Abstract
Polycomb group (PcG) proteins form at least two key complexes, namely polycomb repressive complex 1 and polycomb repressive complex 2. These complexes are involved in the progression of various cancers. Systematic research has not been conducted on the aberrant expression of PcG members in gliomas. Using the Chinese Glioma Genome Atlas data set, PcG expression patterns between normal brain tissues and glioma samples were analyzed, and a PcG-based classifier was then developed using BRB Cox regression and risk-score model. These results were validated in an independent GSE16011 set. A total of six PcGs [chromobox protein homolog (CBX) 6, CBX7, PHD finger protein 1, enhancer of zeste homolog 2 (EZH2), DNA (cytosine-5-)-methyltransferase 3β (DNMT3B) and polyhomeotic-like protein 2] were identified to be associated with glioma grade. Survival analysis then revealed a five-PcG gene signature one protective gene (enhancer of zeste homolog 1) and four risky genes (EZH2, PHD finger protein 19, DNMT3A and DNMT3B), which may identify patients with high risk of poor prognosis of glioma. Multivariate Cox analysis indicated that the five-PcG signature was an independent prognostic biomarker. These findings indicated that a novel prognostic classifier, five-PcG signature, served as an independent prognostic marker for patients with glioma.
Keywords: polycomb repressive complex 1, polycomb repressive complex 2, polycomb group, glioma, cancer progression, enhancer of zeste homolog 2, chromobox protein homolog 7
Introduction
Polycomb group (PcG) genes are epigenetic regulators that form polycomb repressor complexes responsible for gene silencing through post-translational histone modifications (1). The two distinct PcG complexes in mammals are polycomb repressive complex (PRC) 1, which completes gene silencing through histone H2A ubiquitylation, and PRC2, which mediates histone H3 lysine 27 trimethylation (2,3).
PcG proteins were initially identified as transcriptional repressors that control body segmentation in Drosophila (4). PcG proteins have been the focus of investigation in previous cancer studies. For example, enhancer of zeste homolog 2 (EZH2) and BMI1 proto-oncogene polycomb ring finger are upregulated in a number of human cancers, including breast carcinoma, colon carcinoma, liver carcinoma and lung carcinoma, and are associated with tumor aggressiveness (5–9).
However, the PcG expression patterns in gliomas have not been systematically investigated to date. Thus, the Chinese Glioma Genome Atlas (CGGA) database was systematically analyzed for the expression of all PcG genes (10). A total of six PcG genes were identified to be closely associated with glioma grade. Furthermore, survival analysis revealed a five-PcG gene signature; consisting of one protective gene (EZH1) and four risky genes [EZH2, PHD finger protein 19 (PHF19), DNA (cytosine-5-)-methyltransferase 3α (DNMT3A) and DNA (cytosine-5-)-methyltransferase 3β (DNMT3B)], which could identify patients with high risk of poor survival in gliomas regardless of World Health Organization (WHO) grades.
Materials and methods
Patients and samples
Two large gene expression-profiling cohorts of gliomas were used in the present study. The training data from the CGGA (www.cgcg.org.cn) included 58 astrocytomas, 18 oligodendrogliomas, 21 oligoastrocytomas, 8 anaplastic astrocytomas, 11 anaplastic oligodendrogliomas, 15 anaplastic oligoastrocytomas, 4 secary glioblastoma (GBMs) and 85 primary GBMs. A total of five normal brain tissue samples were added, subsequent to informed consent being obtained from patients with severe traumatic brain injury that requires post-trauma surgery, and from patients who had undergone surgery for primary epilepsy. All 220 glioma patients underwent surgical resection between January 2005 and December 2009 and subsequently received radiation therapy and/or alkylating agent-based chemotherapy at Beijing Tiantan Hospital (Beijing, China) (11). Use of the patient data was approved by the institutional review boards of all hospitals involved (Beijing Tiantan Hospital; The First Affiliated Hospital of Nanjing Medical University, Nanjing, China; and Tianjin Medical University General Hospital, Tianjin, China), and written informed consent was obtained from all selected patients (11). Furthermore, the validation dataset GSE16011 was downloaded from a previous study (12). This independent dataset consisted of 284 samples (159 cases of GBM, 52 cases of oligodendrocytoma, 8 cases of pilocytic astrocytoma, 28 cases of oligoastrocytoma, 29 cases of astrocytoma and 8 cases without tumors).
