Abstract
Breast cancer is one of the most common malignant tumors with a high case-fatality rate among women. The present study aimed to investigate the effects of mesenchymal stem cells (MSCs) on breast cancer by exploring the potential underlying molecular mechanisms. The expression profile of GSE43306, which refers to MDA-MB-231 cells with or without a 1:1 ratio of MSCs, was downloaded from Gene Expression Omnibus database for differentially expressed gene (DEG) screening. The Database for Annotation, Visualization and Integrated Discovery was used for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for DEGs. The protein-protein interactional (PPI) network of DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins. The data was subsequently analyzed using molecular complex detection for sub-network mining of modules. Finally, DEGs in modules were analyzed using GO and KEGG pathway enrichment analyses. A total of 291 DEGs including 193 upregulated and 98 downregulated DEGs were obtained. Upregulated DEGs were primarily enriched in pathways including response to wounding (P=5.92×10−7), inflammatory response (P=5.92×10−4) and defense response (P=1.20×10−2), whereas downregulated DEGs were enriched in pathways including the cell cycle (P=7.13×10−4), mitotic cell cycle (P=6.81×10−3) and M phase (P=1.72 ×10−2). The PPI network, which contained 156 nodes and 289 edges, was constructed, and Fos was the hub node with the degree of 29. A total of 3 modules were mined from the PPI network. In total, 14 DEGs in module A were primarily enriched in GO terms, including response to wounding (P=4.77×10−6), wounding healing (P=6.25×10−7) and coagulation (P=1.13 ×10−7), and these DEGs were also enriched in 1 KEGG pathway (complement and coagulation cascades; P=0.0036). Therefore, MSCs were demonstrated to exhibit potentially beneficial effects for breast cancer therapy. In addition, the screened DEGs, particularly in PPI network modules, including FN1, CD44, NGF, SERPINE1 and CCNA2, may be the potential target genes of MSC therapy for breast cancer.
Keywords: mesenchymal stem cells, breast cancer, protein-protein interaction network, beneficial effects
Introduction
Breast cancer is one of the most common malignant tumors and has a high case-mortality rate among women, accounting for ~1/3 cancer cases diagnosed in the United States (1). Breast cancer has a heredity element and frequently occurs during menopause (2). Signs of breast cancer include changes in breast shape and skin, a breast lump and cyst fluid discharged from the nipple (3). At present, the treatments for breast cancer primarily include tumor resection, radiation treatment and chemotherapy (4,5). However, these treatment methods are associated with a high risk of recurrence (6). Therefore, the identification, development and study of novel treatment methods is required.
Mesenchymal stem cells (MSCs) are an important type of adult stem cells, which serve a role in the processes of tumor growth and metastasis (7). In vitro, MSCs are able to arrest the cell cycle progression of tumor cells in the G1 phase and reduce their apoptotic rate (8). In addition, previous studies have revealed that MSCs possess a number of functions, including hematopoietic support, immunoregulation, multilineage differentiation and specific migration (9,10). Studeny et al (11) demonstrated that MSCs are able to form an effective platform for the local production of interferon (IFN)-β, suppressing the process of pulmonary metastasis. Furthermore, MSCs may be used as carriers of a number of therapeutics, including interleukin (IL)-2, IFNs and C-X3-C motif chemokine receptor 1, in order to induce apoptosis and inhibit tumor cell differentiation (12–14).
A previous study revealed that MSCs are able integrate into the tumor-associated stroma and to affect the development of breast cancer (15). In co-culture with breast cancer cells, MSCs secreted a series of factors, including chemokine C-C motif chemokine ligand 2, IL-6 and tissue inhibitor of metalloproteinase 1 (TIMP-1), restricting the growth of cancer cells (16,17). Furthermore, MSCs were able to inhibit the proliferation of breast cancer cells by secreting Dickkopf-related protein 1, a novel inhibitor of Wnt signaling (18). In addition, MSCs are able to arrest cells at the G0/G1 phase of the cell cycle through upregulation of tumor protein 21 (p21) and caspase-3, further inhibiting cell growth (19). Furthermore, breast cancer-associated lymphedema of the arm can be effectively treated using autologous bone MSC transplantation (20).
