Abstract
A variable number tandem repeat polymorphism (VNTR) in the PERIOD 3 (PER3) gene has been associated with heritable sleep and circadian variables, including self-rated chronotypes, polysomnographic (PSG) variables, insomnia, and circadian sleep-wake disorders. This report describes novel molecular and clinical analyses of PER3 VNTR polymorphisms to better define their functional consequences. As the PER3 VNTR is located in the exonic (protein coding) region of PER3, we initially investigated whether both alleles (variants) are transcribed into messenger RNA in human fibroblasts. The VNTR showed bi-allelic gene expression. We next investigated genetic associations in relation to clinical variables in 274 older adult Caucasian individuals. Independent variables included genotypes for the PER3 VNTR as well as a representative set of single nucleotide polymorphisms (SNPs) that tag common variants at the PER3 locus (linkage disequilibrium (LD) between genetic variants < 0.5). In order to comprehensively evaluate variables analyzed individually in prior analyses, dependent measures included PSG total sleep time and sleep latency, self-rated chronotype, estimated with the Composite Scale (CS), and lifestyle regularity, estimated using the Social Rhythm metric (SRM). Initially, genetic polymorphisms were individually analyzed in relation to each outcome variable using analysis of variance (ANOVA). Nominally significant associations were further tested using regression analyses that incorporated individual ANOVA-associated DNA variants as potential predictors and each of the selected sleep/circadian variables as outcomes. The covariates included age, gender, body mass index and an index of medical co-morbidity. Significant genetic associations with the VNTR were not detected with the sleep or circadian variables. Nominally significant associations were detected between SNP rs1012477 and CS scores (p = 0.003) and between rs10462021 and SRM (p = 0.047); rs11579477 and average delta power (p = 0.043) (analyses uncorrected for multiple comparisons). In conclusion, alleles of the VNTR are expressed at the transcript level and may have a functional effect in cells expressing the PER3 gene. PER3 polymorphisms had a modest impact on selected sleep/circadian variables in our sample, suggesting that PER3 is associated with sleep and circadian function beyond VNTR polymorphisms. Further replicate analyses in larger, independent samples are recommended.
Keywords: Circadian, Sleep, genetic, insomnia, Period 3, PER3, association
INTRODUCTION
Sleep and circadian function are intimately related, and their disruption has important health consequences. Environmental zeitgebers clearly play a substantial role in shaping sleep and circadian variation, but the role of hereditary factors is also well-recognized (Daan et al., 1984; Yamazaki et al., 2000). Significant heritability is detectable even when relevant variables are estimated using self-reported questionnaires (Horne & Ostberg, 1976; Klei et al., 2005; Roenneberg et al., 2003). Over the past two decades, individual DNA polymorphisms that contribute to the variation in sleep or circadian functions have also been identified, thus additionally validating a heritable basis for sleep and circadian function (Bamne, 2010; Hall et al., 2008b; Katzenberg et al., 1998; Pagani et al., 2016). Others have also reported genetic associations with sleep disorders such as advanced sleep phase type (ASPT), delayed sleep phase type (DSPT) and Circadian rhythm sleep disorders (CRSDs) (Ebisawa et al., 2001; Hida et al., 2014; Mishima et al., 2005; Takahashi et al., 2008). Specific interest has focused on the PER3 VNTR and its association with diurnal preference, also known as morningness-eveningness (Archer et al., 2003), as well as other circadian and sleep-related variables (Table 1). The VNTR polymorphism consists of a motif of 4 or 5 repeated 54-base pair (bp) nucleotide sequences encoding 18 amino acids; thus the polymorphism could plausibly alter cellular functions of the PER3 gene (Archer et al., 2003).
Table 1.
