Abstract
Fibroblasts and lymphoblastoid cell lines (LCLs) derived from individuals with spinal muscular atrophy (SMA) have been and continue to be essential for translational SMA research. Authentication of cell lines helps ensure reproducibility and rigor in biomedical research. This quality control measure identifies mislabeling or cross-contamination of cell lines and prevents misinterpretation of data. Unfortunately, authentication of SMA cell lines used in various studies has not been possible because of a lack of a reference. In this study, we provide said reference so that SMA cell lines can be subsequently authenticated. We use short tandem repeat (STR) profiling and digital PCR (dPCR), which quantifies SMN1 and SMN2 copy numbers, to generate molecular identity codes for fibroblasts and LCLs that are commonly used in SMA research. Using these molecular identity codes, we clarify the familial relationships within a set of fibroblasts commonly used in SMA research. This study presents the first cell line reference set for the SMA research community and demonstrates its usefulness for re-identification and authentication of lines commonly used as in vitro models for future studies.
Keywords: spinal muscular atrophy, cell line authentication, short tandem repeat profiling, digital PCR, SMN1, SMN2
1. INTRODUCTION
Translational research requires the use of cell culture models of disease which are established from tissue samples from patients. Disease-specific human cell lines must be correctly identified so as to enhance reproducibility and experimental rigor. Misidentification of cell lines can have a major negative impact on translational research [1–3]. Up to 15% of human cell lines are believed to be either misidentified or contaminated with other cell lines [4;5]. Mislabeling, cross-contamination by another cell line or genetic changes caused by repeated passaging can all lead to cell line misidentification [6]. Cell line authentication, which is meant to prevent incorrect interpretation of data generated from misidentified cell lines, is becoming a requirement for federal funding and for publication. We must have a molecular identity code for a given cell line in order to authenticate it.
Short tandem repeat (STR) profiling is one of the most commonly used techniques for generating a specific molecular identity code [7–10]. This assay amplifies a set of polymorphic microsatellite, or STR, markers and then resolves the PCR products by capillary electrophoresis size fractionation [11]. STR profiling has strong discriminatory power even with degraded DNA samples and is able to resolve mixtures of DNA [12]. Furthermore, the data generated is not dependent on one specific testing platform making this assay very powerful for forensic analysis—its original purpose—and for cell line identification or authentication.
While most of the effort in cell line authentication has focused on cancer, identification and authentication of cell lines derived from patients with rare monogenic disorders are equally important to standardize, especially since these biospecimens being so rare are usually shared across many laboratories. One such disorder is spinal muscular atrophy (SMA), an autosomal recessive motor neuron disease that is a leading genetic cause of infant mortality [13;14]. However, as is the case for many neurodegenerative disorders, SMA has a wide range of phenotypes. SMA is caused by the loss of SMN1 (survival motor neuron 1) but retention of the duplicate gene SMN2. The clinical severity of SMA is inversely related to the number of SMN2 copies, i.e. patients with higher SMN2 copy numbers have a less severe clinical presentation of this disease (reviewed in [15]). Many important advances in understanding the molecular pathology of SMA and the impact of SMN2 dosage on severity have been validated using patient-derived cell lines [16–22]. SMA fibroblast lines have been converted into induced pluripotent stem cell lines subsequently used to characterize human-derived SMN1-deleted neural cells in culture [23–26]. Drug discovery efforts to identify small molecule inducers of SMN2 expression and modulators of alternative splicing have relied on the use of SMA patient cell lines [27]. In fact, the discovery of nusinersen (Spinraza, ISIS-SMNRx or ISS-N1), the first FDA-approved SMA drug, was made using SMA fibroblast lines [28].
Due to their importance as in vitro disease models for SMA, it is essential to authenticate patient-derived cell lines in order to ensure reliability and reproducibility. Unfortunately, STR profiles for the commonly used SMA cell lines are not currently available and thus cell line authentication is not possible. Furthermore, accurate copy number determination of SMN2—as measured with array digital PCR (dPCR) [21]—has not yet been reported for specific SMA cell lines. In this study, we generate molecular identity codes based on STR profiling and dPCR copy number measurements for SMA fibroblasts and lymphoblastoid cell lines (LCLs) that are commonly used in SMA research. The set of unique molecular identity codes can serve as a reference to re-identify or authenticate cell lines in future studies which will enhance standards for rigor and reproducibility.
2. MATERIAL AND METHODS
2.1 Cell Lines
Those fibroblast and LCLs cell lines with GM or UMB prefixes were purchased from Coriell Cell Repositories (Camden, NJ) or the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland (Baltimore, MD). The fibroblast lines with a TC prefix were obtained from Johns Hopkins University (Baltimore, MD), those with a KS prefix from the University of Utah (Salt Lake City, UT) and those with an AIDHC prefix from Nemours/Alfred I. duPont Hospital for Children (Wilmington, DE). The fibroblast lines generated at Nemours were either obtained from the Molecular Diagnostics Laboratory or established in the Motor Neuron Diseases Research Laboratory using standard procedures [29].
2.2 Ethics Statement
For those cell lines obtained from non-commercial sources, biospecimens were obtained after written consent or assent and parental permission. This study was approved by the specific Institutional Review Boards. These cell lines were de-identified so that no protected health information is known for these lines. Available information regarding phenotype and genotype is generally limited to gender, SMN1 deletion/mutation status, SMN2 copy number and SMA type.
2.3 Cell Line Maintenance
In this study, we used 24 type I SMA, 23 type II SMA, 11 type III SMA and 26 healthy control fibroblasts and LCLs. The lowest passage number stock vial was used for each cell line. All cell lines were maintained in a humidified 37°C incubator with 5% CO2. Fibroblasts were maintained in Dulbecco’s modified essential medium (DMEM; Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (EquaFETAL; Atlas Biologicals, Fort Collins, CO), 2 mM L-glutamine (Life Technologies) and 1% penicillin-streptomycin (Life Technologies). LCLs were maintained in RPMI-1640 medium (Life Technologies) supplemented with 15% EquaFETAL, 2 mM L-glutamine and 1% penicillin-streptomycin. Cell pellets were collected from each line within 3 passages.