Analysis
Statistical analysis
Student's t-test was used to analyze differences in each two-group comparison, and one-way analysis of variance was used to determine the difference among at least three groups. All data are presented as the mean ± standard error. Kaplan-Meier analysis was employed to assess the survival rate of patients. P<0.05 was considered to be a statistically significant difference Statistical analysis were performed using IBM SPSS, version 19.0, software for Windows (IBM SPSS, Armonk, NY, USA).
Differential gene expression
Significance analysis of microarrays was applied to identify differentially expressed genes between normal brain tissues and glioma samples of all grades. Student's t-test was used to determine significant differences. Genes were considered differentially expressed if they showed a fold-change >1.4 with a false discovery rate of 0.05.
Survival analysis
First, 37 cases and 6 cases with insufficient survival data from the CGGA and GSE16011 data were excluded from survival analysis. Cox's proportional hazard regression analysis was then performed by employing the BRB array tool on the microarray cohort. A permutation test was conducted using 10,000 permutations. A total of 5 PcG genes were associated with survival (P<0.0001). The significant PcG genes were divided into risky and protective types. Risky PcG genes were defined as those genes with a hazard ratio for mortality >1. By contrast, protective PcG genes were defined based on a hazard ratio for mortality <1. Using these five significant PcG genes, a risk-score formula for survival time prediction was constructed according to a linear combination of the expression level of the PcG genes, weighted by the regression coefficient from Cox's univariate regression analysis (13,14). According to this model, patients with high-risk scores are expected to possess poorer survival outcomes compared with patients with low risk scores. The risk scores were calculated as follows: (−1.153 × expression of EZH1) + (0.522 × expression of EZH2) + (1.103 × expression of PHF19) + (1.418 × expression of DNMT3A) + (0.757 × expression of DNMT3B). The 50th percentile risk score was used as the cutoff point, since this divided the training samples into two groups having different survival times with highest significance. Kaplan-Meier survival analysis was used to estimate the survival distributions, and log-rank tests were employed to assess the statistical significance between stratified survival groups using GraphPad Prism 5.0 statistical software (GraphPad Software, Inc., La Jolla, CA, USA). All data were presented as the mean ± standard deviation. P<0.05 (two-tailed) was considered to indicate a statistically significant difference.
Results
Distinctive PcG expression in gliomas
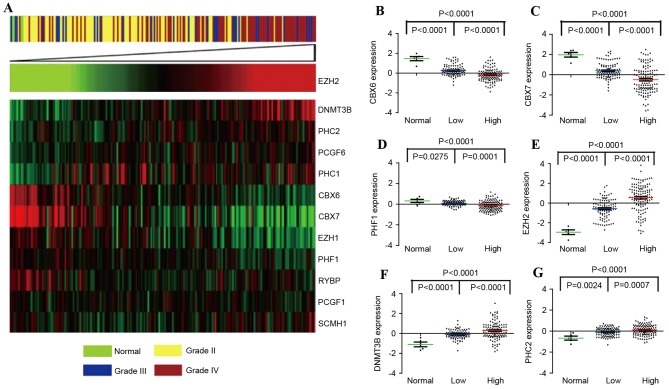
In the present study, PcG expression patterns were initially compared between normal brain tissues and glioma samples of all grades in the CGGA set, using SAM analysis. A total of 12 differentially expressed genes among normal brain tissues and glioma samples were detected (Table I). Since our previous study reported the pro-oncogenic activity of EZH2 in GBMs (15), 11 additional significant genes were clustered according to the level of EZH2 expression. Fig. 1A showed that five PcG genes [EZH2, polyhomeotic homolog (PHC) 1, PHC2, polycomb group ring finger 6 (PCGF6) and DNMT3B] were upregulated, and seven PcG genes (CBX6, CBX7, RING1 and YY1 binding protein, polycomb group ring finger protein 1, PHF1, sex comb on midleg homolog 1 and EZH1) were downregulated. Furthermore, six PcG genes (CBX6, CBX7, PHF1, EZH2, DNMT3B and PHC2) were significantly associated with glioma grade, as shown in Fig. 1B-G (P<0.01). In addition, results were validated in the GSE16011 dataset, and similar results were observed (data not shown).
Table I.