Genetic association studies of PER3 polymorphisms and circadian variables
| Author | Ancestry | Sample size | Association |
|---|---|---|---|
| (Ebisawa et al., 2001) | Japanese |
|
Significant association of PER3 SNPs with DSPS |
| (Archer et al., 2003) | Caucasians |
|
|
| (Johansson et al., 2003) | Caucasians | 159 patients with seasonal affective disorder, 159 matched controls |
G allele of SNP rs10462020 associated with morning preference |
| (Pereira et al., 2005) | Latin Americans | 17 with DSPS 156 Volunteers |
Significant association of PER3 SNPs with DSPS and diurnal preference |
| (Archer et al., 2010) | Caucasians |
|
Significant association of PER3 SNPs with diurnal preference and DSPS |
| (Barclay et al., 2011) | Caucasians | 947 individuals | No association found |
| (Osland et al., 2011) | Caucasians | 432 university students | No significant association |
| (Voinescu & Coogan, 2012) | Caucasians | NA | No significant association |
| (Chellappa et al., 2014) | Caucasians | 18 healthy male volunteers | humans homozygous for the PER35/5 allele are more sensitive to light effects on sleep |
| (Hida et al., 2014) | Japanese | 182 DSPT individuals, 67 free-running type (FRT) individuals, and 925 controls | SNP rs228697 was significantly associated with diurnal preference and the FRT phenotype, the minor allele is more prevalent in evening types than in morning types |
| (Parsons et al., 2014) | Caucasians | 947 individuals | Significant association between PER3 SNP rs10462020 and diurnal preference |
| (Perea et al., 2014) | Latin Americans | 294 undergrad university students | No significant association. |
| (Dmitrzak-Węglarz et al., 2016) | Caucasians |
|
daytime dysfunction with the PER3 SNPs - rs228727, rs228642 and rs10864315), |
| (Viena et al., 2016) | Multiple ethnicities | 205 healthy women | PER3(4/4) genotypes were at greater risk for transient psychological effects (mood and state anxiety) |
Previous publications report on samples that vary by size and by the types of sleep/circadian variables that were analyzed. Furthermore, PER3 polymorphisms other than the VNTR polymorphism have typically not been analyzed. These limitations prevent firm conclusions about the genetic associations at PER3 or about the magnitude of their functional impact. In order to further elucidate the role of PER3 on sleep and circadian function, we conducted two types of analyses. First, we evaluated the VNTR at the transcriptional level, by determining whether both alleles are transcribed. Second, we conducted genetic association studies in relation to several circadian and life style measures. In order to identify genetic associations more comprehensively at the PER3 gene, we examined not only the VNTR, but also a representative set of common single nucleotide polymorphisms. The studies were conducted in an extensively characterized sample of older adults with and without sleep problems.
MATERIALS AND METHODS
Approval for the studies was obtained from the University of Pittsburgh Institutional Review Board (IRB). As required by the IRB, all participants provided written informed consent. Our study conformed to the guidelines of the Declaration of Helsinki as required by this journal (Touitou et al., 2004).
A. Clinical evaluations
The sample comprised older adults whose recruitment and assessment has been described (Hall et al., 2008a). Briefly, participants were drawn from five component projects that comprised the “Aging Well, Sleeping Efficiently: Intervention Studies” (AgeWise) program project (AG020677). Each component projects addressed a specific late-life challenge, including bereavement (Project 1), caregiving (Project 2), insomnia complicated by medical co-morbidity (Project 3), and advancing into the final years of life (Projects 4 and 5). All participants completed a standardized assessment which included demographic data, the 18-item Pittsburgh Sleep Quality Index (PSQI) (Buysse et al., 1989) to characterize self-reported sleep quality; and the Physical Health (PCS) and Mental Health (MCS) Component Summary measures of the Medical Outcomes Survey Short Form 36 (SF-36) (Ware & Sherbourne, 1992) to characterize mental and physical health-related quality of life. Participants also completed a daily sleep diary for 2 weeks, followed by polysomnographic (PSG) sleep studies either in the laboratory or in participants’ homes, using comparable protocols. The PSG montage included bilateral central referential EEG channels (C3 and C4, referenced to A1–A2), electro-oculogram (EOG), submentalis electromyogram (EMG) and electrocardiogram (EKG). Visual sleep stage scoring in 20-second epochs was conducted by PSG technologists trained to reliability using standard scoring criteria (Rechtschaffen & Kales, 1968). PSG recordings were collected and scored prior to the 2007 American Academy of Sleep Medicine PSG scoring criteria. An automated artifact rejection program was run to remove epochs containing EMG artifacts from the EEG record (72) prior to computing power spectral analysis of the EEG during NREM sleep (Brunner et al., 1996). Sleep outcomes derived from visual sleep stage scoring and spectral analysis are operationalized in Table 2. Because laboratory and home recordings were conducted using different PSG systems, we have focused our analyses on relative EEG power, rather than absolute values. Specifically, EEG power in the 0.5–4 Hz band was calculated for each minute of NREM sleep, averaged for all minutes of NREM sleep, and divided by the average total power from 0.5–32 Hz during all minutes of NREM sleep. Venous blood samples for genetic analyses were collected from participants and stored at −80°C.
Table 2.