2.4 Genomic DNA Isolation
Genomic DNA was isolated from cell pellets using the Gentra Puregene Cell Kit (QIAGEN, Germantown, MD) as described previously [21]. Yield was measured with a ND-2000C NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Double-stranded DNA concentrations were measured using the Qubit dsDNA Broad Range Assay (Life Technologies) as recommended by the manufacturer.
2.5 Digital PCR (dPCR)
SMN1 and SMN2 copy numbers for each sample were determined using the QuantStudio 3D Digital PCR System (Life Technologies) as described previously [21]. SMN1 and SMN2 copy numbers were normalized against those for RPPH1, as its copy number does not vary in the human population [30]. The testers were blinded to the subject identities of the cell lines until after analysis.
2.6 Short Tandem Repeat (STR) Profiling
2 ng gDNA were processed for STR profiling using the AmpFISTR Identifiler PCR Amplification kit (Life Technologies) according to the manufacturer’s direction. This kit includes 15 polymorphic microsatellite markers and the XY chromosome-specific marker amelogenin (AMEL). After PCR amplification, the samples were analyzed on the ABI 3130xl Genetic Analyzer (Life Technologies) using the GeneMapper v5.0 software (Life Technologies). The testers were blinded to the subject identities of the cell lines until after analysis.
2.7 Comparison of STR Profiles
To compare the STR profiles of two unrelated samples, an identity score based on the Tanabe algorithm [31]—also known as the Sørensen similarity index—was calculated using the following equation [10;32]:
Two samples are considered identical if their identity score is greater than or equal to 0.95 and are considered to be unrelated if their identity score is less than 0.50.
3. RESULTS
3.1 Cell Line Identification of SMA Fibroblasts and LCLs
To provide a reference standard for cell line authentication of cell lines commonly used in SMA research, we genetically identified 58 SMA and 26 control cell lines—fibroblasts and LCLs—using array dPCR and STR profiling. This collection of cell lines originated from five different sources. SMN1 and SMN2 copy numbers are shown for type I, type II and type III SMA cell lines as well as for healthy controls in Tables 1, 2, 3 and 4, respectively. The SMN1 and SMN2 copy numbers for those cell lines from Coriell Cell Repositories have recently been reported elsewhere [33]. All SMA cell lines tested lacked SMN1 except for the type III SMA fibroblast line AIDHC-SP31 which contains an intragenic missense mutation (SMN1c.38C>G; SMN1p.A2G) [21;34]. As mentioned previously [21], there is a strong inverse relationship between SMN2 copy number and SMA disease severity.
Table 1.
SMN1 and SMN2 Copy Numbers as well as Core STR Profiles for Type I SMA Cell Lines
| copy number | marker alleles | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | Type | SMN1 | SMN2 | AMEL | CSF1PO | D7S820 | D13S317 | D16S539 | TH01 | D5S818 | TPOX | vWA |
| GM00232 | Fib | 0 | 2 | X,Y | 10,12 | 10,11 | 11,12 | 11,13 | 8,9.3 | 13,13 | 8,11 | 15,17 |
| GM10684* | LCL | 0 | 2 | X,X | 10,13 | 9,12 | 8,11 | 11,12 | 11,13 | 7,7 | 8,8 | 17,18 |
| GM23686* | LCL | 0 | 2 | X,X | 10,12 | 10,11 | 10,14 | 11,12 | 7,9 | 11,11 | 8,11 | 15,17 |
| GM23389* | LCL | 0 | 3 | X,Y | 7,10 | 11,12 | 9,9 | 10,12 | 6,6 | 11,13 | 11,12 | 14,15 |
| UMB-1897 | Fib | 0 | 2 | X,Y | 11,12 | 11,11 | 10,13 | 13,13 | 8,9.3 | 12,12 | 11,12 | 16,20 |
| UMB-4648 | Fib | 0 | 2 | X,Y | 11,12 | 10,11 | 8,12 | 12,12 | 9.3,9.3 | 12,13 | 8,8 | 15,17 |
| UMB-4994 | Fib | 0 | 2 | X,X | 11,12 | 11,13 | 9,13 | 11,13 | 9,9.3 | 12,12 | 8,9 | 14,19 |
| AIDHC-SP33 | Fib | 0 | 2 | X,Y | 9,10 | 10,12 | 11,14 | 11,11 | 7,8 | 10,11 | 8,10 | 14,18 |
| AIDHC-SP34 | Fib | 0 | 2 | X,X | 10,11 | 10,11 | 9,14 | 8,13 | 6,9.3 | 11,12 | 8,10 | 16,17 |
| AIDHC-SP35 | Fib | 0 | 2 | X,X | 11,12 | 9,10 | 12,12 | 11,13 | 7,9.3 | 11,12 | 8,11 | 14,18 |
| TC95-2711 | Fib | 0 | 2 | X,X | 11,11 | 10,10 | 11,12 | 11,12 | 7,7 | 9,12 | 8,9 | 16,18 |
| TC95-2806 | Fib | 0 | 2 | X,Y | 10,10 | 9,10 | 12,13 | 13,13 | 6,6 | 12,14 | 8,10 | 17,18 |
| TC95-2817 | Fib | 0 | 2 | X,X | 10,12 | 8,10 | 9,11 | 11,13 | 6,6 | 12,13 | 8,8 | 17,17 |
