Abstract
Breast cancer is the second most common cause of mortality in women; therefore, the identification of novel putative markers is required to improve its diagnosis and prognosis. Selenium is known to protect mammary epithelial cells from oxidative DNA damage, and to inhibit the initiation phase of carcinogenesis by stimulating DNA repair and apoptosis regulation. Consequently, the present study has focused attention on the selenoprotein family and their involvement in breast cancer. The present study performed a global analysis of the seleno-transcriptome expression in human breast cancer MCF-7 and MDA-MB231 cell lines compared with healthy breast MCF-10A cells using reverse transcription-quantitative polymerase chain reaction. The present data revealed the presence of differently expressed genes in MCF-7 and MDA-MB231 cells compared with MCF-10A cells: Four downregulated [glutathione peroxidase (GPX)1, GPX4, GPX5 and GPX7] and three upregulated (deiodinase iodothyronine, type II, GPX2 and GPX3) genes. Additionally, interactomic investigation were performed by the present study to evaluate the association between the downregulated and upregulated genes, and to identify putative HUB nodes, which represent the centers of association between the genes that are capable of direct control over the gene networks. Network analysis revealed that all differentially regulated genes, with the exception of selenoprotein T, are implicated in the same network that presents three HUB nodes interconnected to the selenoprotein mRNAs, including TP53, estrogen receptor 1 and catenin-β1 (CTNNB1). Overall, these data demonstrated for the first time, a profile of seleno-mRNAs specific for human breast cells, indicating that these genes alter their expression on the basis of the ER-positivity or negativity of breast cancer cells.
Keywords: breast cancer, seleno-transcriptome, RT-qPCR, network analysis, HUB genes
Introduction
Human breast cancer is the most common invasive cancer and the second most common cause of mortality in women (1–3). Age, being overweight, early menarche, late menopause, first pregnancy at a late age, the use of postmenopausal hormones, including estrogen and progesterone, and the presence of an inherited mutation in the BRCA1 or BRCA2 breast genes are major risk factors for breast cancer development (4). The growth of breast cancer cells is known to be regulated by estrogen via binding to estrogen receptors (ERs), which induces cell proliferation (5,6) and prevents apoptosis of cells (7,8). Additionally, oxidative stress has been associated with breast cancer development. Oxidative stress may cause DNA damage and induce the overexpression of vascular endothelial growth factor, which is key in tumor angiogenesis and neovascularization (9). Furthermore, evidence suggests that oxidative stress directly affects neoplastic progression and metastasis (9). Consequently, treating oxidative stress in tissues may decrease the metastatic potential of tumors (9).
Selenium (Se) is an important micronutrient that is involved in antioxidant, anti-inflammatory and antiviral mechanisms, and is co-translationally incorporated into proteins in the form of selenocysteine (Sec) (10). Selenoproteins are classified into two groups. The first group are selenoproteins that have incorporated Sec using a specific process, which requires a UGA codon, Sec insertion sequence element and a specific tRNA (tRNA [Ser] Sec) (10). In these selenoproteins, the Sec residue is often observed at the active site, where it is critical for the function of the protein. A total of 25 selenoproteins have been identified in the human genome (11,12). Their biological functions, including glutathione peroxidases (GPXs), deiodinase iodothyronines (DIO) and thioredoxin reductases (TXRs), have been extensively studied, whereas those of other selenoproteins, such as selenoprotein K, remain largely unknown. By contrast, the second group of selenoproteins consists of Se-binding proteins, where Se is bound by specific cysteine residues (13). It has been reported that Se protects mammary epithelial cells from oxidative DNA damage (14,15), inhibits the initiation phase of carcinogenesis, stimulates DNA repair, regulates apoptosis, and prevents angiogenesis (16,17). Furthermore, the association between Se status and breast cancer risk has been documented from clinical observations; Se levels in the sera of patients with breast cancer is decreased compared with healthy control sera (18), and alterations in Se homeostasis aggravate oxidative stress by stimulating tumor progression and metastasis (19).
The present study performed an analysis of the global expression of the seleno-transcriptome family of genes in the human breast cancer MCF-7 and MDA-MB231 cell lines compared with healthy breast MCF-10A cells using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). In addition, the present study performed network studies on the genes identified, in order to evaluate their inter-associations and identify HUB nodes, which are important in controlling associated genes.
Materials and methods
Cell culture
Two human breast cancer cell lines, ER-positive MCF-7 (HTB-22; adenocarcinoma) and ER-negative MDA-MB231 (HTB-26; adenocarcinoma), and human non-cancerous mammary epithelial MCF-10A cell line (CRL-10317; fibrocystic disease) were obtained from Lonza (Verviers, Belgium). MCF-7 and MCF-10A cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Lonza) and MDA-MB231 cells were cultured in RPMI-1640 (Lonza). The medium of all the cells was supplemented with 10% fetal bovine serum (Invitrogen™; Thermo Fisher Scientific, Inc., Waltham, MA, USA), penicillin/streptomycin (100X; Euroclone Ltd, Paignton, UK), Glutamax (100X; Invitrogen™) and non-essential amino acids (100X; Invitrogen™) at 37°C in a humidified atmosphere of 5% CO2. In addition, for MCF-10A cells, DMEM was supplemented with human insulin (10 µg/ml; Thermo Fisher Scientific, Inc.), human epidermal growth factor (20 ng/ml; Thermo Fisher Scientific, Inc.) and hydrocortisone (0.5 µg/ml; Sigma-Aldrich, St. Louis, MO, USA), according to a previously reported procedure (20).