Distribution of demographic and clinical features (n=274)
| Mean (SD) | |
|---|---|
| Age | 73.36 ± 7.2 |
| Gender (male/female) | 98 / 176 |
| Body Mass Index (BMI) | 26.73 (4.16) |
| MOSPCS | 46.81 (9.38) |
| CS score | 42.53 (6.25) |
| SRM score | 4.99 (0.93) |
| Subjective Sleep Quality, total score (PSQI) | 6.04 (4.02) |
| Visually-Scored Sleep Total sleep time (minutes) Sleep latency (minutes) Delta sleep (percent) |
359.23 (61.79) 25.07 (32.74) 3.18 (4.53) |
| Quantitative EEG Average Delta EEG power during NREM sleep (0.05–4 Hz) |
22.57 (10.96) |
Continuous variables are listed as mean (standard deviation). MOSPCS: Medical Outcomes Survey Physical Component Score (scores range from 0–100, with 0 being maximally poor quality of life); CS: Composite Scale (13 items, scores range from 13 (extreme eveningness) to 55 (extreme morningness); SRM: Social Rhythm metric (5 item diary, daily for two weeks); PSQI: Pittsburgh Sleep Quality Index (scores range from 0 (better) to 21 (worse), TOTAL > 5 associated with poor sleep quality): BMI: Basal Metabolic Index.
B. Laboratory assays
Laboratory assays include gene expression and genetic association study as described below:
Gene expression Study
Cell culture assays
Human fibroblasts were obtained following skin punch biopsies in a separate study (D’Aiuto et al., 2014; Das et al., 2015). Briefly, fibroblasts were isolated and cultured in T-25 flasks and 6-well plates containing 10 mL of DMEM media with 10% FBS, 1X antibiotic/anti-mycotic in a tissue culture incubator at 37°C, 5%CO2. At 80% confluency, the cells were trypsinized and stored as a cell pellet at −70°C for further use.
RNA extraction and analysis
RNA samples were extracted from human fibroblast cell lines using Qiagen cell culture DNA mini kits and QIAamp RNA blood mini kits using the manufacturer’s protocol. RNA samples were treated with DNase I (Life Technologies, Inc.) during spin column extraction and before cDNA synthesis. RNA samples were reverse transcribed to cDNA using random hexamer method with SuperScript III First-Strand Synthesis for reverse transcriptase PCR (RT-PCR). cDNA samples were amplified using gDNA PCR primers to check for DNA contamination.
Genetic association study
DNA extraction and analysis
Genomic DNA was extracted from venous blood samples using Qiagen assay kits (https://www.qiagen.com/us/).
SNP selection
In addition to investigating the VNTR, our goal was to select and assay ‘tag’ SNPs that represent common DNA variation at the PER3 gene, i.e., SNPs with reported minor allele frequencies (MAF) greater than 5%. The genomic region selected for the analyses spanned 5 kilobase (kb) upstream and 5 kb downstream of the PER3 coding sequence. Anticipating genotype assay failure, we initially selected and assayed a redundant set of SNPs with linkage disequilibrium (LD, r2) less than 0.8, using software developed by the International HapMap Project (http://www.hapmap.org/). Out of 23 SNPs assayed, only 1 SNP failed quality control standards. To reduce the chances of false positive associations arising from multiple comparisons, we therefore pruned the genotyped SNPs to a set of 10 SNPs at an r2 cut off value of 0.5 (Figure 1).
Figure 1. Linkage Disequilibrium (LD) plot of studied PER3 SNPs in patients with late life insomnia.

Asterisk * indicates a pruned list of genotyped SNPs with r2 cut off value of 0.5.
Genotype assays
The PER3 VNTR was genotyped in genomic DNA or cDNA samples using PCR as described, with minor modifications for RT-PCR (Archer et al., 2003). Briefly, PER3 VNTR PCRs utilized primers specific for DNA (F-TGTCTTTTCATGTGCCCTTACTT and R- TGTCTGGCATTGGAGTTTGA) or for cDNA derived from RNA (F- TTACAGGCAACAATGGCAGT and R- CTGATGCTGCTGAACCAGT). The amplified products were 347 and 401 bps for gDNA PCR, and 267 and 321bp for RT-PCR, respectively. PCR conditions were 94°C for 10 minutes, followed by 35 cycles of 94°C for 30 seconds, 58°C for 30 seconds 72°C for 30 seconds, and a final extension at 72°C for 7 min. All PCR products were resolved on a 2% agarose gels. The PER3 SNPs were genotyped using iPLEX, a multiplexed single-base extension (SBE) method using the MassArray MALDI-TOF MS detection platform (Sequenom Inc., http://www.sequenom.com/getdoc/197b98fa-93f7-40e8-9deb-a8dcfecf899e/iPLEX-brochure_web/).
Quality control for genotype assay
All assays included blind duplicates and negative (distilled water) controls. All genotypes were read blind to clinical status. Assays were repeated in case of ambiguous genotypes. Deviations from Hardy-Weinberg Equilibrium (HWE) were evaluated for each SNP using a global significance threshold of p < 0.005. None of the SNPs were excluded based on Hardy-Weinberg disequilibrium (<0.001) or low genotyping rate (>=2% missing). The mean genotype call rate was 99.2% for SNPS and 83% for the VNTR.