| TC97-2923 | Fib | 0 | 3 | X,Y | 12,12 | 10,11 | 12,13 | 11,12 | 6,9.3 | 11,12 | 9,11 | 15,16 |
| TC97-2932 | Fib | 0 | 2 | X,Y | 10,11 | 9,12 | 9,11 | 11,11 | 7,7 | 12,13 | 8,8 | 14,15 |
| TC97-2947 | Fib | 0 | 2 | X,X | 10,11 | 10,11 | 9,13 | 8,13 | 9,9.3 | 10,11 | 8,8 | 18,18 |
| KS-0007 | Fib | 0 | 2 | X,X | 10,11 | 11,13 | 10,13 | 11,12 | 6,7 | 12,12 | 8,8 | 14,17 |
| KS-0187 | Fib | 0 | 2 | X,X | 10,11 | 8,11 | 10,13 | 11,12 | 7,9.3 | 10,11 | 8,9 | 17,17 |
| KS-0195 | Fib | 0 | 2 | X,Y | 10,11 | 11,13 | 9,10 | 9,12 | 7,9 | 11,14 | 8,8 | 14,15 |
| KS-0196 | Fib | 0 | 2 | X,X | 11,12 | 10,12 | 11,13 | 11,11 | 6,9 | 12,13 | 8,12 | 17,18 |
| KS-0206 | Fib | 0 | 2 | X,Y | 11,12 | 8,10 | 10,11 | 12,12 | 6,9.3 | 10,12 | 9,11 | 14,18 |
| KS-0217 | Fib | 0 | 2 | X,Y | 11,12 | 11,12 | 8,12 | 11,12 | 9,9.3 | 12,13 | 8,8 | 16,17 |
| KS-0251 | Fib | 0 | 2 | X,Y | 11,12 | 6.3,8 | 12,13 | 11,12 | 8,9.3 | 9,11 | 11,11 | 16,17 |
| KS-0315 | Fib | 0 | 2 | X,Y | 10,10 | 8,12 | 9,14 | 11,13 | 9.3,9.3 | 11,11 | 8,8 | 16,17 |
The SMN1 and SMN2 copy numbers for type I SMA fibroblast (Fib) and lymphoblastoid cell lines (LCLs) were measured using array digital PCR. The alleles for the core STR profiles are listed for each cell line. The asterisk (*) denotes those cell lines presumed to be derived from type I SMA patients.
Table 2.
SMN1 and SMN2 Copy Numbers as well as Core STR Profiles for Type II SMA Cell Lines
| copy number | marker alleles | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | Type | SMN1 | SMN2 | AMEL | CSF1PO | D7S820 | D13S317 | D16S539 | TH01 | D5S818 | TPOX | vWA |
| GM03813 | Fib | 0 | 3 | X,Y | 9,10 | 9,10 | 9,11 | 11,12 | 6,7 | 11,13 | 8,8 | 16,17 |
| GM22592 | Fib | 0 | 3 | X,Y | 8,10 | 9,10 | 8,14 | 12,13 | 9,9 | 11,14 | 8,11 | 16,16 |
| AIDHC-SP4 | Fib | 0 | 3 | X,X | 12,12 | 9,9 | 10,14 | 11,12 | 8,9 | 11,12 | 8,8 | 18,19 |
| AIDHC-SP5 | Fib | 0 | 3 | X,X | 11,12 | 11,12 | 11,12 | 9,11 | 6,9 | 11,11 | 8,11 | 17,19 |
| AIDHC-SP22 | Fib | 0 | 3 | X,Y | 11,13 | 11,12 | 8,12 | 11,12 | 6,9 | 10,11 | 8,9 | 16,18 |
| AIDHC-SP23 | Fib | 0 | 3 | X,Y | 11,12 | 10,11 | 11,14 | 9,11 | 9.3,10 | 12,13 | 8,8 | 19,20 |
| AIDHC-SP25 | Fib | 0 | 3 | X,Y | 11,12 | 8,10 | 8,10 | 9,11 | 6,6 | 10,11 | 8,8 | 16,18 |
| AIDHC-SP26 | Fib | 0 | 3 | X,Y | 10,11 | 8,8 | 11,12 | 12,13 | 9,9.3 | 12,13 | 8,8 | 16,18 |
| AIDHC-SP32 | Fib | 0 | 3 | X,Y | 11,12 | 10,12 | 12,12 | 11,15 | 9,9.3 | 12,12 | 8,10 | 15,17 |
| AIDHC-13303 (+DS) | Fib | 0 | 4 | X,Y | 10,12 | 11,11 | 8,12 | 9,12 | 7,9 | 12,13 | 8,8 | 15,16 |
| AIDHC-13010* | Fib | 0 | 3 | X,X | 10,11 | 10,11 | 9,12 | 11,13 | 9.3,9.3 | 11,12 | 8,11 | 18,18 |
| AIDHC-13148 | Fib | 0 | 3 | X,Y | 12,12 | 10,12 | 11,14 | 10,11 | 8,9.3 | 12,13 | 9,9 | 16,18 |
| AIDHC-13822 | Fib | 0 | 3 | X,Y | 11,12 | 11,12 | 12,13 | 11,12 | 6,6 | 11,12 | 11,11 | 17,18 |
| AIDHC-15224* | Fib | 0 | 3 | X,X | 10,11 | 10,11 | 9,12 | 11,13 | 9.3,9.3 | 11,12 | 8,11 | 18,18 |
| TC96-2824 | Fib | 0 | 3 | X,Y | 11,12 | 9,12 | 11,11 | 11,11 | 7,9 | 12,12 | 6,11 | 15,16 |
| TC96-2827 | Fib | 0 | 3 | X,Y | 10,12 | 10,11 | 8,11 | 11,12 | 9.3,9.3 | 9,11 | 8,8 | 16,18 |
| TC96-2831 | Fib | 0 | 3 | X,X | 12,12 | 11,12 | 8,11 | 13,14 | 6,9 | 7,11 | 8,9 | 18,18 |
| TC96-2832 | Fib | 0 | 3 | X,Y | 12,12 | 10,11 | 11,13 | 11,12 | 9,9 | 11,11 | 8,11 | 16,17 |
| TC96-2837 | Fib | 0 | 3 | X,Y | 10,10 | 11,13 | 12,14 | 10,11 | 7,9.3 | 12,13 | 8,11 | 16,18 |
| TC96-2860 | Fib | 0 | 2 | X,X | 9,10 | 8,11 | 12,12 | 9,12 | 8,9.3 | 12,12 | 11,12 | 15,18 |
| TC96-2865 | Fib | 0 | 3 | X,X | 11,12 | 9,10 | 12,12 | 13,14 | 8,9 | 11,13 | 8,9 | 16,16 |
| TC97-2922 | Fib | 0 | 3 | X,X | 10,10 | 8,12 | 11,11 | 11,12 | 6,9 | 12,12 | 11,12 | 18,19 |
| KS-0151 | Fib | 0 | 3 | X,Y | 10,11 | 9,10 | 11,11 | 9,10 | 7,9.3 | 11,12 | 8,8 | 16,16 |
| KS-0303 | Fib | 0 | 3 | X,X | 11,12 | 12,12 | 10,12 | 9,12 | 7,9.3 | 12,12 | 8,11 | 16,18 |
The SMN1 and SMN2 copy numbers for type II SMA fibroblast (Fib) cell lines were measured using array digital PCR. The alleles for the core STR profiles are listed for each cell line. AIDHC-13303 was derived from an individual with type II SMA and Down syndrome (DS). The asterisk (*) denotes those cell lines which were derived from the same type II SMA patient.