RNA preparation and RT-qPCR
Total RNA was extracted from MCF-7, MDA-MB231 and MCF-10A cells using the RNAeasy Mini kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocol. The extracted RNA was dissolved in diethyl pyrocarbonate treated water, and its concentration and purity were assessed by measurement of optical density at 260⁄280 nm, using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Inc., Wilmington, DE, USA). In total, 2 µg of total RNA of each sample was reverse-transcribed with SuperScript VILO cDNA Synthesis kit (Invitrogen™), according to the manufacturer's protocol, and subsequently diluted to 1:20 with nuclease-free water (Ambion®; Thermo Fisher Scientific, Inc.). RT were performed in a Veriti® Thermal Cycler (Thermo Fisher Scientific, Inc.) with the following thermal cycling conditions: 25°C for 10 min, followed by 42°C for 60 min, 85°C for 5 min and chilling at 4°C. The RT products were used to perform a qPCR in order to evaluate the expression level of seleno-transcripts. Sequences for mRNAs from a nucleotide data bank (National Center for Biotechnology Information, U.S. National Library of Medicine, Bethesda MD, USA) were used to design primer pairs for qPCR (Primer Express software v.3.0.1; Applied Biosystems®; Thermo Fisher Scientific, Inc.) (Table I). Oligonucleotides were synthesized by Sigma-Aldrich. The efficiency of each primer pair was calculated according to standard method curves, using the equation E=10−1/slope. Five serial dilutions were set up to determine Cq values and reaction efficiencies for all primer pairs. Standard curves were generated for each oligonucleotide pair using Cq values vs. the logarithm of each dilution factor. qPCR assays were run on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems®). Total RNA (2 µg) was reverse-transcribed using SuperScript VILO cDNA Synthesis kit (Invitrogen™), according to the manufacturer's protocol. In total, 10 ng cDNA was amplified in a total volume of 25 µl containing 1X SYBR Green PCR Master Mix (Applied Biosystems®) and 300 nM of forward and reverse primers. The thermal cycling conditions were as follows: 5 min of denaturation at 95°C followed by 44 cycles at 95°C for 30 sec, and 60°C for 1 min. An extra cycle was used for melting curve analysis (95°C for 15 sec, 60°C for 15 sec and 95°C for 15 sec) to verify the presence of a single product. Each assay included a no-template control for each primer pair. To capture intra-assay variability, all RT-qPCR reactions were performed in triplicate. For all RT-qPCR experiments, the data from each cDNA samples were normalized using β-actin mRNA as an endogenous level (21). Sample ΔCq values were calculated as the difference between the means of selenoprotein markers Cq and housekeeping assay Cq from the same sample. 2∆∆Cq values were determined in order to define the fold change of selenoprotein expression levels in tumor cells compared to the non-cancerous MCF-10A cells (22). These data were also confirmed by Relative Expression Software Tool (www.gene-quantification.com/rest.html) based on the Pfaffl method (23,24). Fold expression >1x indicated a significance between selenoprotein expression levels.
Table I.
Parameters for reverse transcription-polymerase chain reaction analysis.
| Gene | Melting temperature, °C | Annealing temperature, °C | Primer sequence 5′-3′ | Bp |
|---|---|---|---|---|
| DIO1 | 59.8 | 61.0 | F, AGCTTACTCTGGCTTTGCCGA | 21 |
| R, TATTACCCGTCTTCTCGCCCA | 21 | |||
| DIO2 | 59.8 | 60.0 | F, CTTACTCTGGCTTTGCCGAGA | 21 |
| R, CAGGATGTTCCGCTTGACTCT | 21 | |||
| DIO3 | 59.8 | 60.0 | F, GGTAGTTTCCCCCGCTTGTTT | 21 |
| R, TTTAGGTGCTGCTTTGAGGCC | 21 | |||
| GPX1 | 59.8 | 60.0 | F, TTATGACCGACCCCAAGCTCA | 21 |
| R, ATGTCAATGGTCTGGAAGCGG | 21 | |||
| GPX2 | 57.3 | 58.0 | F, GGAGAATGAACCCAAGCGAA | 20 |