However, certain opposing studies have revealed that MSCs are able to promote breast cancer metastasis (21,22). Therefore, the objective of the current study was to further examine this controversial issue by analyzing the underlying molecular mechanisms of the effect of MSCs in breast cancer. In the present study, to further explore the molecular mechanisms of MSCs in breast cancer, the GSE43306 gene expression dataset was downloaded from the Gene Expression Omnibus (GEO) database for differentially expressed gene (DEG) screening, pathway enrichment analysis and protein-protein interaction (PPI) network construction. Finally, PPI network modules were screened and analyzed.
Materials and methods
Microarray data
The GSE43306 expression profile dataset was downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). This dataset was collected using Illumina HiSeq 2000 (23). A total of nine tissue samples consisting of MDA-MB-231 cells comprised this dataset, including five samples supplemented with a 1:1 ratio of bone marrow MSCs (M) to MDA-MB-231 cells and four samples without MSC supplementation (A), which were compared in the current study to elucidate the effect of MSCs in breast cancer.
Data pre-processing and DEG analysis
The raw data were converted into a FASTQ format, subsequently the Next Generation Sequencing Quality Control Toolkit (24) was used for quality control and filtering of high-quality reads. Reads with ≥20 bases (70% read length) were selected as high-quality reads. By default, the high-quality reads were aligned to the full Human Genome (version 19) (https://www.ncbi.nlm.nih.gov/genome/51) using TopHat2 (25). Based on the results of comparison and genome annotation profiles, gene expression levels were calculated and genes with an expression value of 0 were removed. Subsequently, a gene expression matrix was produced through the removal of repeated gene symbols. The NOISeq R/Bioc package (bioconductor.org) (26) was applied to screen for DEGs between the M and A groups with the threshold of q≥0.05.
Functional and pathway enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery, which uses analytical tools to extract biological functions for numerous genes (27), was used for Gene Ontology (GO) (28) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (29) functional and pathway enrichment analysis, respectively, of the DEGs identified. The cut-off criteria were determined as P<0.05 and an enriched gene count >2.
PPI network construction
A PPI network of the DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, with a threshold combined score of >0.4. In the PPI network, the nodes and edges (lines) represent proteins and their interactions, respectively. Nodes with an average node connective degree ≥9 were regarded as hub proteins (30).
PPI network analysis
The PPI network was analyzed using Molecular Complex Detection (31), which mines for sub-network modules. The sub-networks were screened with the following default cut-off thresholds: Degree, 2; node score, 0.2; K-core, 2; maximum depth, 100. Subsequently, the nodes with the average node degree ≥3 and combined score (consisting of neighborhood, fusion, co-occurrence, co-expression, experimental database and text mining) were screened, and DEGs in these modules were used for pathway enrichment analysis.
Results
Screening of differentially expressed genes
Following pre-processing of the GSE43306 dataset, a total of 291 DEGs, including 193 upregulated and 98 downregulated DEGs, were identified (data not shown).
Functional and pathway enrichment analysis
As presented in Table I, upregulated DEGs were primarily enriched in the following two types of GO terms: The first type included wound response (P=5.92×10−7), the inflammatory response (P=5.92×10−4) and the immune response (P=1.20×10−2); and the second type included wound healing (P=4.97×10−6), regulation of body fluid levels (P=6.82×10−4), coagulation (P=6.83×10−4), blood coagulation (P=6.83×10−4) and hemostasis (P=9.23×10−4). Simultaneously, the upregulated DEGs were enriched in a number of other functions, including the mitogen-activated protein kinase signaling pathway (P=4.56×10−2) and hematopoietic cell lineage (P=2.13×10−2). The downregulated DEGs were also enriched in two types of GO term, as follows: The first type included the cell cycle (P=7.13×10−4), mitotic cell cycle (P=6.81×10−3), M phase (P=1.72×10−2), M phase of the mitotic cell cycle (P=1.93×10−2), cell cycle phase (P=4.08×10−2) and cell cycle process (P=4.41×10−2); the second type included RNA processing (P=3.35×10−3), RNA splicing (P=9.61×10−3), mRNA processing (P=1.57×10−2) and mRNA metabolic process (P=2.70×10−2).
Table I.