Table 3.
SMN1 and SMN2 Copy Numbers as well as Core STR Profiles for Type III SMA Cell Lines
| copy number | marker alleles | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | Type | SMN1 | SMN2 | AMEL | CSF1PO | D7S820 | D13S317 | D16S539 | TH01 | D5S818 | TPOX | vWA |
| GM23255 | LCL | 0 | 3 | X,Y | 9,12 | 11,12 | 8,11 | 9,11 | 6,9 | 10,13 | 8,11 | 14,14 |
| AIDHC-SP24 | Fib | 0 | 4 | X,Y | 11,11 | 8,12 | 9,11 | 9,12 | 6,8 | 11,12 | 8,8 | 15,17 |
| AIDHC-SP27 | Fib | 0 | 4 | X,Y | 11,12 | 8,12 | 6,12 | 11,12 | 7,9 | 11,12 | 10,11 | 16,20 |
| AIDHC-SP31 | Fib | 1* | 1 | X,Y | 8,11 | 10,11 | 11,11 | 11,11 | 7,8 | 12,12 | 8,8 | 15,18 |
| AIDHC-00138 | Fib | 0 | 4 | X,Y | 9,12 | 9,10 | 6,12 | 9,11 | 6,9 | 11,12 | 8,11 | 16,19 |
| AIDHC-13219 | Fib | 0 | 4 | X,X | 11,11 | 8,12 | 8,9 | 9,11 | 6,8 | 11,11 | 8,8 | 15,17 |
| TC96-2873 | Fib | 0 | 3 | X,X | 11,12 | 8,9 | 11,14 | 10,11 | 6,6 | 11,12 | 11,11 | 17,18 |
| TC96-2906 | Fib | 0 | 4 | X,Y | 10,12 | 9,10 | 10,12 | 11,13 | 7,9.3 | 11,12 | 10,12 | 16,17 |
| KS-0064 | Fib | 0 | 4 | X,Y | 11,11 | 8,11 | 10,12 | 11,12 | 6,7 | 11,12 | 8,9 | 17,17 |
| KS-0123 | Fib | 0 | 4 | X,Y | 11,11 | 8,11 | 10,13 | 11,13 | 6,9.3 | 12,12 | 8,8 | 17,17 |
| KS-0330 | Fib | 0 | 3 | X,Y | 11,11 | 11,12 | 11,12 | 12,12 | 6,9.3 | 11,12 | 11,11 | 18,18 |
The SMN1 and SMN2 copy numbers for type III SMA fibroblast (Fib) and lymphoblastoid cell lines (LCLs) were measured using array digital PCR. The alleles for the core STR profiles are listed for each cell line.
AIDHC-SP31 contains an intragenic missense mutation on SMN1 (c.38C>G; SMN1p.A2G).
Table 4.