| R, CAGGTTTGTCACAGCCAGTGAT | 22 | |||
| GPX3 | 59.8 | 60.0 | F, TCTCATCCCATGTCCACCATG | 21 |
| R, TGCATCCATTTGTGCCAGG | 19 | |||
| GPX4 | 59.8 | 60.0 | F, AGAGATCAAAGAGTTCGCCGC | 21 |
| R, TCTTCATCCACTTCCACAGCG | 21 | |||
| GPX5 | 57.9 | 58.0 | F, TCCTTCCACGACAATGGTTCA | 21 |
| R, TGTGACTGTGACCCCATTGCT | 21 | |||
| GPX6 | 59.8 | 61.0 | F, CAGAAACCCCACCTCACATGA | 21 |
| R, TGCCATGACCTGAATGCACT | 20 | |||
| GPX7 | 57.9 | 56.0 | F, TTGGTCCCATCATTCTTGTGG | 21 |
| R, GGCTGGTGATTCACTGGTCAA | 21 | |||
| SELI | 56.7 | 59.0 | F, AAAGGCCAGGTTCCCAGAA | 19 |
| R, CAATCCTGCTGCAGTCCAAGT | 21 | |||
| SELK | 57.3 | 59.0 | F, AATCAATCATCTGCGTGGCC | 20 |
| R, TGGTCAGCCTTCCACTTCTTG | 21 | |||
| SELM | 57.9 | 61.0 | F, TCACGCAGGACATTCCATTCT | 21 |
| R, CCTGCACTAGCGCATTGATCT | 21 | |||
| SELS | 59.8 | 56.0 | F, CAGCTGCTCGACTGAAAATGC | 21 |
| R, GCATGCTGTCCCACATTTCAA | 21 | |||
| SELT | 57.9 | 58.0 | F, TCAATCCCACACCATCGATCA | 21 |
| R, ACAACGAGCCTGCCAAGAAAG | 21 | |||
| SEP15 | 59.8 | 59.0 | F, ATCGGAGGCATGCAGAGAGTT | 21 |
| R, TCTGCAATCAGGATCCAGCTG | 21 | |||
| SEPHS2 | 57.3 | 60.0 | F, CGGCTCGCTTTTGTTCTGAA | 20 |
| R, TCGCGGCTTGTCAATGATC | 19 | |||
| SEPX1 | 59.8 | 61.0 | F, AGCGGCTGTTGCTCCATAACT | 21 |
| R, ATTTCAGCATCACCCACCCTC | 21 | |||
| SEPW1 | 59.8 | 60.0 | F, GTTTATTGTGGCGCTTGAGGC | 21 |
| R, CCATCACTTCAAAGAACCCGG | 21 | |||
| TRXR1 | 57.9 | 60.0 | F, CACAATTGGAATCCACCCTGT | 21 |
| R, GGTTTGCAGTCTTGGCAACA | 20 | |||
| TRXR2 | 57.9 | 62.0 | F, AGGACATTTGCTGGTCGAAGC | 21 |
| R, GGAATCCCCTGGAAAAACGTT | 21 | |||
| β-actin | 59.8 | 61.0 | F, GGTGGCTTTTAGGATGGCAAG | 21 |
| R, ACTGGAACGGTGAAGGTGACA | 21 |
DIO, deiodinase iodothyronine; GPX, glutathione peroxidase; SEL/SEP, selenoprotein; SEPH52, selenophosphate synthetase 2; TRXR, thioredoxin reductase; F, forward; R, reverse.
Bioinformatics analysis
Network analysis was performed by Ingenuity Pathway Analysis (IPA) program (Ingenuity®; Qiagen, Inc.), using the same procedure reported previously (25). Briefly, IPA builds and investigates transcriptional networks to identify regulatory events from signaling to transcriptional effects.
Results and Discussion
RT-qPCR evaluations on human breast cancer and non-cancerous cell line
The gene expression profiles of human breast cancer MCF-7 and MDA-MB231 cells in comparison with non-cancerous breast MCF-10A cells were assessed using RT-qPCR. The results revealed that there were four downregulated (GPX1, GPX4, GPX5 and GPX7) and eight upregulated [DIO1, DIO2, DIO3, selenoprotein T (SELT), selenoprotein W1 (SEPW1), selenoprotein X1 (SEPX1), GPX2 and GPX3] genes in MCF-7 cells, and five downregulated (GPX1, GPX4, GPX5, GPX6 and GPX7) and four upregulated [DIO2, selenoprotein 15 (SEP15), GPX2 and GPX3] genes in MDA-MB231 cells compared with non-cancerous cells (Fig. 1). In particular, four downregulated (GPX1, GPX4, GPX5 and GPX7) and three upregulated (DIO2, GPX2 and GPX3) genes were common between the two breast cancer cell lines. However, it cannot be excluded that this result may be due to intrinsic phenotypic differences between the two breast cancer cell lines, since MCF7 cells are ER-positive and MDA-MB231 cells are ER-negative. Furthermore, MDA-MB231 cells are known to constitutively express mutated tumor protein (TP) 53 gene, whereas MCF-7 cells have wild-type TP53; therefore, MDA-MB231 cells are characterized with a more malignant phenotype (26).
Figure 1.
Differences in the expression levels of (A) DIO, (B) seleno-mRNAs and (C) GPXs in human breast cancer MCF-7 (black) and MDA-MB231 (grey) cells compared with human breast non-cancerous MCF-10A cells. Fold changes >±1 were considered significant. *P<0.05, **P<0.01 and ***P<0.001. DIO, deiodinase iodothyronine; GPX, glutathione peroxidase; SEL/SEP, selenoprotein; SEPH52, selenophosphate synthetase 2; TRXR, thioredoxin reductase.