Top two types of functional and pathway enrichment analysis for upregulated and downregulated DEGs through GO and KEGG.
| Category | GO or KEGG term | Description | Number of nodes | P-value | Enrichment score |
|---|---|---|---|---|---|
| Up 1 | 3.792 | ||||
| BP | GO:0009611 | Wound response | 21 | 5.918×10−7 | |
| BP | GO:0006954 | Inflammatory response | 12 | 5.948×10−4 | |
| BP | GO:0006952 | Defense response | 14 | 1.199×10−2 | |
| Up 2 | 3.567 | ||||
| BP | GO:0042060 | Wound healing | 12 | 4.970×10−6 | |
| BP | GO:0050878 | Regulation of body fluid levels | 8 | 6.818×10−4 | |
| BP | GO:0050817 | Coagulation | 7 | 6.834×10−4 | |
| BP | GO:0007596 | Blood coagulation | 7 | 6.834×10−4 | |
| BP | GO:0007599 | Hemostasis | 7 | 9.229×10−4 | |
| P | hsa04010 | MAPK signaling pathway | 8 | 4.562×10−2 | |
| P | hsa04640 | Hematopoietic cell lineage | 5 | 2.132×10−2 | |
| Down 1 | 1.758 | ||||
| BP | GO:0007049 | Cell cycle | 12 | 7.129×10−4 | |
| BP | GO:0000278 | Mitotic cell cycle | 7 | 6.808×10−3 | |
| BP | GO:0000279 | M phase | 6 | 1.724×10−2 | |
| BP | GO:0000087 | M phase of mitotic cell cycle | 5 | 1.932×10−2 | |
| BP | GO:0022403 | Cell cycle phase | 6 | 4.085×10−2 | |
| BP | GO:0022402 | Cell cycle process | 7 | 4.415×10−2 | |
| Down 2 | 1.758 | ||||
| BP | GO:0006396 | RNA processing | 9 | 3.355×10−3 | |
| BP | GO:0008380 | RNA splicing | 6 | 9.611×10−3 | |
| BP | GO:0006397 | mRNA processing | 6 | 1.566×10−2 | |
| BP | GO:0016071 | mRNA metabolic process | 6 | 2.702×10−2 | |
| BP | GO:0000375 | RNA splicing, via transesterification reactions | 4 | 3.303×10−2 | |
| BP | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as a nucleophile | 4 | 3.303×10−2 | |
| BP | GO:0000398 | Nuclear mRNA splicing, via the spliceosome | 4 | 3.303×10−2 |
Up, upregulated; down, downregulated; BP, biological process; P, pathway; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; DEG, differentially expressed gene.
PPI network
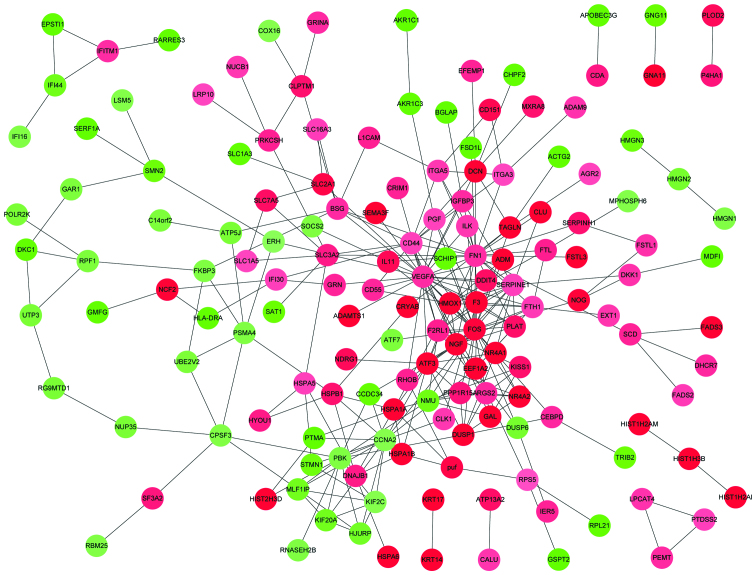
The PPI network constructed contained 156 proteins and 289 interactions (Fig. 1). Based on the average degree of the nodes, the following 12 nodes with degrees ≥9 were obtained: Fos proto-oncogene AP-1 transcription factor subunit (FOS), vascular endothelial growth factor A (VEGFA), fibronectin 1 (FN1), cluster of differentiation 44 (CD44), nerve growth factor (NGF), activating transcription factor 3 Serpin Family E Member 1 (SERPINE1), cyclin A2 (CCNA2), PBZ binding kinase, tissue factor F3, heme oxygenase-1 and ferritin heavy chain 1. Among them, FOS and VEGFA were the hub proteins with the highest node degree (29).