SMN1 and SMN2 Copy Numbers as well as Core STR Profiles for Control Cell Lines Commonly Used in SMA Research
| copy number | marker alleles | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | Type | SMN1 | SMN2 | AMEL | CSF1PO | D7S820 | D13S317 | D16S539 | TH01 | D5S818 | TPOX | vWA |
| GM00409 | Fib | 3 | 1 | X,Y | 10,13 | 8,10 | 11,11 | 11,13 | 8,9 | 11,11 | 11,11 | 15,17 |
| GM00498 | Fib | 2 | 2 | X,Y | 10,11 | 8,8 | 11,12 | 8,12 | 9.3,9.3 | 11,12 | 8,11 | 19,19 |
| GM02036 | Fib | 2 | 1 | X,X | 12,13 | 8,11 | 8,12 | 9,12 | 6,6 | 12,13 | 9,11 | 17,19 |
| GM03814 | Fib | 1 | 5 | X,X | 9,10 | 10,10 | 11,12 | 11,11 | 6,7 | 12,13 | 8,11 | 14,16 |
| GM03815 | Fib | 1 | 1 | X,Y | 10,12 | 9,10 | 9,11 | 11,12 | 6,9.3 | 11,12 | 8,12 | 14,17 |
| GM23687 | LCL | 1 | 2 | X,X | 10,11 | 10,12 | 9,14 | 11,11 | 7,9 | 11,12 | 8,8 | 17,19 |
| GM23688 | LCL | 1 | 2 | X,Y | 10,12 | 10,11 | 10,11 | 9,12 | 6,9 | 7,11 | 8,11 | 15,16 |
| AIDHC-SC1 | Fib | 2 | 2 | X,Y | 11,12 | 11,12 | 11,12 | 12,12 | 7,8 | 11,12 | 11,11 | 14,15 |
| AIDHC-SC2 | Fib | 2 | 2 | X,Y | 9,10 | 8,11 | 11,11 | 9,12 | 6,9 | 10,11 | 10,10 | 15,16 |
| AIDHC-NMC1 | Fib | 2 | 2 | X,Y | 10,12 | 10,11 | 11,12 | 11,12 | 6,9 | 12,12 | 8,11 | 16,17 |
| AIDHC-NMC2 | Fib | 1 | 3 | X,Y | 10,10 | 12,12 | 10,11 | 10,12 | 6,7 | 11,11 | 8,8 | 14,18 |
| AIDHC-NMC3 | Fib | 2 | 1 | X,Y | 9,11 | 9,11 | 9,11 | 10,11 | 6,7 | 12,13 | 8,10 | 14,18 |
| AIDHC-NMC5 | Fib | 2 | 3 | X,Y | 11,12 | 9,11 | 8,13 | 11,13 | 8,8 | 11,11 | 11,11 | 12,18 |
| AIDHC-NMC7 | Fib | 2 | 0 | X,Y | 11,12 | 10,10 | 11,11 | 8,13 | 6,8 | 11,11 | 11,12 | 14,15 |
| AIDHC-NMC8 | Fib | 3 | 2 | X,Y | 11,11 | 10,11 | 11,12 | 11,12 | 9.3,9.3 | 11,11 | 8,9 | 18,19 |
| AIDHC-NFC1 | Fib | 2 | 2 | X,X | 10,11 | 10,11 | 10,11 | 9,9 | 9.3,9.3 | 12,13 | 8,8 | 17,17 |
| AIDHC-NFC2 | Fib | 2 | 2 | X,X | 11,11 | 9,10 | 11,11 | 12,13 | 9.3,10 | 11,14 | 8,8 | 16,17 |
| AIDHC-NFC3 | Fib | 2 | 2 | X,X | 11,13 | 10,13 | 11,11 | 11,12 | 6,9.3 | 12,12 | 8,9 | 18,18 |
| AIDHC-NFC4 | Fib | 2 | 2 | X,X | 12,12 | 11,13 | 9,15 | 8,9 | 6,6 | 11,13 | 11,11 | 15,17 |
| AIDHC-NFC5 | Fib | 2 | 2 | X,X | 11,11 | 9,11 | 12,13 | 11,12 | 6,9.3 | 11,12 | 8,8 | 14,17 |
| AIDHC-NFC6 | Fib | 2 | 2 | X,X | 11,12 | 11,12 | 12,13 | 9,13 | 9,9.3 | 11,12 | 8,8 | 16,18 |
| AIDHC-NFC7 | Fib | 2 | 2 | X,X | 12,12 | 10,12 | 12,14 | 11,12 | 9.3,9.3 | 11,13 | 8,11 | 15,17 |
| AIDHC-NFC8 | Fib | 2 | 3 | X,X | 11,13 | 7,9 | 10,13 | 11,13 | 6,9.3 | 9,9 | 11,11 | 18,19 |
| AIDHC-14338 | Fib | 2 | 2 | X,Y | 10,11 | 8,12 | 9,11 | 9,11 | 7,9 | 11,11 | 8,11 | 17,19 |
| AIDHC-14697 | Fib | 2 | 1 | X,X | 10,10 | 12,12 | 9,11 | 12,12 | 7,9.3 | 11,13 | 9,11 | 15,15 |
| AIDHC-14531 | Fib | 6 | 2 | X,X | 7,11 | 10,11 | 8,12 | 9,12 | 7,9 | 10,12 | 9,9 | 16,16 |
The SMN1 and SMN2 copy numbers for healthy control fibroblast (Fib) and lymphoblastoid cell lines (LCLs) were measured using array digital PCR. The alleles for the core STR profiles are listed for each cell line.
We analyzed the STR profiles of all of the samples using 16 different loci. To protect the identity of the subjects, 9 microsatellite markers at different loci, hereafter referred to as the core loci, were used to report on the identity of a given sample [32]. These core loci are AMEL (X:p22.1-22.3; Y:p11.2), CSF1PO (5q33.3-34), D7S820 (7q11.21-22), D13S317 (13q22-31), D16S539 (16q24-qter), TH01 (11p15.5), D5S818 (5q21-31), TPOX (2p23-2pter) and vWA (12p12-pter). The core STR profiles for type I, type II and type III SMA cell lines as well as for healthy controls are provided as a reference for cell line authentication in Tables 1, 2, 3 and 4.
Using all 16 markers, the STR profiles revealed that each cell line is derived from a unique individual, except for AIDHC-13010 and AIDHC-15224. Comparison of the STR profiles of these two lines yielded an identity score of 1.00 showing that AIDHC-13010 and AIDHC-15224 arose from the same subject. Upon unblinding of the data, it was revealed that indeed these two fibroblast lines are derived from the same type II SMA patient but were established from separate skin samples collected at different times.