Recent advances in cancer research reveal that tumor development cannot be understood only through genetic mutations of cancerous cells; gene and protein inter-associations, which are associated with and regulate metabolic processes, should be also considered (27). Therefore, to understand the role of the seleno-transcriptome in breast cancer, the present study focused on the functional role of the components of this family. In general, DIO1, DIO2, and DIO3 constitute a group of dimeric integral membrane thioredoxin fold-containing proteins, which activate or inactivate the thyroid hormone, depending on their action on the phenolic or the tyrosyl ring of iodothyronines. In addition, it is well known that the thyroid hormone, as well as estrogens, acts via nuclear receptors, and the mRNA level of the hormone is altered in breast cancer tissues (28). It is also known that thyroid hormone receptors bind to an estrogen response element of target gene promoters, which affects the estrogen-dependent gene transcription (29). A study on breast cancer cell lines have revealed that the thyroid hormone stimulates cell growth and division in the ER-positive MCF-7 cell line (30). This highlights that the thyroid hormone is clearly associated with the expression of the three DIOs; therefore explaining their overexpression in the MCF-7 cells observed in the present study.
The GPX family is composed of seven components known to catalyze the reduction of H2O2 or organic hydroperoxides to water or corresponding alcohols using reduced glutathione as an electron donor. In mammalian tissues, there are five major Se-dependent GPX isozymes: GPX1 are located in the cytosol, nucleus and mitochondria of erythrocytes, and the liver, lung and kidney; GPX2 in the cytosol and nucleus, and the gastrointestinal tract; GPX3 in the mitochondria of several organs, including the kidney, lung, epididymis, breast, heart and muscle; GPX4 in the nucleus, cytosol and mitochondria of various tissues; and GPX6 in the olfactory epithelium. By contrast, GPX5, secreted in the epididymis, does not have Sec at the active site, and GPX7, recently described as a novel phospholipid hydroperoxide glutathione peroxidase, incorporates cysteine instead of Sec in the conserved catalytic motif (31). The best characterized member of the GPX family is GPX1, which uses reducing equivalents from glutathione to detoxify peroxides (32) and is decreased in tumor cells compared with their normal counterparts (33). Therefore, the present results for GPX1 in the two breast cancer cell lines are in agreement with data reported for other cancers (32,33). GPX5, which is androgen-regulated (34), was demonstrated to be associated with overall survival among patients with non-small cell lung cancer (35), but no data about its possible involvement in breast cancer has been reported. Therefore, the present study suggests for the first time the role of GPX5 in breast cancer. In addition, for GPX4 and GPX7, the present results confirmed the general observations that they are clearly inter-associated from a functional point of view, and this may explain why the two genes are downregulated in the present experiments. These two enzymes present similar structural features and are involved in countering the effects of oxidative damage particularly to membrane lipids, and they often have a reduced expression in cancer (34). In detail, GPX4 has been demonstrated to have a reduced expression in a number of cell lines, including breast cancer MCF-7, colon cancer HT29 and ovarian cancer A2780 cell lines (36), while the absence of non-Sec phospholipid hydroperoxide glutathione peroxidase complex with GPX6 expression has been identified in the majority of breast cancer cell lines (37). The potential role in alleviating oxidative stress induced by dietary consumption of fatty acids has been suggested for GPX7 in breast cancer cells (38). By contrast, in the present study, GPX2 and GPX3 were upregulated in ER-positive MCF-7 and ER-negative MDA-MB231 cells. This may be explained by their known effects in cancer. GPX2 is required in healthy tissues to maintain normal self-renewing of the gastrointestinal epithelium and to reduce inflammatory processes. Therefore, GPX2 should inhibit carcinogenesis; however, once a cell has been programmed to proliferate in an uncontrolled way, GPX2 supports the growth of cells by inhibiting apoptosis (39). This appears to be consistent with the present results. GPX3 was also identified as upregulated in the present study; therefore, it may be considered that GPX3 is a major scavenger of reactive oxygen species produced during normal metabolism or following oxidative insult. In addition, GPX3 is downregulated by promoter hyper-methylation in several types of human cancer, which suggests its function as a tumor suppressor (40). However, GPX3 has also been revealed to be highly upregulated in epithelial ovarian carcinoma, where it was proposed as a molecular marker that was highly specific for clear cell carcinoma (41). In addition, GPX3 directly targets the ERα gene in white adipose tissue, for which it was proposed as an important mediator of the estrogen effects in association with fat mass (42). Considering the link between visceral fat and breast cancer initiation and progression (43), it is reasonable to observe an overexpression of GPX3 in breast cancer cells. However, more specific studies are required to deeply understand the specific role of GPX3 in breast cancer.
SEP15, SEPW1 and SELT proteins share a similar structural feature; they have a thioredoxin-like domain in common. However, SEPW1 has been revealed to be involved in cell cycle progression (44), where it facilitates the G1 to S-phase transition by downregulating the expression of the cyclin-dependent kinase inhibitor p21 via controlling the levels of TP53 transcription factor (44). In the literature, it is also reported that SELT is involved in cell adhesion, the expression of several oxidoreductase genes, cell structural organization, redox regulation and cell anchorage. However, it has also been revealed that SEPW1 may functionally compensate for SELT expression (45). Therefore, these data demonstrate that SEPW1 and SELT are inter-associated and may explain why they were upregulated in the present study. SEP15 is involved in redox function, as well as carcinogenesis (46). It has been demonstrated that SEP15 has a role in allelic loss during breast cancer development (47), which is in agreement with the present data.