Figure 1.
DEG protein-protein interaction network. Red and green nodes represent upregulated and downregulated DEGs, respectively. The shades of color represent the strength of this regulation. DEG, differentially expressed gene.
Analysis of PPI network modules
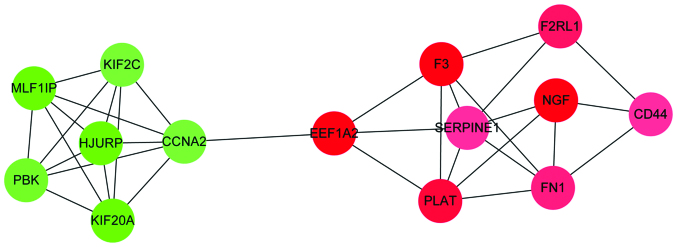
Through module analysis of the PPI network, a total of 3 modules, including modules A, B and C, were obtained (data not shown). The number of nodes in modules A, B and C were 14, 5 and 6, respectively. While the number of interactions in modules A, B and C were 33, 8 and 9, respectively. Module A is illustrated in Fig. 2. Amongst the three modules, module A had the highest enriched score. Therefore, module A was further analyzed with GO functional and KEGG pathway enrichment analyses, respectively (Table II). The 14 DEGs in module A were primarily enriched in GO terms such as wound response (P=4.77×10−6), wound healing (P=6.25×10−7) and coagulation (P=1.13×10−7). In addition, these DEGs were enriched in one KEGG pathway, the complement and coagulation cascade (P=0.0036).
Figure 2.
Protein-protein interactions network of module A DEGs. Red and green nodes represent upregulated and downregulated DEGs, respectively. The shades of color represent the strength of this regulation. DEG, differentially expressed gene.
Table II.
Top 10 enriched GO functions and KEGG pathways for DEGs in module A.
| Category | GO or KEGG term | Description | Number of nodes | P-value |
|---|---|---|---|---|
| BP | GO:0009611 | Response to wounding | 7 | 4.770×10−6 |
| BP | GO:0042060 | Wound healing | 6 | 6.251×10−7 |
| BP | GO:0050817 | Coagulation | 4 | 1.127×10−4 |
| BP | GO:0007596 | Blood coagulation | 4 | 1.127×10−4 |
| BP | GO:0007599 | Hemostasis | 4 | 1.335×10−4 |
| BP | GO:0050878 | Regulation of body fluid levels | 4 | 2.937×10−4 |
| BP | GO:0006954 | Inflammatory response | 4 | 3.285×10−3 |
| BP | GO:0006952 | Defense response | 4 | 1.901×10−2 |
| BP | GO:0007049 | Cell cycle | 4 | 3.492×10−2 |
| BP | GO:0042127 | Regulation of cell proliferation | 4 | 3.620×10−2 |
| P | hsa04610 | Complement and coagulation cascades | 3 | 3.647×10−3 |
BP, biological process; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially expressed gene.
Discussion
The effect of MSCs on the development and progression of tumors, including breast cancer, remains a subject of debate (19). It is important to study the effects of MSCs on tumor growth in order to develop novel therapies for the treatment of cancer (32). In the current study, a PPI network was constructed from DEGs in breast cancer. An important module with 14 nodes, including FN1, CD44, NGF, SERPINE1 and CCNA2, was identified through mining for modules in this network.