Distinctive PcG expression in gliomas.
| A, Positive genes | |||||||
|---|---|---|---|---|---|---|---|
| Row | Gene ID | Gene name | Score, d | Numerator, r | Denominator, s+s0 | Fold change | q-value, % |
| 13 | EZH2 | −2.69779 | 3.513838 | 2.83718 | 0.80743 | 7.146217 | 0 |
| 23 | PHC1 | −0.9938 | 1.958637 | 0.965258 | 0.492821 | 1.952413 | 0 |
| 10 | DNMT3B | −1.11903 | 1.925787 | 0.980461 | 0.509122 | 1.973095 | 0 |
| 22 | PCGF6 | −0.45277 | 1.610712 | 0.57532 | 0.357184 | 1.490008 | 0 |
| 24 | PHC2 | −0.37176 | 1.308672 | 0.538816 | 0.411727 | 1.452779 | 4.95495495 |
| B, Negative genes | |||||||
| Row | Gene ID | Gene name | Score, d | Numerator, r | Denominator, s+s0 | Fold change | q-value, % |
| 6 | CBX6 | 1.684784 | −3.07648 | −1.61744 | 0.525745 | 0.325912 | 0 |
| 7 | CBX7 | 1.84724 | −2.93163 | −2.33339 | 0.795935 | 0.198417 | 0 |
| 32 | RYBP | 0.615774 | −1.95196 | −0.84237 | 0.431549 | 0.557728 | 0 |
| 26 | PHF1 | 0.139365 | −1.55231 | −0.62273 | 0.401164 | 0.649441 | 0 |
| 18 | PCGF1 | 0.186895 | −1.31428 | −0.4946 | 0.376323 | 0.70976 | 0 |
| 12 | EZH1 | 0.46613 | −1.25352 | −0.68906 | 0.549698 | 0.620258 | 0 |
| 33 | SCMH1 | 0.625105 | −1.19795 | −0.52556 | 0.438716 | 0.694689 | 0 |
EZH2, enhancer of zeste homolog 2; PHC1, polyhomeotic homolog 1; DNMT3B, DNA (cytosine-5-)-methyltransferase 3β; PCGF6, polycomb group ring finger 6; PHC2, polyhomeotic homolog 2; CBX6, chromobox protein homolog 6; CBX7, chromobox protein homolog 7; RYBP, ring1 and YY1 binding protein; PHF1, PHD finger protein 1; PCGF1, polycomb group ring finger protein 1; EZH1, enhancer of zeste homolog 1; SCMH1, sex comb on midleg homolog 1.
Figure 1.
Differentially expressed genes between normal brain tissues and gliomas of increasing malignancy grades. (A) Heat map of a total of 12 genes identified as significantly different between normal brain tissues and gliomas by significance analysis of microarrays, sorted by level of EZH2 expression. Levels of (B) CBX6 (C) CBX7, (D) PHF1, (E) EZH2, (F) DNMT3B and (G) PHC2 were analyzed in different glioma tissues of Chinese Glioma Genome Atlas data. DNMT3B, DNA (cytosine-5-)-methyltransferase 3β; EZH1, enhancer of zeste homolog 1; EZH2, enhancer of zeste homolog 2; PCGF6, polycomb group ring finger 6; PHC1, polyhomeotic homolog 1; PHC2, polyhomeotic homolog 2; CBX6, chromobox protein homolog 6; PHF1, PHD finger protein 1; PCGF1, polycomb group ring finger protein 1; RYBP, RING1 and YY1 binding protein; SCMH1, sex comb on midleg homolog 1.
Identification of five PcG genes and their association with the survival of patients
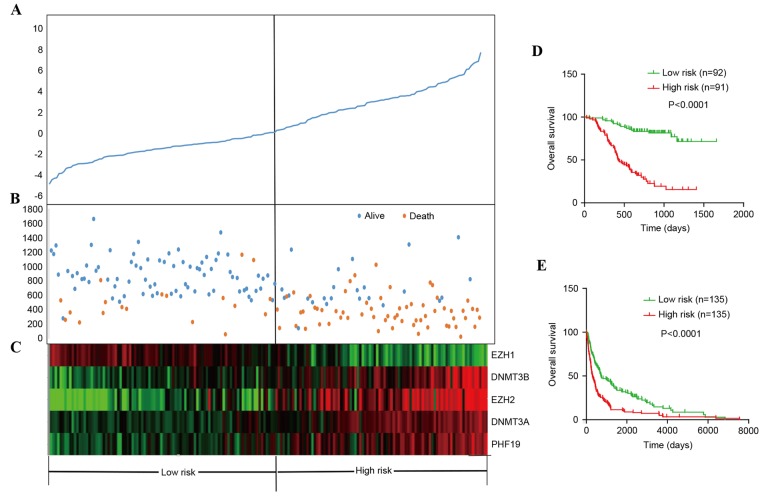
To investigate the prognostic ability of the PcG, Cox proportional hazard regression of all PcG genes in 183 CGGA patients with glioma was performed by BRB array tools using the permutation test method (16). In total, five PcG genes (EZH1, EZH2, PHF19, DNMT3A and DNMT3B) were significantly associated with patient survival (permutation, P<0.01; Table II). Risky PcG genes (EZH2, PHF19, DNMT3A and DNMT3B) were defined as those genes with a hazard ratio for mortality >1. By contrast, genes with a hazard ratio for mortality <1 were defined as protective PcG genes (EZH1). The five PcG genes were used to construct a signature by using the risk-score method. The distribution of PcG gene expression, patient risk scores and the survival status of 183 CGGA patients are shown in Fig. 2A-C. Patients with low-risk scores tended to express high levels of protective PcG genes (EZH1), whereas patients with high-risk scores tended to express high levels of risky PcG genes (EZH2, PHF19, DNMT3A and DNMT3B).