Statistical Analysis
Study 1 outcomes rely on qualitative data; no statistical analyses were conducted. In view of a large number of DNA polymorphisms and clinical variables for Study 2, we adopted a two-stage analysis approach. Each polymorphism was initially analyzed in relation to each clinical variable using analysis of variance (ANOVA). Genetic associations that attained statistical significance (p=0.05) were next analyzed using regression analyses with the clinical variable of interest as the outcome and the genotype for the polymorphism of interest as the independent variable, along with covariates.
RESULTS
Gene expression study
VNTR and allelic expression
Among 7 fibroblast cell lines analyzed for PER3 VNTR, 4 were homozygous for the 4-repeat allele (4/4), 1 was homozygous for the 5-repeat allele (5/5) and 2 were heterozygous (4/5). The fibroblast cell lines were not synchronized before harvesting RNA. The ages for fibroblast donors range from 29–67 years with a mean age of 51 (SD=13). Inspection of the genomic DNA and cDNA bands after PCR amplification and electrophoresis of three representative heterozygous samples indicated that the heterozygous cell lines showed robust amplification for both alleles, indicating bi-allelic expression (Figure 2).
Figure 2. Bi-alleleic expression of PER3 in human fibroblasts.
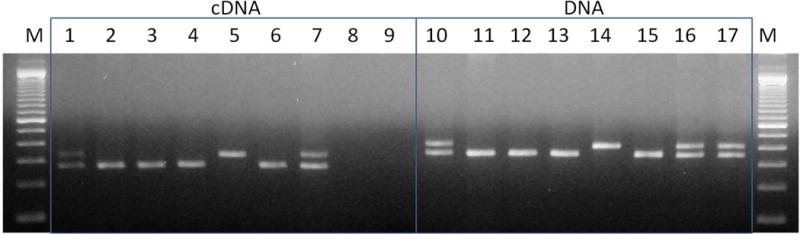
A panel of 7 individuals was selected based on genomic DNA analyses. Amplified cDNA with PER3 Exon 18 primers (Lanes 1–7) and amplified products from the respective genomic DNA samples from the same individuals (Lanes 10–16). Negative control (lane 8) and no-RT control (lane 9) are also shown for the cDNA samples. Positive control genomic DNA, heterozygous for the PER3 VNTR, derived from another blood sample is shown in lane 17.
M-100 bp DNA ladder.
Genetic association study
Sample characteristics
The sample was composed of 98 men and 176 women, mean age 73.36 years (standard deviation, SD, 7.2 years). All the participants reported Caucasian ancestry. The sample included 79 older adults with insomnia, 83 without sleep complaints, 73 caregivers and 39 bereaved adults. The circadian and sleep variables used for analyses are summarized in Table 2.
Genetic association analyses
Older adults with insomnia, good sleeper controls, caregivers and bereaved adults showed no significant differences in genotype frequencies for either VNTRs or the SNPs that were assayed (data not shown). Therefore, separate ANOVAs were conducted for each outcome of interest and each polymorphism initially in the entire sample (10 outcomes, 11 polymorphisms, including the VNTR). Several nominally significant associations were detected: rs1012477 with total CS score (p = 0.035); rs10462021 and rs1012477 with SRM (p = 0.017); rs228642 with total sleep time (p = 0.009); rs1012477 with sleep latency (SL) (p = 0.002); rs228729 (p = 0.039), rs10462021 (p = 0.051) and VNTR (p = 0.049) with average relative delta power.
Regression analyses were then conducted for these variables as outcomes with the associated SNPs as predictors and the following covariates: age, gender, BMI, and SF-36 PCS and MCS scores (Table 3 and Supplementary Table 1). Regression analyses showed nominally significant association with the following SNPs, uncorrected for multiple comparisons: rs1012477 with CS score (p = 0.003); rs10462021 with SRM (p = 0.047); rs228642 with PSG total sleep time (p = 0.016); average relative g_delta power with rs11579477 (p = 0.043).
Table 3.
Regression analyses for significant genetic associations.