FN1 encodes the protein fibronectin 1, which is involved in cell adhesion and migration processes, including wound healing, embryogenesis, blood coagulation, host pathogenic defense and metastasis (33). FN1 is upregulated during the chondrogenic differentiation of MSCs and in various metastatic chondrosarcomas (34). In addition, FN1 expression has been demonstrated to be closely associated with various migration processes, including wound healing, embryogenesis and the metastasis of cancer cells (35). In addition, in the PC3 prostate cancer cell line, FN1 was able to interact with the cell membrane reporter(s), inhibiting focus formation and tumorigenesis (36). Similarly, the mRNA expression of FN1 in renal cancer cells is significantly increased compared with that in normal renal tissue (37). In the current study, FN1 was enriched in mRNA metabolism signaling pathways, which indicated that FN1 may have an important inhibitive role in the development of breast cancer via increased mRNA or protein expression levels.
CD44, which encodes a receptor for hyaluronic acid, participates in various cellular functions including hematopoiesis, lymphocyte activation and tumor cell metastasis (38). Furthermore, CD44, which is widely used as a MSC marker, has been demonstrated to serve a role in the migration of MSCs via migration assays and small interfering RNA experiments (39). In addition, during the development and progression of breast cancer, hyaluronic acid-CD44 signaling was able to inhibit breast cancer cell metastasis through epithelial-stromal interactions (40). The results of the current study are in agreement with these previous studies, demonstrating that CD44 was upregulated in cancer tissue samples supplemented with MDA-MB-231 cells and MSCs. Furthermore, the results of the current study suggest that CD44 serves a role in the metastasis of breast cancer, as CD44 was observed to be enriched in the pathway of hemostasis. In addition, these results indicate that CD44 is a potential therapeutic target for the treatment of breast cancer with MSCs.
NGF is a member of the NGF-β family that encodes a secreted protein with nerve growth stimulating activity (41). In a previous study of glioma, MSCs were able to produce NGF, thus having an antitumor effect (42). In addition, NGF has been demonstrated to be an important regulator of breast cancer progression, inhibiting the progression of breast cancer through interactions with the p75 neurotrophin receptor and p140 tropomyosin receptor kinase A (43). In the current study, NGF was upregulated in samples supplemented with MSCs, suggesting that is a potential target gene for the treatment of breast cancer with MSCs.
SERPINE1, a member of the serine proteinase inhibitor superfamily, encodes a protein that inhibits urokinase-type plasminogen activator (uPA) (44). In breast cancer, tumor severity may be associated with polymorphism of the plasminogen activator inhibitor type 1 4G/5G gene (45). In addition, uPA has been revealed to be associated with poor patient prognosis and tumor metastasis in breast cancer via various signaling pathways, including extracellular matrix breakdown (46). In addition, uPA may stimulate MSC migration via the ERK signaling pathway (47). These results indicate that uPA is a potential therapeutic target for MSC-mediated breast cancer treatment.
CCNA2 is a member of the highly conserved cyclin family, which binds to and activates cyclin-dependent kinases (CDKs) 1 and 2 in order to promote G1/S and G2/M cell cycle progression (48). Lysine-specific demethylase 1 promotes the development and aggressiveness of breast cancer through regulating CCNA2 expression levels (49). In addition, overexpression of CCNA2 was demonstrated to be associated with the poor prognosis of patients with breast cancer (50). However, few studies have investigated the expression of CCNA2 in breast cancer cell samples supplemented with MSCs. In the current study, CCNA2 was identified to be a downregulated in breast cancer cell samples supplemented with MSCs compared with cells without supplementation. Furthermore, CCNA2 was demonstrated to be enriched in the cell cycle signaling pathway and was a node in the PPI network constructed for the DEGs. These results suggest that CCNA2 may be a potential target gene for the treatment of breast cancer with MSCs.
In conclusion, the results of the present study indicate that MSCs have beneficial effects for the treatment breast cancer. The DEGs identified, particularly those in PPI network modules, including FN1, CD44, NGF, SERPINE1 and CCNA2, may be the potential target genes for the treatment of breast cancer with MSCs. However, these results require confirmation through further study.
Acknowledgements
The present study was supported by the Technology and Applied Basic Research Program of Yunnan province (grant no. 2012FB165), the Health and Technology program of Yunnan province (grant no. 2016NS010) and the Yunnan provincial doctoral graduate student award.