Table II.
Five PcG genes.
| Gene ID | FDR | Permutation P-value | Hazard ration | Coefficient |
|---|---|---|---|---|
| EZH1 | <0.0001 | <0.0001 | 0.315 | 1.153 |
| DNMT3B | <0.0001 | <0.0001 | 4.312 | 1.418 |
| EZH2 | <0.0001 | <0.0001 | 1.686 | 0.522 |
| PHF19 | <0.0001 | <0.0001 | 3.017 | 1.103 |
| DNMT3A | <0.0001 | <0.0001 | 2.133 | 0.757 |
EZH1, enhancer of zeste homolog 1; DNMT3B, DNA (cytosine-5-)-methyltransferase 3β; EZH2, enhancer of zeste homolog 2; PHF19, PHD finger protein 19; DNMT3A, DNA (cytosine-5-)-methyltransferase 3α; FDR, false discovery rate.
Figure 2.
PcG risk-score analysis of 183 patients and validation of the five-PcG signature for survival prediction by the CGGA and GSE16011 set. (A) The prognostic five-PcG signature risk-score distribution. (B) Survival status and time of patients. (C) Heat map of the five-PcG expression profiles. Rows represent PcG genes, and columns represent patients. Overall survival of (D) 183 patients in the CGGA set and (E) 270 patients in the GSE16011 set. PcG, polycomb group; CGGA, Chinese Glioma Genome Atlas; EZH1, enhancer of zeste homolog 1; EZH2, enhancer of zeste homolog 2; DNMT3A, DNA (cytosine-5-)-methyltransferase 3; DNMT3B, DNA (cytosine-5-)-methyltransferase 3α; PHF19, PHD finger protein 19.
The risk score formula, obtained from the training set, was then used to classify 183 patients in the CGGA set and 270 patients in the GSE16011 set into high- and low-risk groups, using the same cutoff point (the median risk score), and to predict their survival. In the CGGA set, the patients in the low-risk groups had longer overall survival than those in the high-risk groups (P<0.0001; Fig. 2D). Furthermore, in the GSE16011 data, the patients in the high-risk groups had shorter overall survival compared with those in the low-risk groups (P<0.0001; Fig. 2E).
To explore whether the PcG signature is an independent prognostic factor in patients with glioma, Cox's univariate regression analysis was conducted using the clinical characteristics of the CGGA and GSE16011 data. As shown in Table III, the high-risk score group was significantly associated with grades (P<0.0001) and not associated with gender distribution. In the CGGA set, the five-PcG signature [hazard ratio (HR), 1.109; P=0.033], grades (HR, 2.186; P<0.0001) and age (HR, 1.038; P<0.0001) were independent prognostic factors associated with overall survival. Similarly, the five-PcG signature (HR, 1.178; P<0.0001) and grades (HR, 2.786; P<0.0001) were also identified in the GSE16011 set.
Table III.
