in intestinal epithelial cells. J Biol Chem. 1997;272(14):9424–35. doi: 10.1074/jbc.272.14.9424. [DOI] [PubMed] [Google Scholar]
- 41.Suga K, et al. Down-regulation of protein kinase C-alpha detected in human colorectal cancer. Biochem Mol Biol Int. 1998;44(3):523–8. doi: 10.1080/15216549800201552. [DOI] [PubMed] [Google Scholar]
- 42.Oster H, Leitges M. Protein kinase C alpha but not PKCzeta suppresses intestinal tumor formation in ApcMin/+ mice. Cancer Res. 2006;66(14):6955–63. doi: 10.1158/0008-5472.CAN-06-0268. [DOI] [PubMed] [Google Scholar]
- 43.Garg R, et al. Protein kinase C and cancer: what we know and what we do not. Oncogene. 2014;33(45):5225–37. doi: 10.1038/onc.2013.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Payne SR, Kemp CJ. Tumor suppressor genetics. Carcinogenesis. 2005;26(12):2031–45. doi: 10.1093/carcin/bgi223. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26(57):7825–32. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- 46.Hollander MC, et al. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nature reviews Cancer. 2011;11(4):289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belot A, et al. Protein kinase cdelta deficiency causes mendelian systemic lupus erythematosus with B cell-defective apoptosis and hyperproliferation. Arthritis Rheum. 2013;65(8):2161–71. doi: 10.1002/art.38008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuehn HS, et al. Loss-of-function of the protein kinase C delta (PKCdelta) causes a B-cell lymphoproliferative syndrome in humans. Blood. 2013;121(16):3117–25. doi: 10.1182/blood-2012-12-469544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salzer E, et al. B-cell deficiency and severe autoimmunity caused by deficiency of protein kinase C delta. Blood. 2013;121(16):3112–6. doi: 10.1182/blood-2012-10-460741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiykim A, et al. Potentially Beneficial Effect of Hydroxychloroquine in a Patient with a Novel Mutation in Protein Kinase Cdelta Deficiency. J Clin Immunol. 2015;35(6):523–6. doi: 10.1007/s10875-015-0178-9. [DOI] [PubMed] [Google Scholar]
- 51.Bernatsky S, et al. An international cohort study of cancer in systemic lupus erythematosus. Arthritis and rheumatism. 2005;52(5):1481–90. doi: 10.1002/art.21029. [DOI] [PubMed] [Google Scholar]
- 52.Kataoka K, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47(11):1304–15. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 53.Basu A, Pal D. Two faces of protein kinase Cdelta: the contrasting roles of PKCdelta in cell survival and cell death. ScientificWorldJournal. 2010;10:2272–84. doi: 10.1100/tsw.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mauro LV, et al. PKC Delta (PKCdelta) promotes tumoral progression of human ductal pancreatic cancer. Pancreas. 2010;39(1):e31–41. doi: 10.1097/MPA.0b013e3181bce796. [DOI] [PubMed] [Google Scholar]
- 55.Symonds JM, et al. Protein kinase C delta is a downstream effector of oncogenic K-ras in lung tumors. Cancer Res. 2011;71(6):2087–97. doi: 10.1158/0008-5472.CAN-10-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hafeez BB, et al. Genetic ablation of PKC epsilon inhibits prostate cancer development and metastasis in transgenic mouse model of prostate adenocarcinoma. Cancer Res. 2011;71(6):2318–27. doi: 10.1158/0008-5472.CAN-10-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allen-Petersen BL, et al. Protein kinase Cdelta is required for ErbB2-driven mammary gland tumorigenesis and negatively correlates with prognosis in human breast cancer. Oncogene. 2014;33(10):1306–15. doi: 10.1038/onc.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKiernan E, et al. Protein kinase Cdelta expression in breast cancer as measured by real-time PCR, western blotting and ELISA. Br J Cancer. 2008;99(10):1644–50. doi: 10.1038/sj.bjc.6604728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graybill C, et al. Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J Biol Chem. 2012;287(25):21003–11. doi: 10.1074/jbc.M112.360495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai LC, et al. Zeta Inhibitory Peptide Disrupts Electrostatic Interactions That Maintain Atypical Protein Kinase C in Its Active Conformation on the Scaffold p62. J Biol Chem. 2015;290(36):21845–56. doi: 10.1074/jbc.M115.676221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galvez AS, et al. Protein kinase Czeta represses the interleukin-6 promoter and impairs tumorigenesis in vivo. Mol Cell Biol. 2009;29(1):104–15. doi: 10.1128/MCB.01294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linch M, et al. A cancer-associated mutation in atypical protein kinase Ciota occurs in a substrate-specific recruitment motif. Sci Signal. 2013;6(293):ra82. doi: 10.1126/scisignal.2004068. [DOI] [PubMed] [Google Scholar]
- 63.Parker PJ, et al. Atypical protein kinase Ciota as a human oncogene and therapeutic target. Biochem Pharmacol. 2014;88(1):1–11. doi: 10.1016/j.bcp.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Justilien V, et al. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell. 2014;25(2):139–51. doi: 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ali SA, et al. Protein Kinase Ciota Drives a NOTCH3-dependent Stem-like Phenotype in Mutant KRAS Lung Adenocarcinoma. Cancer Cell. 2016;29(3):367–78. doi: 10.1016/j.ccell.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kusne Y, et al. Targeting aPKC disables oncogenic signaling by both the EGFR and the proinflammatory cytokine TNFalpha in glioblastoma. Sci Signal. 2014;7(338):ra75. doi: 10.1126/scisignal.2005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verbeek DS, et al. PKC gamma mutations in spinocerebellar ataxia type 14 affect C1 domain accessibility and kinase activity leading to aberrant MAPK signaling. J Cell Sci. 2008;121(Pt 14):2339–49. doi: 10.1242/jcs.027698. [DOI] [PubMed] [Google Scholar]
- 68.Alfonso SI, et al. Gain-of-function mutations in protein kinase Calpha (PKCalpha) may promote synaptic defects in Alzheimer’s disease. Sci Signal. 2016;9(427):ra47. doi: 10.1126/scisignal.aaf6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kubo M, et al. A nonsynonymous SNP in PRKCH (protein kinase C eta) increases the risk of cerebral infarction. Nat Genet. 2007;39(2):212–7. doi: 10.1038/ng1945. [DOI] [PubMed] [Google Scholar]
- 70.Takata Y, et al. Genetic association between the PRKCH gene encoding protein kinase Ceta isozyme and rheumatoid arthritis in the Japanese population. Arthritis Rheum. 2007;56(1):30–42. doi: 10.1002/art.22262. [DOI] [PubMed] [Google Scholar]
- 71.Goto Y, et al. PRKCH gene polymorphism is associated with the risk of severe gastric atrophy. Gastric Cancer. 2010;13(2):90–4. doi: 10.1007/s10120-009-0542-7. [DOI] [PubMed] [Google Scholar]
- 72.Don AS, Zheng XF. Recent clinical trials of mTOR-targeted cancer therapies. Rev Recent Clin Trials. 2011;6(1):24–35. doi: 10.2174/157488711793980147. [DOI] [PubMed] [Google Scholar]
- 73.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18(1):64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gould CM, et al. The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase C through a conserved PXXP motif in the C-terminal tail. J Biol Chem. 2009;284(8):4921–35. doi: 10.1074/jbc.M808436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11(6):859–71. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]