3.2 Using STR Profiling to Determine Familial Relationships Between SMA Cell Lines
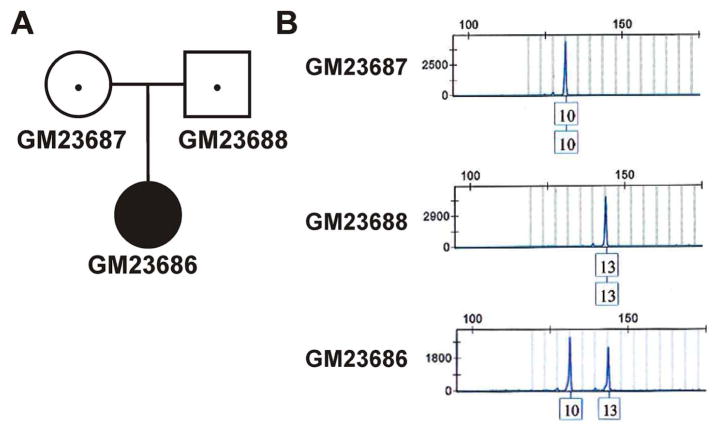
STR profiling can be used to determine the filial relationship between cell lines within a family. To show a familial relationship, each marker locus of the child will share one allele with each parent. Within the NIGMS Collection, there is one SMA family (#3042; Figure 1A) where the affected, presumably with type I SMA, child (GM23686) and her parents (GM23687 and GM23688) are represented as LCLs. GM23686, GM23687 and GM23688 cells have SMN1:SMN2 copy numbers of 0:2, 1:2 and 1:2, respectively (Tables 1 and 4) confirming that GM23686 has SMA (loss of SMN1 and at least 1 copy of SMN2) and that GM23687 and GM23688 are carriers (1 copy of SMN1). GM23686, as expected, shared an allele for each of the 16 marker loci with both GM23687 and GM23688. GM23687 and GM23688 are not related based on their identity score (0.32). Allelic sharing between the parents and the affected child is shown at the D8S1179 (Figure 1B), as a representative example of the allele sharing observed at all 16 loci.
Figure 1. Identification of familial relationships within family #3042 of the NIGMS Cell Collection using the D8S1179 microsatellite marker.
(A) Pedigree analysis of family #3042. GM23686 was diagnosed with type I SMA. (B) Electrophoretograms showing the D8S1179 alleles for GM23686, GM23687 and GM23688.
3.3 Using Array Digital PCR and STR Profiling to Verify Identity of SMA Cell Lines
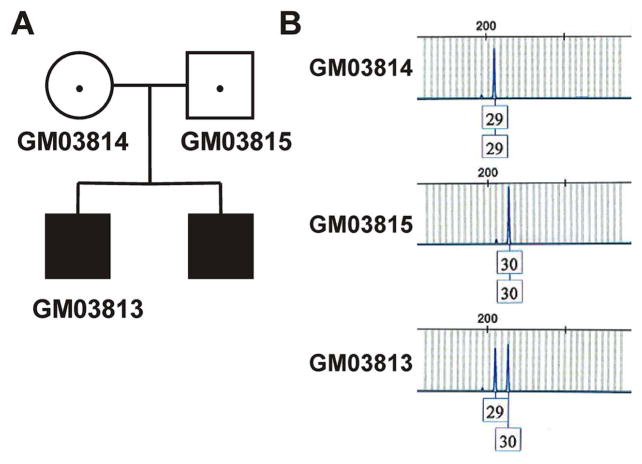
While fibroblasts derived from family #553 in the NIGMS Collection are some of the most widely used cell culture models for SMA (Figure 2A), there has been some controversy about the identity of some of the cell models originating from this family. GM03813 fibroblasts were derived from a type II male SMA patient and GM03814 and GM03815 originate from his carrier parents [35]. However, Wan and colleagues [36] concluded, based on their snRNP assembly data, that GM03815 was in fact derived from the affected brother of GM03813 rather than his father.
Figure 2. Identification of familial relationships within family #553 of the NIGMS Cell Collection using the D21S11 microsatellite marker.
(A) Pedigree analysis of family #553. GM03813 was diagnosed with type II SMA and had a brother with the same SMA phenotype. (B) Electrophoretograms showing the D21S11 alleles for GM03813, GM03814 and GM03815.
To resolve this reported discrepancy, we used array digital PCR as well as STR profiling to clarify the filial relationships between these cell lines. GM03813, GM03814 and GM03815 cells have SMN1:SMN2 copy numbers of 0:3, 1:5 and 1:1, respectively (Tables 2 and 4). Copy number measurements rule out mislabeling of the cell lines and demonstrate that GM03813 is indeed the affected individual and GM03815 is a carrier. The full STR profiles demonstrate that the parental cell lines are from different, unrelated individuals (identity score = 0.42) thereby ruling out GM03815 being the child of GM03814. GM03813 shared an allele for each marker locus with both GM03814 and GM03815 proving that he is the child of these parental cell lines. As an example, the inheritance pattern of the D21S11 marker is shown in Figure 2B. Based on these results, GM03815 is the carrier father of GM03813 and not its sibling as reported in Wan et al. [36].
4. DISCUSSION
Human cell lines have driven biomedical research by serving as cell culture models for disease. Misidentification of cell lines by mislabeling or cross-contamination is problematic and costly for researchers [2]. It is, therefore, essential to authenticate these cell lines prior to starting any experimental studies so as to ensure reproducibility and rigor. In fact, the National Institutes of Health Office of Extramural Research is now requiring authentication of cell lines used in biomedical research as part of their efforts to enhance reproducibility ([37]; Notice Number: NOT-OD-15-103). Cancer cell lines have received the most attention with respect to cell line authentication [4] but it is equally important to authenticate cell lines derived from individuals with rare monogenic disorders. Using dPCR and STR profiling, we generated unique molecular identity codes for SMA fibroblasts and LCLs that are commonly used in this field of research.
STR profiling is the most widely used assay for cell line identification and authentication. In fact, it is the standard tool used by the American National Standards Institute (ANSI) and the American Type Culture Collection (ATCC) [12]. Single nucleotide polymorphism (SNP) profiling has also been used for cell line authentication [38;39]. When comparing these two approaches, Yu et al. [10] observe higher identity scores between two cell lines using SNP profiling when compared against STR profiling. The differences observed between these assays are likely due to the greater number of potentially informative sites using SNP profiling. While microsatellite instability may also more adversely affect STR analysis than SNP analysis [40], none of the STR markers used in this study are prone to microsatellite instability. Additionally, STR profiling is less costly to run than SNP arrays which is a primary reason for STR profiling being the standard tool for cell line authentication.