Clinical characteristics of the in CGGA and GSE16011 data set.
| Characteristic | Patients with high-risk PcG signature, % | Patients with low-risk PcG signature, % | P-value |
|---|---|---|---|
| A, CGGA data set (n=183) | |||
| Total, n | 92 | 91 | |
| Age, years (mean ± SD) | 38.25±10.90 | 45.78±12.99 | 0.015 |
| Gender | 0.549 | ||
| Male | 50 | 54 | |
| Female | 42 | 37 | |
| Grade | <0.0001 | ||
| Low | 59 | 4 | |
| High | |||
| III | 13 | 20 | |
| IV | 20 | 67 | |
| B, GSE16011 data set (n=270) | |||
| Total, n | 135 | 135 | |
| Age, years (mean ± SD) | 47.12±14.55 | 52.88±14.93 | 0.972 |
| Gender | 0.52 | ||
| Male | 92 | 87 | |
| Female | 43 | 48 | |
| Grade | <0.0001 | ||
| Low | 59 | 4 | |
| High | |||
| III | 13 | 20 | |
| IV | 20 | 67 | |
PcG, polycomb group; CGGA, Chinese glioma genome atlas; SD, standard deviation.
PcG signature can predict survival of patients within WHO grades and histological subgroups
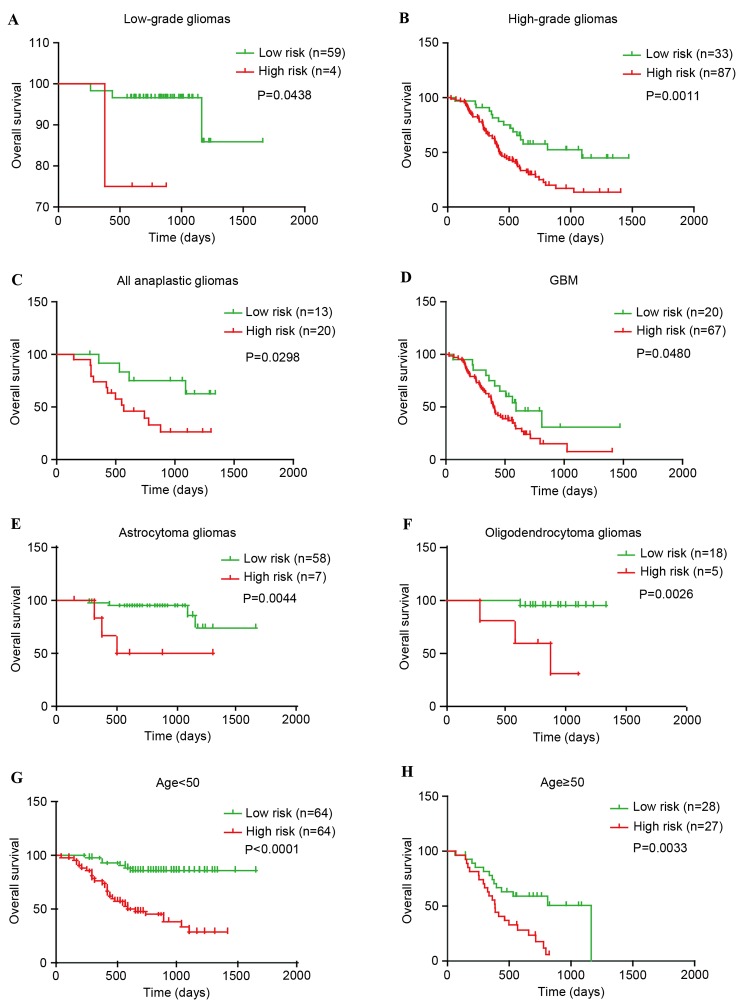
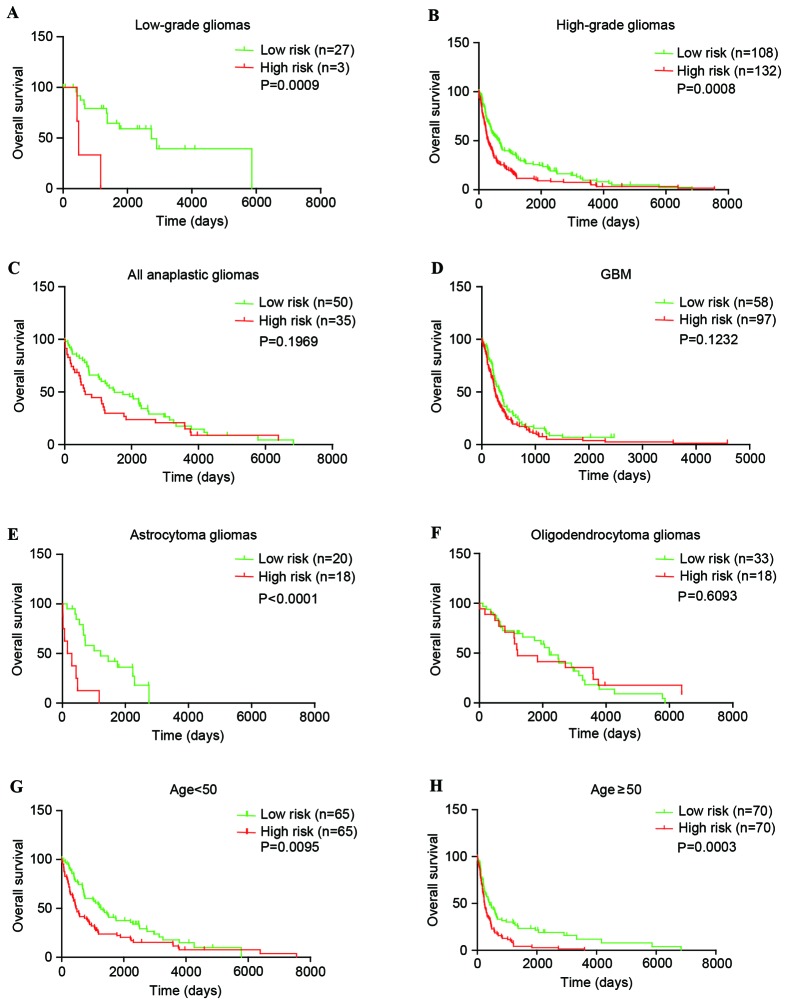
The present study explored whether the PcG signature can distinguish high-risk vs. low-risk groups of patients within each grade and predict their survival. In the CGGA set, for low-grade gliomas, the PcG signature was significantly associated with overall survival of patients (P=0.0438). For high-grade gliomas, the results were similar (P=0.0011). Furthermore, subsequent analysis revealed that the PcG signature was also significantly associated with overall survival of anaplastic and GBM patients (P=0.0298 and P=0.0408; Fig. 3A-D). In addition, similar results (P=0.0009, low-grade gliomas and P=0.0008, high-grade gliomas) were observed in the GSE16011 set. Although the P-value was >0.05, the high-risk survival curve lies below the low-risk curve in anaplastic and GBM patients (Fig. 4A-D).
Figure 3.
The five-polycomb group signature was tightly associated with prognosis within WHO grades, histological subgroups and each age group of gliomas in the Chinese glioma genome atlas set. (A) Low-grade gliomas (n=63). (B) High-grade gliomas (n=120). (C) Anaplastic gliomas (n=33). (D) GBM (n=87). (E) Astrocytoma gliomas (n=65). (F) Oligodendroglioma gliomas (n=23). (G) Younger group (age, <50 years; n=128). (H) Elder group (age, ≥50 years; n=55). GBM, glioblastoma; WHO, World Health Organization.