By contrast, SEPX1 belongs to the methionine sulfoxide reductase B (MSRB) family, and for this reason it is also termed MSRB1 (48). High levels of its transcript were detected in human breast cancer MCF7 cells, whereas lower levels were detected in highly aggressive MDA-MB231 cells (48). The present data are in clear agreement with this previous study.
Network analysis
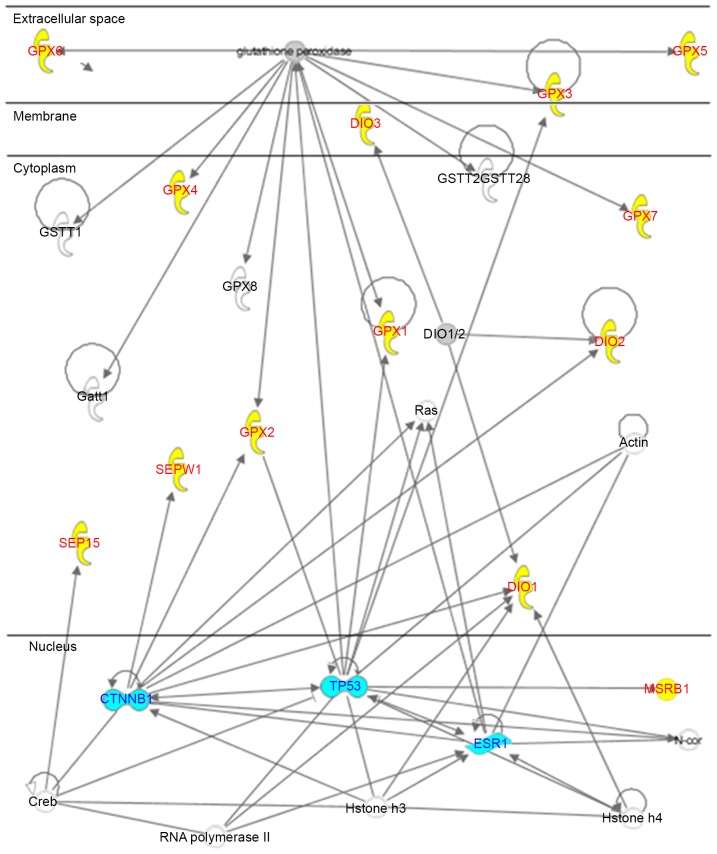
To evaluate the inter-associations between these downregulated and upregulated genes, as well as to identify putative HUB nodes capable of exerting direct control over them, the present study used an IPA algorithm. Fig. 2 reveals that all the differentially regulated genes, with the exception of SELT, are implicated in the same network that presents three HUB nodes interconnected to the selenoprotein mRNAs, including TP53, estrogen receptor 1 (ESR1) and catenin-β1 (CTNNB1).
Figure 2.
Network analysis of genes identified in human breast cancer cells. Downregulated and upregulated genes, yellow; HUB genes, blue; other genes, white.
In detail, analyzing the associations between downregulated and upregulated genes using the IPA algorithm (Fig. 2) revealed the following: ESR1 binds TP53 and CTNNB1; TP53 binds GPX family and SEPX1; and CTNNB1 binds DIO1, DIO2, GPX2, SEPW1 and SEP15 through cAMP response element-binding protein (CREB). In general, CTNNB1 regulates the coordination of cell-cell adhesion and gene transcription, and acts in the Wnt signaling pathway as an intracellular signal. However, CTNNB1 is also known to bind ESR1 during estrogen signaling (49), and to increase the transcriptional activity of human TP53 (50), which is the most frequently mutated gene in human cancer and is often referred to as ‘the guardian of the genome’ (44). In response to genomic stresses, TP53 causes cell cycle arrest to allow time for genomic damage to be repaired prior to cell division, or induces apoptosis to eliminate irreparably damaged cells (44). In addition, TP53 activation is affected by the cellular transcription factor CREB, which binds SEP15 (51,52). However it is inactivated by expression of SEPX1-mRNA, which also involves the oncogenic mutant HRAS protein (53). Furthermore, the literature reports that CTNNB1 is involved in the expression of human SEPW1 (54), DIO1 (55), GPX2 and DIO2 mRNA by binding to human T-cell signaling factor (56).
Overall, the network analysis suggests a reasonable role for CTNNB1 as an upstream regulator, since it is an important link connecting the DIOs, the GPXs, the seleno-mRNAs belonging to the thioredoxin-like family, TP53 and the ER, which are important in ER-positive MCF-7 cells.