| Standardized Beta | Unstandardized B | t | Sig. | 95.0% Confidence Interval for unstandardized B | |||
|---|---|---|---|---|---|---|---|
| Outome | SNP / VNTR | Lower Bound | Upper Bound | ||||
| CS | rs1012477 | −0.031 | −0.439 | −.465 | .642 | −2.302 | 1.423 |
| SRM | rs10462021 | 0.137 | 0.235 | 1.997 | .047 | .003 | .468 |
| SRM | rs1012477 | −0.029 | −0.060 | −.426 | .670 | −.337 | .217 |
| PSG sleep time | rs228642 | −0.172 | −15.674 | −2.440 | .016 | −28.349 | −3.000 |
| PSG sleep latency | rs1012477 | 0.076 | 5.692 | 1.049 | .295 | −5.011 | 16.395 |
| average delta power | rs11579477 | 0.155 | 9.992 | 2.041 | .043 | .310 | 19.673 |
| average delta power | rs1012477 | −0.022 | −0.514 | −.291 | .771 | −4.010 | 2.981 |
| average relative delta | rs228729 | 0.030 | 0.003 | .166 | .869 | −.036 | .043 |
| average relative delta | rs10462021 | −0.029 | −0.004 | −.293 | .770 | −.033 | .025 |
| average relative delta | PER3_vntr | 0.163 | 0.017 | .893 | .374 | −.021 | .056 |
Each regression analysis included the following covariates: age, gender, BMI, and index of medical problems. The data relating to the covariates are shown in Supplementary Table 1. avg_relativedelta: Relative delta power/minute; delta power/total power from 0.5–32 Hz
DISCUSSION
Our studies indicate bi-allelic expression of the PER3 VNTR, suggesting a functional role for both alleles in heterozygous individuals. We also analyzed representative common genetic variations at the PER3 locus in relation to a set of clinically relevant circadian and sleep-related variables. Nominally significant associations were detected between rs1012477 and CS scores. Modest associations were also detected between this SNP, as well as rs10462021, and SRM scores. Separately, associations were also detected for rs11579477 with PSG sleep time. In support of earlier studies, our analyses suggest that PER3 is associated with a range of sleep and circadian characteristics, including chronotype, the stability of daily social and activity rhythms, total sleep time and the amount of “deep” NREM sleep, indicated by relative delta power.
Though the PER3 VNTR was not significantly associated with any of the sleep or circadian variables analyzed in our sample, the RT-PCR analyses indicate bi-allelic transcription of the PER3 VNTR and thus lend credence to the published associations between this polymorphism and functional sleep outcomes. Recently, Hasan and colleagues (Hasan et al., 2014) reported on ‘humanized’ mice expressing VNTR alleles of exon 18 from the human PER3 gene. There were no significant differences at baseline between the wild type mice and the mice bearing any of the three VNTR genotypes with regard to circadian rhythmicity or sleep timing, but mice bearing both alleles of the VNTR showed deficits in sleep homeostasis estimated through theta power during wake electroencephalography (EEG) and delta power during sleep. These intriguing results could not be investigated in our sample, as data relating to waking quantitative EEG and sleep deprivation were not available.
The circadian and sleep characteristics associated with PER3 SNPs in this study are themselves associated with important health outcomes. For instance, a more “evening” chronotype, indicated in our study by higher CS score, has been associated with alcohol and substance abuse, depression and other psychiatric disorders, hypertension, and type 2 diabetes (reviewed in Partonen, 2015)(Partonen, 2015). The low regularity of daily rhythms indicated by lower SRM scores have been associated with conditions such as rapid-cycling bipolar disorder (Ashman et al., 1999), the onset of bipolar mood episodes,(Malkoff-Schwartz et al., 1998; Malkoff-Schwartz et al., 2000) and bereavement-related depression.(Brown et al., 1996) Conversely, greater regularity of daily rhythms, indicated by higher SRM scores, has been associated with reduced likelihood of recurrence in bipolar disorder(Frank et al., 2005) (see Grandin et al., 2006 for review) (Grandin et al., 2006). Total sleep time, or sleep duration, has been associated with a wide range of health outcomes, including obesity, hypertension, depression, and mortality (Hirshkowitz et al.; Watson et al., 2015). Although most of these associations are derived from habitual self-reported sleep duration, rather than PSG sleep duration as reported here, objective sleep duration is associated with similar health risks, particularly when associated with self-reported insomnia (Vgontzas et al., 2013). Likewise, reduced “deep” NREM sleep, indexed by relative delta power, is associated with glucose homeostasis and risk for type 2 diabetes (Tasali et al., 2008). Thus, associations between specific PER3 SNPs and sleep/circadian variables suggest that a common genetic pathway may underlie observed sleep/circadian-health outcomes.
Some shortcomings of the present study should be noted. None of the genetic associations would remain significant if corrections for multiple comparisons were applied. Consistent associations have been difficult to identify for any of the published associations with the circadian gene variants, most likely because the magnitude of the genetic associations with individual polymorphisms is relatively small. Thus, even our moderately large sample might be under-powered to detect such associations. Although we encountered relatively high failure rate when assaying VNTR genotypes, it is worth noting that SNP rs228729 was assayed in over 90% of the samples. As it is in tight LD with the VNTR, it would be feasible to impute the VNTR genotypes. Another limitation is the sample is restricted to older individuals.
In conclusion, we report that both alleles of the PER3 VNTR are transcribed. In support of earlier studies, our analyses suggest but do not conclusively prove that Per3 is associated with a range of sleep and circadian characteristics. Further replicative analyses in well-powered samples are recommended.