Figure 4.
The five-polycomb group signature was tightly associated with prognosis within WHO grades, histological subgroups and each age group of gliomas in the GSE16011 set. (A) Low-grade gliomas (n=30). (B) High-grade gliomas (n=240). (C) Anaplastic gliomas (n=85). (D) GBM (n=155). (E) Astrocytoma gliomas (n=38). (F) Oligodendroglioma gliomas (n=51). (G) Younger group (age, <50 years; n=130). (H) Elder group (age, ≥50 years; n=140). GBM, glioblastoma; WHO, World Health Organization.
The patients with glioma (with the exception of GBM patients) were then stratified using the histological subtype of O and A. In the CGGA set, the PcG signature was significantly associated with overall survival of O and A patients (P=0.0026 and P=0.0044; Fig. 3E-F). However, in the GSE16011 set, the results were complex (P<0.0001 for A and P=0.6093 for O; Fig. 4E-F). These findings indicated that the PcG signature may almost predict patient survival within WHO grades and histological subgroups.
Finally, additional analysis was conducted on the association between the PcG signature and patient age. The entire CGGA patients (n=183; Fig. 3G-H) and GSE16011 patients (n=270; Fig. 4G-H) were stratified into an elder group (age >50) or a younger group (age ≤50). Within each age group, the high-risk survival curve lay below the low-risk curve.
Discussion
In mammals, PcG proteins play an important role in numerous aspects of development, whereas PcG deregulation may lead to oncogenesis (15). Our previous study reported that EZH2 is a negative prognostic factor and exhibits pro-oncogenic activity in GBM (15). BMI1 and CBX7 have been reported as oncogenes or tumor suppressor genes in different tumors (17–20). Nevertheless, the mechanism for PcG gene mediating gene repression remains unknown. Thus, to assess the role of PcG genes in gliomas, the CGGA data was analyzed for aberrant expression of all PRC1, PRC2 and PRC2-associated genes.