Conclusion
The present study revealed a profile of seleno-mRNAs specific for human breast cells, and demonstrated that these genes alter their expression on the basis of the ER-positivity or negativity of breast cancer cells. In addition, the present study identified HUB nodes that represent the association centers and control the genes, and this data suggested the potential role of CTNNB1 in breast cancer cells. These data are notable, since there is little data reported in the literature concerning the evaluation of the entire seleno-transcriptome in cancer cell lines and tissues; the literature only reports a few seleno-mRNAs (57). Recently, the present authors evaluated the global expression of the seleno-transcriptome in human hepatocellular carcinoma HepG2 and Huh7 cells compared with normal human hepatocytes. That study revealed seleno-mRNAs that are specific to liver cancer in the absence of viral infection or genetic mutations (57).
Novel studies are required to evaluate the seleno-transcriptome in bioptic tissues of breast cancer patients to confirm the results obtained by the present study, and to suggest novel markers to improve the diagnosis and prognosis of this cancer.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008 GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Ko BS, Noh WC, Kang SS, Park BW, Kang EY, Paik NS, Yang JH, Ahn SH, Korean Breast Cancer Society Changing patterns in the clinical characteristics of Korean breast cancer from 1996–2010 using an online nationwide breast cancer database. J Breast Cancer. 2012;15:393–400. doi: 10.4048/jbc.2012.15.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leclere B, Molinié F, Trétarre B, Stracci F, Daubisse-Marliac L, M; GRELL Working Group Colonna. Trends in incidence of breast cancer among women under 40 in seven European countries: A GRELL cooperative study. Cancer Epidemiol. 2013;37:544–549. doi: 10.1016/j.canep.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Althuis MD, Brogan DD, Coates RJ, Daling JR, Gammon MD, Malone KE, Schoenberg JB, Brinton LA. Breast cancers among very young premenopausal women (United States) Cancer Causes Control. 2003;14:151–160. doi: 10.1023/A:1023006000760. [DOI] [PubMed] [Google Scholar]
- 5.Prall OW, Sarcevic B, Musgrove EA, Watts CK, Sutherland RL. Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin dependent kinase inhibitor association with cyclin E-Cdk2. J Biol Chem. 1997;272:10882–10894. doi: 10.1074/jbc.272.16.10882. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland RL, Prall OW, Watts CK, Musgrove EA. Estrogen and progestin regulation of cell cycle progression. J Mammary Gland Biol Neoplasia. 1998;3:63–72. doi: 10.1023/A:1018774302092. [DOI] [PubMed] [Google Scholar]
- 7.Kyprianou N, English HF, Davidson NE, Isaacs JT. Programmed cell death during regression of the MCF-7 human breast cancer following estrogen ablation. Cancer Res. 1991;51:162–166. [PubMed] [Google Scholar]
- 8.Wang TT, Phang JM. Effects of estrogen on apoptotic pathways in human breast cancer cell line MCF-7. Cancer Res. 1995;55:2487–2489. [PubMed] [Google Scholar]
- 9.Guo CH, Hsia S, Chen PC. Distribution of selenium and oxidative stress in breast tumor-bearing mice. Nutrients. 2013;5:594–607. doi: 10.3390/nu5020594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 11.Davis CD, Tsuji PA, Milner JA. Selenoproteins and cancer prevention. Ann Rev Nutr. 2012;32:73–95. doi: 10.1146/annurev-nutr-071811-150740. [DOI] [PubMed] [Google Scholar]
- 12.Rayman MP. Selenium and human health. Lancet. 2012;379:1256–1268. doi: 10.1016/S0140-6736(11)61452-9. [DOI] [PubMed] [Google Scholar]
- 13.Fang W, Goldberg ML, Pohl NM, Bi X, Tong C, Xiong B, Koh TJ, Diamond AM, Yang W. Functional and physical interaction between the selenium-binding protein 1 (SBP1) and the glutathione peroxidase 1 selenoprotein. Carcinogenesis. 2010;31:1360–1366. doi: 10.1093/carcin/bgq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae I, Fan S, Meng Q, Rih JK, Kim HJ, Kang HJ, Xu J, Goldberg ID, Jaiswal AK, Rosen EM. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Res. 2004;64:7893–7909. doi: 10.1158/0008-5472.CAN-04-1119. [DOI] [PubMed] [Google Scholar]
- 15.Borek C. Dietary antioxidants and human cancer. Integr Cancer Ther. 2004;3:333–341. doi: 10.1177/1534735404270578. [DOI] [PubMed] [Google Scholar]
- 16.Jackson MI, GF Combs Jr. Selenium and anticarcinogenesis: Underlying mechanisms. Curr Opin Clin Nutr Metab Care. 2008;11:718–726. doi: 10.1097/MCO.0b013e3283139674. [DOI] [PubMed] [Google Scholar]
- 17.Chen YC, Sosnoski DM, Gandhi UH, Novinger LJ, Prabhu KS, Mastro AM. Selenium modifies the osteoblast inflammatory stress response to bone metastatic breast cancer. Carcinogenesis. 2009;30:1941–1948. doi: 10.1093/carcin/bgp227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Saez JB, Senra-Varela A, Pousa-Estevez L. Selenium in breast cancer. Oncology. 2003;64:227–231. doi: 10.1159/000069312. [DOI] [PubMed] [Google Scholar]