SMN2 is an endogenous genetic modifier of SMA disease severity in that higher SMN2 copy numbers are found in SMA patients with milder clinical phenotypes [15]. Because of this inverse relationship, SMN2 copy number is being used as a criterion for inclusion in SMA clinical trials. dPCR can accurately measure copy number variations found in many pediatric-onset disorders including SMA [41]. We and others have demonstrated that dPCR provides an accurate and reliable measurement of SMN2 copy number [21;42]. Because of the importance of SMN2 in translational and clinical SMA research, its copy number needs to be accurately measured in cell culture models for SMA. For this reason, we included SMN2 copy number—as well as SMN1 for our non-SMA control lines—as part of our molecular identity code for each cell line.
In addition to ensuring correct molecular identity of a cell line, it is important to have the correct clinical information about this line. There are currently no molecular markers that can definitively identify the clinical severity of SMA although there is a very strong, but not absolute, inverse relationship between SMN2 copy number and clinical severity [15]. As a result, clinical severity information must be provided by the depositor of the cell line in question. This information needs to also be correctly curated by the depositor and anyone using this cell line. For example, GM03813 SMA fibroblasts have been used by many groups as a cell culture model for SMA. Unfortunately, this cell line has been mislabeled as a type I SMA fibroblast in the vendor’s database and this incorrect identification has been reflected in multiple studies, including [23;43]. GM03813 cells were actually derived from a three year-old male diagnosed with type II SMA [35] and our SMN2 copy number analysis performed using dPCR corroborate the clinical findings by revealing a higher SMN2 copy number than expected of a patient with type I SMA. In addition, both STR profiling and dPCR analysis of GM03813 and GM03815 cell lines helped resolve a controversy regarding the familial relationship between these two lines [36], i.e. GM03815 is the carrier father of, as opposed to the affected brother of, GM03813. We propose that when working with related SMA cell lines, they be correctly authenticated by STR profiling and dPCR as well as correctly identified with respect to their curated clinical severities.
In summary, we have developed a two-step approach to generate molecular identity codes for human fibroblast and LCLs used in SMA research. This approach uses dPCR to accurately measure SMN1 and SMN2 copy numbers and STR profiling to provide a unique molecular identity code for a human cell line. The molecular identity codes generated by this cell line identification approach will serve as a tool to ensure experimental rigor in SMA research.
HIGHLIGHTS.
SMA cell line authentification is essential for rigor and reproducibility
STR profiling and dPCR can generate molecular identity codes for SMA cell lines
GM03815 is the father of, as opposed to the brother of, GM03813
Acknowledgments
We would like to thank Dr. Vicky Funanage, Susan Kirwin and Priscilla Moses of the Molecular Diagnostics Laboratory for providing some of the fibroblast lines from their archive, Dr. Diana Corao-Uribe and Janell Raber of the Nemours Biobank for obtaining skin biopsies for some of the fibroblast lines and Dr. Ron Zielke and John Cottrell of the NICHD Brain and Tissue Bank for Neurodevelopmental Disorders at the University of Maryland for providing some of the fibroblast lines. The fibroblast lines generated for Johns Hopkins University were obtained from the Intellectual and Developmental Disabilities Research Center at Kennedy Krieger Institute (Baltimore, MD).
This study was supported by funds from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) Institutional Development Award (IDeA) Centers of Biomedical Research Excellence [grant numbers P20GM103464 and P30GM114736 to MERB and KSC], IDeA Networks of Biomedical Research Excellence [grant number P20GM103446 to MERB and KSC] and the Nemours Foundation. Cell lines from SMA patients contributed by KJS were supported by funds from the NIH [grant number R01HD054599]. Cell lines from SMA patients contributed by TOC were supported by funds from the NIH [U54HD079123-01A1].
ABBREVIATIONS
- dPCR
digital PCR
- LCL
lymphoblastoid cell line
- SMA
spinal muscular atrophy
- SMN1
survival motor neuron 1
- SMN2
survival motor neuron 2
- SNP
single nucleotide polymorphism
- STR
short tandem repeat
Footnotes
AUTHORS CONFLICTS OF INTEREST
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441–8. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 2.Freedman LP, Gibson MC, Ethier SP, Soule HR, Neve RM, Reid YA. Reproducibility: changing the policies and culture of cell line authentication. Nat Methods. 2015;12:493–7. doi: 10.1038/nmeth.3403. [DOI] [PubMed] [Google Scholar]
- 3.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: good news and bad news. Sci Transl Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 4.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RAF, et al. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 5.Masters JR. End the scandal of false cell lines. Nature. 2012;492:186. doi: 10.1038/492186a. [DOI] [PubMed] [Google Scholar]
- 6.Reid YA. Characterization and authentication of cancer cell lines: an overview. Methods Mol Biol. 2011;731:35–43. doi: 10.1007/978-1-61779-080-5_4. [DOI] [PubMed] [Google Scholar]
- 7.Masters JR, Thomson JA, Daly-Burns B, Reid YA, Dirks WG, Packer P, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci U S A. 2001;98:8012–7. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nims RW, Sykes G, Cottrill K, Ikonomi P, Elmore E. Short tandem repeat profiling: part of an overall strategy for reducing the frequency of cell misidentification. In Vitro Cell Dev Biol Animal. 2010;46:811–9. doi: 10.1007/s11626-010-9352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barallon R, Bauer SR, Butler J, Capes-Davis A, Dirks WG, Elmore E, et al. Recommendation of short tandem profiling for authenticating human cell lines, stem cells and tissues. In Vitro Cell Dev Biol Animal. 2010;46:727–32. doi: 10.1007/s11626-010-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu M, Selvaraj SK, Liang-Chu MMY, Aghajani S, Busse M, Yuan J, et al. A resource for cell line authentication, annotation and quality control. Nature. 2015;520:307–11. doi: 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- 11.Oldroyd NJ, Urquhart AJ, Kimpton CP, Millican ES, Watson SK, Downes T, et al. A highly discriminating octoplex short tandem repeat polymerase chain reaction system suitable for human individual identification. Electrophoresis. 1995;16:334–7. doi: 10.1002/elps.1150160155. [DOI] [PubMed] [Google Scholar]