The survey revealed that CBX6, CBX7 and PHF1 were downregulated with ascending malignancy grades in glioma. CBX7 was the primary CBX protein assembled in PRC1, which was reported to be decreased in the majority of human malignant neoplasia, including thyroid (21), pancreatic (19), colon (22), lung (23), gastric (24), bladder (25) and breast (26) carcinomas. In addition, upregulation of CBX7 expression may affect the cell cycle, cell proliferation and the epithelial mesenchymal transition in carcinoma cells of different origins (19,27,28). PHF1 is a component of the PRC2 complex, which is essential for H3K27 methylation and Hox gene expression (29). Results of the present study revealed that PHF1 expression was significantly downregulated, and that PHF1 may function as a tumor suppressor in glioma (P<0.01). CBX6 is a polycomb group protein and a component of the PRC1 complex (30). However, few studies showed expression changes of CBX6 in tumors (10,31,32).
An additional finding was that EZH2, DNMT3B and PHC2 were increased with the elevation of glioma malignancy. EZH2 was overexpressed and reported to be an important prognostic marker in several human cancers, including ovarian carcinoma (33), gastric cancer (6) and glioma (15). In GBM, EZH2 bound to signal transducer and activator of transcription 3 (STAT3), leading to enhanced STAT3 activity and promoted tumorigenicity of glioblastoma stem-like cells (34). In colon, gastric, breast and prostate cancers, EZH2 repressed E-cadherin through histone H3 Lys27 trimethylation, consequently promoting tumor progression, invasiveness and metastasis (35,36). DNMT3B is one of the three known DNA methyltransferases with catalytic activity, and is responsible for the de novo methylation of DNA (37). DNMT3B level is increased in various cancer tissues and cell lines, indicating that it has an important role in tumorigenesis (38). PHC2 is a component of PRC1, which is expressed as two isoforms (phc2a and phc2b) and may be associated with CBX proteins (39). However, the role of PHC2 in cancer has not been investigated clearly.
Furthermore, the five-PcG genes (EZH1, EZH2, PHF19, DNMT3A and DNMT3B) that were significantly associated with patient survival were identified. It was then identified that the five-PcG signature was an independent prognostic factor, and it can predict patient survival time within WHO grades and histological subgroups.
In summary, the present study demonstrated that PcG may play a critical role during progression from normal brain tissues to high-grade gliomas. Furthermore, these findings highlight the potential value of the five-PcG signature as a useful prognostic biomarker and therapeutic target.
Acknowledgements
The present study was supported by grants from the National High Technology Research and Development Program of China (863) (grant no. 2012AA02A508), the National Natural Science Foundation of China (grant nos. 81472362, 81372709 and 81302185), Jiangsu Province's Natural Science Foundation (grant no. 20131019) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD.
References
- 1.Mathews LA, Crea F, Farrar WL. Epigenetic gene regulation in stem cells and correlation to cancer. Differentiation. 2009;78:1–17. doi: 10.1016/j.diff.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 3.Gil J, O'Loghlen A. PRC1 complex diversity: Where is it taking us? Trends Cell Biol. 2014;24:632–641. doi: 10.1016/j.tcb.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/S0959-437X(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 5.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsukawa Y, Semba S, Kato H, Ito A, Yanagihara K, Yokozaki H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva J, García V, García JM, Peña C, Domínguez G, Díaz R, Lorenzo Y, Hurtado A, Sánchez A, Bonilla F. Circulating Bmi-1 mRNA as a possible prognostic factor for advanced breast cancer patients. Breast Cancer Res. 2007;9:R55. doi: 10.1186/bcr1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li DW, Tang HM, Fan JW, Yan DW, Zhou CZ, Li SX, Wang XL, Peng ZH. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. J Cancer Res Clin Oncol. 2010;136:997–1006. doi: 10.1007/s00432-009-0745-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Luo XD, Da CL, Xin Y. Clinicopathological significance of B-cell-specific Moloney murine leukemia virus insertion site 1 expression in gastric carcinoma and its precancerous lesion. World J Gastroenterol. 