Acknowledgments
This study was funded in part by grants from the National Institute of Aging (2 P01 AG020677-06A1) and the National Institutes of Health (MH093246, D43 TW009114, MH63480, and D43TW008302).
Footnotes
DECLARATION OF INTEREST STATEMENT:
The authors declare no conflicts of interest.
References
- Archer SN, Carpen JD, Gibson M, Lim GH, Johnston JD, Skene DJ, von Schantz M. Polymorphism in the PER3 promoter associates with diurnal preference and delayed sleep phase disorder. Sleep. 2010;33:695–701. doi: 10.1093/sleep/33.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, Arendt J, von Schantz M. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–415. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- Ashman SB, Monk TH, Kupfer DJ, Clark CH, Myers FS, Frank E, Leibenluft E. Relationship between social rhythms and mood in patients with rapid cycling bipolar disorder. Psychiatry Res. 1999;86:1–8. doi: 10.1016/s0165-1781(99)00019-0. [DOI] [PubMed] [Google Scholar]
- Bamne MN, Mansour H, Monk TH, Buysse DJ, Nimgaonkar VL. Approaches to unravel the genetics of sleep. Sleep medicine reviews. 2010;14:397–404. doi: 10.1016/j.smrv.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay NL, Eley TC, Mill J, Wong CC, Zavos HM, Archer SN, Gregory AM. Sleep quality and diurnal preference in a sample of young adults: associations with 5HTTLPR, PER3, and CLOCK 3111. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:681–690. doi: 10.1002/ajmg.b.31210. [DOI] [PubMed] [Google Scholar]
- Brown LF, Reynolds CF, Monk TH, Prigerson HG, Dew MA, Houck PR, Mazumdar S, Buysse DJ, Hoch CC, Kupfer DJ. Social rhythm stability following late-life spousal bereavement: associations with depression and sleep impairment. Psychiatry Res. 1996;62:161–169. doi: 10.1016/0165-1781(96)02914-9. [DOI] [PubMed] [Google Scholar]
- Brunner DP, Vasko RC, Detka CS, Monahan JP, Reynolds CF, Kupfer DJ. Muscle artifacts in the sleep EEG: automated detection and effect on all-night EEG power spectra. J Sleep Res. 1996;5:155–164. doi: 10.1046/j.1365-2869.1996.00009.x. [DOI] [PubMed] [Google Scholar]
- Buysse DJ, Reynolds CF, 3rd, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res. 1989;28:193–213. doi: 10.1016/0165-1781(89)90047-4. [DOI] [PubMed] [Google Scholar]
- Chellappa SL, Viola AU, Schmidt C, Bachmann V, Gabel V, Maire M, Reichert CF, Valomon A, Landolt HP, Cajochen C. Light modulation of human sleep depends on a polymorphism in the clock gene Period3. Behav Brain Res. 2014;271:23–29. doi: 10.1016/j.bbr.2014.05.050. [DOI] [PubMed] [Google Scholar]
- D’Aiuto L, Zhi Y, Kumar Das D, Wilcox MR, Johnson JW, McClain L, MacDonald ML, Di Maio R, Schurdak ME, Piazza P, Viggiano L, Sweet R, Kinchington PR, Bhattacharjee AG, Yolken R, Nimgaonka VL. Large-scale generation of human iPSC-derived neural stem cells/early neural progenitor cells and their neuronal differentiation. Organogenesis. 2014;10:365–377. doi: 10.1080/15476278.2015.1011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S, Beersma DG, Borbely AA. Timing of human sleep: recovery process gated by a circadian pacemaker. Am J Physiol. 1984;246:R161–183. doi: 10.1152/ajpregu.1984.246.2.R161. [DOI] [PubMed] [Google Scholar]
- Das DK, Tapias V, D’Aiuto L, Chowdari KV, Francis L, Zhi Y, Ghosh BA, Surti U, Tischfield J, Sheldon M, Moore JC, Fish K, Nimgaonkar V. Genetic and morphological features of human iPSC-derived neurons with chromosome 15q11.2 (BP1–BP2) deletions. Mol Neuropsychiatry. 2015;1:116–123. doi: 10.1159/000430916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrzak-Węglarz M, Pawlak J, Wiłkość M, Miechowicz I, Maciukiewicz M, Ciarkowska W, Zaremba D, Hau J. Chronotype and sleep quality as a subphenotype in association studies of clock genes in mood disorders. Acta Neurobiol Exp (Wars) 2016;76:32–42. doi: 10.21307/ane-2017-003. [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Uchiyama M, Kajimura N, Mishima K, Kamei Y, Katoh M, Watanabe T, Sekimoto M, Shibui K, Kim K, Kudo Y, Ozeki Y, Sugishita M, Toyoshima R, Inoue Y, Yamada N, Nagase T, Ozaki N, Ohara O, Ishida N, Okawa M, Takahashi K, Yamauchi T. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank E, Kupfer DJ, Thase ME, Mallinger AG, Swartz HA, Fagiolini AM, Grochocinski V, Houck P, Scott J, Thompson W, Monk T. Two-year outcomes for interpersonal and social rhythm therapy in individuals with bipolar I disorder. Arch Gen Psychiatry. 2005;62:996–1004. doi: 10.1001/archpsyc.62.9.996. [DOI] [PubMed] [Google Scholar]