References
- 1.DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61:408–418. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 2.Breast International Group (BIG) 1–98 Collaborative Group. Thürlimann B, Keshaviah A, Coates AS, Mouridsen H, Mauriac L, Forbes JF, Paridaens R, Castiglione-Gertsch M, Gelber RD, et al. A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353:2747–2757. doi: 10.1056/NEJMoa052258. [DOI] [PubMed] [Google Scholar]
- 3.Nuhu A, Aliyu S, Musa A. Management of breast lumps in Maiduguri, Nigeria. Sahel Medical Journal. 2014;17:50. doi: 10.4103/1118-8561.134475. [DOI] [Google Scholar]
- 4.Ahn S, Moon H, Kim J, et al. P5-23-01: The impact of primary tumor resection on the survival of patients with stage IV breast cancer according to molecular subtype. Cancer Res. 2011;71 doi: 10.1158/0008-5472.SABCS11-P5-23-01. [DOI] [Google Scholar]
- 5.Solin LJ, Gray R, Goldstein LJ, Recht A, Baehner FL, Shak S, Badve S, Perez EA, Shulman LN, Martino S, et al. Prognostic value of biologic subtype and the 21-gene recurrence score relative to local recurrence after breast conservation treatment with radiation for early stage breast carcinoma: Results from the Eastern Cooperative Oncology Group E2197 study. Breast Cancer Res Treat. 2012;134:683–692. doi: 10.1007/s10549-012-2072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Darby S, McGale P, Correa C, Taylor C, Arriagada R, Clarke M, Cutter D, Davies C, Ewertz M, et al. Effect of radiotherapy after breast-conserving surgery on 10-year recurrence and 15-year breast cancer death: Meta-analysis of individual patient data for 10,801 women in 17 randomised trials. Lancet. 2011;378:1707–1716. doi: 10.1016/S0140-6736(11)61629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waterman RS, Henkle SL, Betancourt AM. Mesenchymal stem cell 1 (MSC1)-based therapy attenuates tumor growth whereas MSC2-treatment promotes tumor growth and metastasis. PLoS One. 2012;7:e45590. doi: 10.1371/journal.pone.0045590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramasamy R, Lam EW, Soeiro I, Tisato V, Bonnet D, Dazzi F. Mesenchymal stem cells inhibit proliferation and apoptosis of tumor cells: Impact on in vivo tumor growth. Leukemia. 2007;21:304–310. doi: 10.1038/sj.leu.2404489. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Armstrong MA, Li G. Mesenchymal stem cells in immunoregulation. Immunol Cell Biol. 2006;84:413–421. doi: 10.1111/j.1440-1711.2006.01458.x. [DOI] [PubMed] [Google Scholar]
- 10.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Studeny M, Marini FC, Dembinski JL, Zompetta C, Cabreira-Hansen M, Bekele BN, Champlin RE, Andreeff M. Mesenchymal stem cells: Potential precursors for tumor stroma and targeted-delivery vehicles for anticancer agents. J Natl Cancer Inst. 2004;96:1593–1603. doi: 10.1093/jnci/djh299. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Lin X, Zhao J, Shi W, Zhang H, Wang Y, Kan B, Du L, Wang B, Wei Y, et al. A tumor-selective biotherapy with prolonged impact on established metastases based on cytokine gene-engineered MSCs. Mol Ther. 2008;16:749–756. doi: 10.1038/mt.2008.3. [DOI] [PubMed] [Google Scholar]
- 13.Seo KW, Lee HW, Oh YI, Ahn JO, Koh YR, Oh SH, Kang SK, Youn HY. Anti-tumor effects of canine adipose tissue-derived mesenchymal stromal cell-based interferon-β gene therapy and cisplatin in a mouse melanoma model. Cytotherapy. 2011;13:944–955. doi: 10.3109/14653249.2011.584864. [DOI] [PubMed] [Google Scholar]