- 19.Mates JM, Segura JA, Alonso FJ, Marquez J. Sulphur-containing non enzymatic antioxidants: Therapeutic tools against cancer. Front Biosci (Schol Ed) 2012;4:722–748. doi: 10.2741/s296. [DOI] [PubMed] [Google Scholar]
- 20.Rothwell DG, Li Y, Ayub M, Tate C, Newton G, Hey Y, Carter L, Faulkner S, Moro M, Pepper S, et al. Evaluation and validation of a robust single cell RNA-amplification protocol through transcriptional profiling of enriched lung cancer initiating cells. BMC Genomics. 2014;15:1129. doi: 10.1186/1471-2164-15-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peirce SK, Chen WY, Chen WY. Quantification of prolactin receptor Mrna in multiple human tissues and cancer cell lines by real time RT-PCR. J Endocrinol. 2001;171:R1–R4. doi: 10.1677/joe.0.171R001. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costantini S, Capone F, Maio P, Guerriero E, Colonna G, Izzo F, Castello G. Cancer biomarker profiling in patients with chronic hepatitis C virus, liver cirrhosis and hepatocellular carcinoma. Oncol Rep. 2013;29:2163–2168. doi: 10.3892/or.2013.2378. [DOI] [PubMed] [Google Scholar]
- 26.Bando H, Toi M, Kitada K, Koike M. Genes commonly upregulated by hypoxia in human breast cancer cells MCF-7 and MDA-MB-231. Biomed Pharmacother. 2003;57:333–340. doi: 10.1016/S0753-3322(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 27.Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, Shukla M, Chesnick B, Kadan M, Papp E, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva JM, Dominguez G, González-Sancho JM, García JM, Silva J, García-Andrade C, Navarro A, Muñoz A, Bonilla F. Expression of thyroid hormone receptor/erbA genes is altered in human breast cancer. Oncogene. 2002;21:4307–4316. doi: 10.1038/sj.onc.1205534. [DOI] [PubMed] [Google Scholar]
- 29.Hall LC, Salazar EP, Kane SR, Liu N. Effects of thyroid hormones on human breast cancer cell proliferation. J Steroid Biochem Mol Biol. 2008;109:57–66. doi: 10.1016/j.jsbmb.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Debski MG, Pachucki J, Ambroziak M, Olszewski W, Bar-Andziak E. Human breast cancer tissue expresses high level of type 1 5′-deiodinase. Thyroid. 2007;17:3–10. doi: 10.1089/thy.2006.0012. [DOI] [PubMed] [Google Scholar]
- 31.Margis R, Dunand C, Teixeira FK, Margis-Pinheiro M. Glutathione peroxidase family-an evolutionary overview. FEBS J. 2008;275:3959–3970. doi: 10.1111/j.1742-4658.2008.06542.x. [DOI] [PubMed] [Google Scholar]
- 32.Bera S, Weinberg F, Ekoue DN, Ansenberger-Fricano K, Mao M, Bonini MG, Diamond AM. Natural allelic variations in glutathione peroxidase-1 affect its subcellular localization and function. Cancer Res. 2014;74:5118–5126. doi: 10.1158/0008-5472.CAN-14-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang HP, Schafer FQ, Goswami PC, Oberley LW, Buettner GR. Phospholipid hydroperoxide glutathione peroxidase induces a delay in G1 of the cell cycle. Free Radic Res. 2003;37:621–630. doi: 10.1080/1071576031000088283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brigelius-Flohé R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830:3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Sun Z, Cunningham JM, Aubry MC, Wampfler JA, Croghan GA, Johnson C, Wu D, Aakre JA, Molina J, et al. Genetic variations in multiple drug action pathways and survival in advanced stage non-small cell lung cancer treated with chemotherapy. Clin Cancer Res. 2011;17:3830–3840. doi: 10.1158/1078-0432.CCR-10-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding WQ, Lind SE. Phospholipid hydroperoxide glutathione peroxidase plays a role in protecting cancer cells from docosahexaenoic acid-induced cytotoxicity. Mol Cancer Ther. 2007;6:1467–1474. doi: 10.1158/1535-7163.MCT-06-0608. [DOI] [PubMed] [Google Scholar]
- 37.Rodrigues P, de Marco G, Furriol J, Mansego ML, Pineda-Alonso M, Gonzalez-Neira A, Martin-Escudero JC, Benitez J, Lluch A, Chaves FJ, Eroles P. Oxidative stress in susceptibility to breast cancer study in Spanish population. BMC Cancer. 2014;14:861. doi: 10.1186/1471-2407-14-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Utomo A, Jiang X, Furuta S, Yun J, Levin DS, Wang YC, Desai KV, Green JE, Chen PL, Lee WH. Identification of a novel putative non-selenocysteine containing phospholipid hydroperoxide glutathione peroxidase (NPGPx) essential for alleviating oxidative stress generated from polyunsaturated fatty acids in breast cancer cells. J Biol Chem. 2004;279:43522–43529. doi: 10.1074/jbc.M407141200. [DOI] [PubMed] [Google Scholar]