- Grandin LD, Alloy LB, Abramson LY. The social zeitgeber theory, circadian rhythms, and mood disorders: review and evaluation. Clin Psychol Rev. 2006;26:679–694. doi: 10.1016/j.cpr.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Hall M, Buysse DJ, Nofzinger EA, Reynolds CF, Thompson W, Mazumdar S, Monk TH. Financial strain is a significant correlate of sleep continuity disturbances in late-life. Biol Psychol. 2008a;77:217–222. doi: 10.1016/j.biopsycho.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MH, Muldoon MF, Jennings JR, Buysse DJ, Flory JD, Manuck SB. Self-reported sleep duration is associated with the metabolic syndrome in midlife adults. Sleep. 2008b;31:635–643. doi: 10.1093/sleep/31.5.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S, van der Veen DR, Winsky-Sommerer R, Hogben A, Laing EE, Koentgen F, Dijk DJ, Archer SN. A human sleep homeostasis phenotype in mice expressing a primate-specific PER3 variable-number tandem-repeat coding-region polymorphism. FASEB J. 2014;28:2441–2454. doi: 10.1096/fj.13-240135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida A, Kitamura S, Katayose Y, Kato M, Ono H, Kadotani H, Uchiyama M, Ebisawa T, Inoue Y, Kamei Y, Okawa M, Takahashi K, Mishima K. Screening of clock gene polymorphisms demonstrates association of a PER3 polymorphism with morningness-eveningness preference and circadian rhythm sleep disorder. Sci Rep. 2014;4:6309. doi: 10.1038/srep06309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshkowitz M, Whiton K, Albert SM, Alessi C, Bruni O, DonCarlos L, Hazen N, Herman J, Katz ES, Kheirandish-Gozal L, Neubauer DN, O’Donnell AE, Ohayon M, Peever J, Rawding R, Sachdeva RC, Setters B, Vitiello MV, Ware JC, Adams Hillard PJ. National Sleep Foundation’s sleep time duration recommendations: methodology and results summary. Sleep Health: Journal of the National Sleep Foundation. 2015;1:40–43. doi: 10.1016/j.sleh.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. International Journal of Chronobiology. 1976;4:97–110. [PubMed] [Google Scholar]
- Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, Kieseppa T, Lichtermann D, Praschak-Rieder N, Neumeister A, Nilsson LG, Kasper S, Peltonen L, Adolfsson R, Schalling M, Partonen T. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology. 2003;28:734–739. doi: 10.1038/sj.npp.1300121. [DOI] [PubMed] [Google Scholar]
- Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, Mignot E. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- Klei L, Reitz P, Miller M, Wood J, Maendel S, Gross D, Waldner T, Eaton J, Monk TH, Nimgaonkar VL. Heritability of morningness-eveningness and self-report sleep measures in a family based sample of 521 Hutterites. Chronobiology International. 2005;22:1041–1054. doi: 10.1080/07420520500397959. [DOI] [PubMed] [Google Scholar]
- Malkoff-Schwartz S, Frank E, Anderson B, Sherrill JT, Siegel L, Patterson D, Kupfer DJ. Stressful life events and social rhythm disruption in the onset of manic and depressive bipolar episodes: a preliminary investigation. Arch Gen Psychiatry. 1998;55:702–707. doi: 10.1001/archpsyc.55.8.702. [DOI] [PubMed] [Google Scholar]
- Malkoff-Schwartz S, Frank E, Anderson BP, Hlastala SA, Luther JF, Sherrill JT, Houck PR, Kupfer DJ. Social rhythm disruption and stressful life events in the onset of bipolar and unipolar episodes. Psychol Med. 2000;30:1005–1016. doi: 10.1017/s0033291799002706. [DOI] [PubMed] [Google Scholar]
- Mishima K, Tozawa T, Satoh K, Saitoh H, Mishima Y. The 3111T/C polymorphism of hClock is associated with evening preference and delayed sleep timing in a Japanese population sample. Am J Med Genet B Neuropsychiatr Genet. 2005;133:101–104. doi: 10.1002/ajmg.b.30110. [DOI] [PubMed] [Google Scholar]