- 12.Almeida JL, Cole KD, Plant AL. Standards for cell line authentication and beyond. PLoS Biol. 2016;14:e1002476. doi: 10.1371/journal.pbio.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 14.Kolb SJ, Kissel JT. Spinal muscular atrophy. Neurol Clin. 2015;33:831–46. doi: 10.1016/j.ncl.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butchbach MER. Copy number variations in the Survival Motor Neuron genes: implications for spinal muscular atrophy and other neurodegenerative diseases. Front Mol Biosci. 2016;3:7. doi: 10.3389/fmolb.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–65. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 17.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–9. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 18.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–14. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–65. [PMC free article] [PubMed] [Google Scholar]
- 20.Kolb SJ, Gubitz AK, Olszewski RF, Jr, Ottinger E, Sumner CJ, Fischbeck KH, et al. A novel cell immunoassay to measure survival of motor neurons protein in blood cells. BMC Neurol. 2006;6:6. doi: 10.1186/1471-2377-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stabley DL, Harris AW, Holbrook J, Chubbs NJ, Lozo KW, Crawford TO, et al. SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol Genet Genomic Med. 2015;3:248–57. doi: 10.1002/mgg3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crawford TO, Paushkin S, Kobayashi DT, Forrest SJ, Joyce CL, Finkel RS, et al. Evaluation of SMN protein, transcript and copy number in the Biomarkers for Spinal Muscular Atrophy (BforSMA) clinical study. PLoS ONE. 2012;7:e33572. doi: 10.1371/journal.pone.0033572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–80. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGivern JV, Patitucci TN, Nord JA, Barabas MEA, Stucky CL, Ebert AD. Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia. 2013;61:1418–28. doi: 10.1002/glia.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwab AJ, Ebert AD. Sensory neurons do not induce motor neuron loss in a human stem cell model of spinal muscular atrophy. PLoS ONE. 2014;9:e103112. doi: 10.1371/journal.pone.0103112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sareen D, Ebert AD, Heins BM, McGivern JV, Ornelas L, Svendsen CN. Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS ONE. 2012;7:e39113. doi: 10.1371/journal.pone.0039113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherry JJ, Kobayashi DT, Lynes MM, Naryshkin NN, Tiziano FD, Zaworksi PG, et al. Assays for the identification and prioritization of drug candidates for spinal muscular atrophy. Assay Drug Dev Technol. 2014;12:315–41. doi: 10.1089/adt.2014.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333–46. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villegas J, McPhaul M. Establishment and culture of human skin fibroblasts. Curr Protoc Mol Biol. 2005;(Unit 28.3):28.3.1–28.3.9. doi: 10.1002/0471142727.mb2803s71. [DOI] [PubMed] [Google Scholar]
- 30.Baer M, Nilsen TW, Costigan C, Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990;18:97. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tiss Cult Res Commun. 1999;18:329–38. [Google Scholar]
- 32.Capes-Davis A, Reid YA, Kline MC, Storts DR, Strauss E, Dirks WG, et al. Match criteria for human cell lines authentication: where do we draw the line? Int J Cancer. 2013;132:2510–9. doi: 10.1002/ijc.27931. [DOI] [PubMed] [Google Scholar]
- 33.Butchbach MER. SMN1 and SMN2 copy numbers of commercially available spinal muscular atrophy fibroblast and lymphoblastoid cell lines. In: Sumner CJ, Paushkin S, Ko CP, editors. Spinal Muscular Atrophy: Disease Mechanisms and Therapy. 1. Elsevier; 2017. p. 431. [Google Scholar]
- 34.Wang W, DiMatteo D, Funanage VL, Scavina M. Increased susceptibility of spinal muscular atrophy fibroblasts to camptothecin-induced cell death. Mol Genet Metab. 2005;85:38–45. doi: 10.1016/j.ymgme.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Scudiero DA, Polinsky RJ, Brumback RA, Tarone RE, Nee LE, Robbins JH. Alzheimer disease fibroblasts are hypersensitive to the lethal effects of a DNA-damaging chemical. Mutat Res. 1986;159:125–31. doi: 10.1016/0027-5107(86)90121-1. [DOI] [PubMed] [Google Scholar]
- 36.Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, et al. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–51. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorsch JR, Collins FS, Lippincott-Schwartz J. Fixing problems with cell lines. Science. 2014;346:1452–3. doi: 10.1126/science.1259110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castro R, Dirks WG, Fähnrich S, Hotz-Wagenblatt A, Pawlita M, Schmitt M. High-throughput SNP-based authentication of human cell lines. Int J Cancer. 2013;132:308–14. doi: 10.1002/ijc.27675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang-Chu MMY, Yu M, Haverty PM, Koeman J, Ziegle J, Lee M, et al. Human biosample authentication using the high-throughput, cost-effective SNPtrace system. PLoS ONE. 2015;10:e0116218. doi: 10.1371/journal.pone.0116218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Much M, Buza N, Hui P. Tissue identity testing of cancer by short tandem repeat polymorphism: pitfalls of interpretation in the presence of microsatellite instability. Hum Pathol. 2014;45:549–55. doi: 10.1016/j.humpath.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 41.Butchbach MER. Applicability of digital PCR to the investigation of pediatric-onset genetic disorders. Biomol Detect Quantif. 2016;10:9–14. doi: 10.1016/j.bdq.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong Q, Bhattacharya S, Kotsopoulos S, Olson J, Taly V, Griffiths AD, et al. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip. 2011;11:2167–74. doi: 10.1039/c1lc20126c. [DOI] [PubMed] [Google Scholar]
- 43.Thurmond J, Butchbach MER, Palomo M, Pease B, Rao M, Bedell L, et al. Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy. J Med Chem. 2008;51:449–69. doi: 10.1021/jm061475p. [DOI] [PubMed] [Google Scholar]