- 39.Brigelius-Flohé R, Müller M, Lippmann D, Kipp AP. The yin and yang of nrf2-regulated selenoproteins in carcinogenesis. Int J Cell Biol. 2012;2012:486147. doi: 10.1155/2012/486147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Jin J, Ying J, Sun M, Cui Y, Zhang L, Xu B, Fan Y, Zhang Q. Frequent epigenetic suppression of tumor suppressor gene glutathione peroxidase by promoter hypermethylation and its clinical implication in clear cell renal cell carcinoma. Int J Mol Sci. 2015;16:10636–10649. doi: 10.3390/ijms160510636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hough CD, Cho KR, Zonderman AB, Schwartz DR, Morin PJ. Coordinately up-regulated genes in ovarian cancer. Cancer Res. 2001;61:3869–3876. [PubMed] [Google Scholar]
- 42.Lundholm L, Putnik M, Otsuki M, Andersson S, Ohlsson C, Gustafsson JA, Dahlman-Wright K. Effects of estrogen on gene expression profiles in mouse hypothalamus and white adipose tissue: Target genes include glutathione peroxidase 3 and cell death-inducing DNA fragmentation factor, alpha-subunit-like effector A. J Endocrinol. 2008;196:547–557. doi: 10.1677/JOE-07-0277. [DOI] [PubMed] [Google Scholar]
- 43.Rohan TE, Heo M, Choi L, Datta M, Freudenheim JL, Kamensky V, Ochs-Balcom HM, Qi L, Thomson CA, Vitolins MZ, et al. Body fat and breast cancer risk in postmenopausal women: A longitudinal study. J Cancer Epidemiol. 2013;2013:754815. doi: 10.1155/2013/754815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkes WC, Alkan Z. Delayed cell cycle progression from SEPW1 depletion is p53- and p21-dependent in MCF-7 breast cancer cells. Biochem Biophys Res Commun. 2011;413:36–40. doi: 10.1016/j.bbrc.2011.08.032. [DOI] [PubMed] [Google Scholar]
- 45.Sengupta A, Carlson BA, Labunskyy VM, Gladyshev VN, Hatfield DL. Selenoprotein T deficiency alters cell adhesion and elevates selenoprotein W expression in murine fibroblast cells. Biochem Cell Biol. 2009;87:953–961. doi: 10.1139/O09-064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kasaikina MV, Fomenko DE, Labunskyy VM, Lachke SA, Qiu W, Moncaster JA, Zhang J, Wojnarowicz MW, Jr, Natarajan SK, Malinouski M, et al. 15-kDa selenoprotein (Sep15) knockout mice: Roles of Sep15 in redox homeostasis and cataract development. J Biol Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nasr MA, Hu YJ, Diamond AM. Allelic loss at the SEP15 locus in breast cancer. Cancer Therapy. 2003;1:293–298. [Google Scholar]
- 48.De Luca A, Sacchetta P, Nieddu M, Di Ilio C, Favaloro B. Important roles of multiple Sp1 binding sites and epigenetic modifications in the regulation of the methionine sulfoxide reductase B1 (MsrB1) promoter. BMC Mol Biol. 2007;8:39. doi: 10.1186/1471-2199-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kouzmenko AP, Takeyama K, Ito S, Furutani T, Sawatsubashi S, Maki A, Suzuki E, Kawasaki Y, Akiyama T, Tabata T, Kato S. Wnt/beta-catenin and estrogen signaling converge in vivo. J Biol Chem. 2004;279:40255–40258. doi: 10.1074/jbc.C400331200. [DOI] [PubMed] [Google Scholar]
- 50.Damalas A, Ben-Ze'ev A, Simcha I, Shtutman M, Leal JF, Zhurinsky J, Geiger B, Oren M. Excess beta-catenin promotes accumulation of transcriptionally active p53. EMBO J. 1999;18:3054–3063. doi: 10.1093/emboj/18.11.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ariumi Y, Kaida A, Lin JY, Hirota M, Masui O, Yamaoka S, Taya Y, Shimotohno K. HTLV-1 tax oncoprotein represses the p53-mediated trans-activation function through coactivator CBP sequestration. Oncogene. 2000;19:1491–1499. doi: 10.1038/sj.onc.1203450. [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson MB, Xiao G, Kumar A, LaPlant Q, Renthal W, Sikder D, Kodadek TJ, Nestler EJ. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J Neurosci. 2009;29:7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boiko AD, Porteous S, Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–252. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin YM, Ono K, Satoh S, Ishiguro H, Fujita M, Miwa N, Tanaka T, Tsunoda T, Yang KC, Nakamura Y, Furukawa Y. Identification of AF17 as a downstream gene of the beta-catenin/T-cell factor pathway and its involvement in colorectal carcinogenesis. Cancer Res. 2001;61:6345–6309. [PubMed] [Google Scholar]
- 55.Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SP. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz DR, Wu R, Kardia SL, Levin AM, Huang CC, Shedden KA, Kuick R, Misek DE, Hanash SM, Taylor JM, et al. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63:2913–2922. [PubMed] [Google Scholar]
- 57.Guariniello S, Di Bernardo G, Colonna G, Cammarota M, Castello G, Costantini S. Evaluation of the selenotranscriptome expression in two hepatocellular carcinoma cell lines. Anal Cell Pathol (Amst) 2015;2015:419561. doi: 10.1155/2015/419561. [DOI] [PMC free article] [PubMed] [Google Scholar]