- Osland TM, Bjorvatn BR, Steen VM, Pallesen S. Association study of a variable-number tandem repeat polymorphism in the clock gene PERIOD3 and chronotype in Norwegian university students. Chronobiol Int. 2011;28:764–770. doi: 10.3109/07420528.2011.607375. [DOI] [PubMed] [Google Scholar]
- Pagani L, St Clair PA, Teshiba TM, Service SK, Fears SC, Araya C, Araya X, Bejarano J, Ramirez M, Castrillón G, Gomez-Makhinson J, Lopez MC, Montoya G, Montoya CP, Aldana I, Navarro L, Freimer DG, Safaie B, Keung LW, Greenspan K, Chou K, Escobar JI, Ospina-Duque J, Kremeyer B, Ruiz-Linares A, Cantor RM, Lopez-Jaramillo C, Macaya G, Molina J, Reus VI, Sabatti C, Bearden CE, Takahashi JS, Freimer NB. Genetic contributions to circadian activity rhythm and sleep pattern phenotypes in pedigrees segregating for severe bipolar disorder. Proc Natl Acad Sci U S A. 2016;113:E754–761. doi: 10.1073/pnas.1513525113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Lester KJ, Barclay NL, Archer SN, Nolan PM, Eley TC, Gregory AM. Polymorphisms in the circadian expressed genes PER3 and ARNTL2 are associated with diurnal preference and GNβ3 with sleep measures. J Sleep Res. 2014;23:595–604. doi: 10.1111/jsr.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partonen T. Chronotype and Health Outcomes. Current Sleep Medicine Reports. 2015;1:205–211. [Google Scholar]
- Perea CS, Niño CL, López-León S, Gutiérrez R, Ojeda D, Arboleda H, Camargo A, Adan A, Forero DA. Study of a Functional Polymorphism in the PER3 Gene and Diurnal Preference in a Colombian Sample. Open Neurol J. 2014;8:7–10. doi: 10.2174/1874205X01408010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira DS, Tufik S, Louzada FM, Benedito-Silva AA, Lopez AR, Lemos NA, Korczak AL, D’Almeida V, Pedrazzoli M. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- Rechtschaffen A, Kales AA. A Manual of Standardized Terminology, Techniques, and Scoring System for Sleep Stages of Human Subjects. Bethesda: National Institute of Neurological Diseases and Blindness; 1968. [Google Scholar]
- Roenneberg T, Wirz-justice A, Merrow M. Life between clocks: daily temporal patterns of human chronotypes. J Biol Rhythms. 2003;18:80–90. doi: 10.1177/0748730402239679. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasali E, Leproult R, Ehrmann DA, Van Cauter E. Slow-wave sleep and the risk of type 2 diabetes in humans. Proc Natl Acad Sci U S A. 2008;105:1044–1049. doi: 10.1073/pnas.0706446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou Y, Portaluppi F, Smolensky MH, Rensing L. Ethical principles and standards for the conduct of human and animal biological rhythm research. Chronobiol Int. 2004;21:161–170. doi: 10.1081/cbi-120030045. [DOI] [PubMed] [Google Scholar]
- Vgontzas AN, Fernandez-Mendoza J, Liao D, Bixler EO. Insomnia with objective short sleep duration: the most biologically severe phenotype of the disorder. Sleep medicine reviews. 2013;17:241–254. doi: 10.1016/j.smrv.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viena TD, Gobin CM, Fins AI, Craddock TJ, Tartar A, Tartar JL. A PER3 Polymorphism Interacts with Sleep Duration to Influence Transient Mood States in Women. J Circadian Rhythms. 2016;14:3. doi: 10.5334/jcr.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinescu BI, Coogan AN. A variable-number tandem repeat polymorphism in PER3 is not associated with chronotype in a population with self-reported sleep problems. Sleep and Biological Rhythms. 2012;10:23–26. [Google Scholar]
- Ware JE, Jr, Sherbourne CD. The MOS 36-item short-form health survey (SF-36). I. Conceptual framework and item selection. Medical care. 1992;30:473–483. [PubMed] [Google Scholar]
- Watson NF, Badr MS, Belenky G, Bliwise DL, Buxton OM, Buysse D, Dinges DF, Gangwisch J, Grandner MA, Kushida C, Malhotra RK, Martin JL, Patel SR, Quan SF, Tasali E, Panel CC. Joint Consensus Statement of the American Academy of Sleep Medicine and Sleep Research Society on the Recommended Amount of Sleep for a Healthy Adult: Methodology and Discussion. J Clin Sleep Med. 2015;11:931–952. doi: 10.5664/jcsm.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. Order. [DOI] [PubMed] [Google Scholar]