- 14.Xin H, Sun R, Kanehira M, Takahata T, Itoh J, Mizuguchi H, Saijo Y. Intratracheal delivery of CX3CL1-expressing mesenchymal stem cells to multiple lung tumors. Mol Med. 2009;15:321–327. doi: 10.2119/molmed.2009.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molloy AP, Martin FT, Dwyer RM, Griffin TP, Murphy M, Barry FP, O'Brien T, Kerin MJ. Mesenchymal stem cell secretion of chemokines during differentiation into osteoblasts, and their potential role in mediating interactions with breast cancer cells. Int J Cancer. 2009;124:326–332. doi: 10.1002/ijc.23939. [DOI] [PubMed] [Google Scholar]
- 17.Honczarenko M, Le Y, Swierkowski M, Ghiran I, Glodek AM, Silberstein LE. Human bone marrow stromal cells express a distinct set of biologically functional chemokine receptors. Stem cells. 2006;24:1030–1041. doi: 10.1634/stemcells.2005-0319. [DOI] [PubMed] [Google Scholar]
- 18.Ho IA, Chan KY, Ng WH, Guo CM, Hui KM, Cheang P, Lam PY. Matrix Metalloproteinase 1 is necessary for the migration of human bone marrow-derived mesenchymal stem cells toward human glioma. Stem Cells. 2009;27:1366–1375. doi: 10.1002/stem.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu YR, Yuan Y, Wang XJ, Wei LL, Chen YN, Cong C, Li SF, Long D, Tan WD, Mao YQ, et al. The growth inhibitory effect of mesenchymal stem cells on tumor cells in vitroin vivo. Cancer Biol Ther. 2008;7:245–251. doi: 10.4161/cbt.7.2.5296. [DOI] [PubMed] [Google Scholar]
- 20.Hou C, Wu X, Jin X. Autologous bone marrow stromal cells transplantation for the treatment of secondary arm lymphedema: A prospective controlled study in patients with breast cancer related lymphedema. Jpn J Clin Oncol. 2008;38:670–674. doi: 10.1093/jjco/hyn090. [DOI] [PubMed] [Google Scholar]
- 21.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 22.De Luca A, Lamura L, Gallo M, Maffia V, Normanno N. Mesenchymal stem cell-derived interleukin-6 and vascular endothelial growth factor promote breast cancer cell migration. J Cell Biochem. 2012;113:3363–3370. doi: 10.1002/jcb.24212. [DOI] [PubMed] [Google Scholar]
- 23.Zhang XH, Jin X, Malladi S, Zou Y, Wen YH, Brogi E, Smid M, Foekens JA, Massagué J. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154:1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel RK, Jain M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarazona S, Furió-Tarí P, Turrà D, Pietro AD, Nueda MJ, Ferrer A, Conesa A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Rosenfeld R, Simon I, Nau GJ, Bar-Joseph Z. A probabilistic generative model for GO enrichment analysis. Nucleic acids Res. 2008;36:e109. doi: 10.1093/nar/gkn434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2:e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu W, Xu W, Jiang R, Qian H, Chen M, Hu J, Cao W, Han C, Chen Y. Mesenchymal stem cells derived from bone marrow favor tumor cell growth in vivo. Exp Mol Pathol. 2006;80:267–274. doi: 10.1016/j.yexmp.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Soikkeli J, Podlasz P, Yin M, Nummela P, Jahkola T, Virolainen S, Krogerus L, Heikkilä P, von Smitten K, Saksela O, Hölttä E. Metastatic outgrowth encompasses COL-I, FN1, and POSTN up-regulation and assembly to fibrillar networks regulating cell adhesion, migration, and growth. Am J Pathol. 2010;177:387–403. doi: 10.2353/ajpath.2010.090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malchenko S, Seftor EA, Nikolsky Y, Hasegawa SL, Kuo S, Stevens JW, Poyarkov S, Nikolskaya T, Kucaba T, Wang M, et al. Putative multifunctional signature of lung metastases in dedifferentiated chondrosarcoma. Sarcoma. 2012;2012:820254. doi: 10.1155/2012/820254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helleman J, Jansen MP, Ruigrok-Ritstier K, van Staveren IL, Look MP, Meijer-van Gelder ME, Sieuwerts AM, Klijn JG, Sleijfer S, Foekens JA, Berns EM. Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin Cancer Res. 2008;14:5555–5564. doi: 10.1158/1078-0432.CCR-08-0555. [DOI] [PubMed] [Google Scholar]
- 36.Ifon ET, Pang A, Johnson W, Cashman K, Zimmerman S, Muralidhar S, Chan WY, Casey J, Rosenthal LJ. U94 alters FN1 and ANGPTL4 gene expression and inhibits tumorigenesis of prostate cancer cell line PC3. Cancer Cell Int. 2005;5:19. doi: 10.1186/1475-2867-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waalkes S, Atschekzei F, Kramer MW, Hennenlotter J, Vetter G, Becker JU, Stenzl A, Merseburger AS, Schrader AJ, Kuczyk MA, Serth J. Fibronectin 1 mRNA expression correlates with advanced disease in renal cancer. BMC Cancer. 2010;10:503. doi: 10.1186/1471-2407-10-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Q, Zhou Y, Ma B, Chen X, Wen Y, Liu Y, Fan Q, Qiu X. The clinical value of CXCR4, HER2 and CD44 in human osteosarcoma: A pilot study. Oncol Lett. 2012;3:797–801. doi: 10.3892/ol.2012.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, Mitsuhashi N, Klein A, Barsky LW, Weinberg K, Barr ML, Demetriou A, Wu GD. The role of the hyaluronan receptor CD44 in mesenchymal stem cell migration in the extracellular matrix. Stem Cells. 2006;24:928–935. doi: 10.1634/stemcells.2005-0186. [DOI] [PubMed] [Google Scholar]
- 40.Lopez JI, Camenisch TD, Stevens MV, Sands BJ, McDonald J, Schroeder JA. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 2005;65:6755–6763. doi: 10.1158/0008-5472.CAN-05-0863. [DOI] [PubMed] [Google Scholar]
- 41.Ullrich A, Gray A, Berman C, Dull TJ. Human β-nerve growth factor gene sequence highly homologous to that of mouse. Nature. 1983;303:821–835. doi: 10.1038/303821a0. [DOI] [PubMed] [Google Scholar]
- 42.Hamada H, Kobune M, Nakamura K, Kawano Y, Kato K, Honmou O, Houkin K, Matsunaga T, Niitsu Y. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Sci. 2005;96:149–156. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adriaenssens E, Vanhecke E, Saule P, Mougel A, Page A, Romon R, Nurcombe V, Le Bourhis X, Hondermarck H. Nerve growth factor is a potential therapeutic target in breast cancer. Cancer Res. 2008;68:346–351. doi: 10.1158/0008-5472.CAN-07-1183. [DOI] [PubMed] [Google Scholar]
- 44.Ulisse S, Baldini E, Sorrenti S, D'Armiento M. The urokinase plasminogen activator system: A target for anti-cancer therapy. Curr Cancer Drug Targets. 2009;9:32–71. doi: 10.2174/156800909787314002. [DOI] [PubMed] [Google Scholar]
- 45.Castelló R, España F, Vázquez C, Fuster C, Almenar SM, Aznar J, Estellés A. Plasminogen activator inhibitor-1 4G/5G polymorphism in breast cancer patients and its association with tissue PAI-1 levels and tumor severity. Thromb Res. 2006;117:487–492. doi: 10.1016/j.thromres.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 46.Han B, Nakamura M, Mori I, Nakamura Y, Kakudo K. Urokinase-type plasminogen activator system and breast cancer (Review) Oncol Rep. 2005;14:105–112. [PubMed] [Google Scholar]
- 47.Pulukuri SM, Gorantla B, Dasari VR, Gondi CS, Rao JS. Epigenetic upregulation of urokinase plasminogen activator promotes the tropism of mesenchymal stem cells for tumor cells. Mol Cancer Res. 2010;8:1074–1083. doi: 10.1158/1541-7786.MCR-09-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo KW, Lee SR, Bhandari DR, Roh KH, Park SB, So AY, Jung JW, Seo MS, Kang SK, Lee YS, Kang KS. OCT4A contributes to the stemness and multi-potency of human umbilical cord blood-derived multipotent stem cells (hUCB-MSCs) Biochem Biophys Res Commun. 2009;384:120–125. doi: 10.1016/j.bbrc.2009.04.094. [DOI] [PubMed] [Google Scholar]
- 49.Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 50.Husdal A, Bukholm G, Bukholm I. The prognostic value and overexpression of cyclin A is correlated with gene amplification of both cyclin A and cyclin E in breast cancer patient. Cell Oncol. 2006;28:107–116. doi: 10.1155/2006/721919. [DOI] [PMC free article] [PubMed] [Google Scholar]