2009;15:2145–2150. doi: 10.3748/wjg.15.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, Warden C, Zou Z, Neman J, Krueger JS, Jain A, Jandial R, Chen M. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One. 2013;8:e80970. doi: 10.1371/journal.pone.0080970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan W, Zhang W, You G, Zhang J, Han L, Bao Z, Wang Y, Liu Y, Jiang C, Kang C, et al. Molecular classification of gliomas based on whole genome gene expression: A systematic report of 225 samples from the Chinese glioma cooperative group. Neuro Oncol. 2012;14:1432–1440. doi: 10.1093/neuonc/nos263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB, Kloosterhof NK, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 13.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Q, Sun J. Cox survival analysis of microarray gene expression data using correlation principal component regression. Stat Appl Genet Mol Biol. 2007;6:Article 16. doi: 10.2202/1544-6115.1153. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Qin JJ, Voruganti S, Nag S, Zhou J, Zhang R. Polycomb group (PcG) proteins and human cancers: Multifaceted functions and therapeutic implications. Med Res Rev. 2015;35:1220–1267. doi: 10.1002/med.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cenci T, Martini M, Montano N, D'Alessandris QG, Falchetti ML, Annibali D, Savino M, Bianchi F, Pierconti F, Nasi S, et al. Prognostic relevance of c-Myc and BMI1 expression in patients with glioblastoma. Am J Clin Pathol. 2012;138:390–396. doi: 10.1309/AJCPRXHNJQLO09QA. [DOI] [PubMed] [Google Scholar]
- 18.Xu CR, Lee S, Ho C, Bommi P, Huang SA, Cheung ST, Dimri GP, Chen X. Bmi1 functions as an oncogene independent of Ink4A/Arf repression in hepatic carcinogenesis. Mol Cancer Res. 2009;7:1937–1945. doi: 10.1158/1541-7786.MCR-09-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karamitopoulou E, Pallante P, Zlobec I, Tornillo L, Carafa V, Schaffner T, Borner M, Diamantis I, Esposito F, Brunner T, et al. Loss of the CBX7 protein expression correlates with a more aggressive phenotype in pancreatic cancer. Eur J Cancer. 2010;46:1438–1444. doi: 10.1016/j.ejca.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 20.Scott CL, Gil J, Hernando E, Teruya-Feldstein J, Narita M, Martínez D, Visakorpi T, Mu D, Cordon-Cardo C, Peters G, et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc Natl Acad Sci USA. 2007;104:5389–5394. doi: 10.1073/pnas.0608721104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pallante P, Federico A, Berlingieri MT, Bianco M, Ferraro A, Forzati F, Iaccarino A, Russo M, Pierantoni GM, Leone V, et al. Loss of the CBX7 gene expression correlates with a highly malignant phenotype in thyroid cancer. Cancer Res. 2008;68:6770–6778. doi: 10.1158/0008-5472.CAN-08-0695. [DOI] [PubMed] [Google Scholar]
- 22.Pallante P, Terracciano L, Carafa V, Schneider S, Zlobec I, Lugli A, Bianco M, Ferraro A, Sacchetti S, Troncone G, et al. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur J Cancer. 2010;46:2304–2313. doi: 10.1016/j.ejca.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Forzati F, Federico A, Pallante P, Abbate A, Esposito F, Malapelle U, Sepe R, Palma G, Troncone G, Scarfò M, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122:612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, Zhang Y. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45:17–23. doi: 10.1007/s00535-009-0135-6. [DOI] [PubMed] [Google Scholar]
- 25.Hinz S, Kempkensteffen C, Christoph F, Krause H, Schrader M, Schostak M, Miller K, Weikert S. Expression parameters of the polycomb group proteins BMI1, SUZ12, RING1 and CBX7 in urothelial carcinoma of the bladder and their prognostic relevance. Tumour Biol. 2008;29:323–329. doi: 10.1159/000170879. [DOI] [PubMed] [Google Scholar]
- 26.Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, Esposito F, Iaccarino A, Troncone G, Fusco A. Identification of a new pathway for tumor progression: MicroRNA-181b up-regulation and CBX7 down-regulation by HMGA1 protein. Genes Cancer. 2010;1:210–224. doi: 10.1177/1947601910366860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forzati F, Federico A, Pallante P, Fedele M, Fusco A. Tumor suppressor activity of CBX7 in lung carcinogenesis. Cell Cycle. 2012;11:1888–1891. doi: 10.4161/cc.20022. [DOI] [PubMed] [Google Scholar]
- 28.Zhang XW, Zhang L, Qin W, Yao XH, Zheng LZ, Liu X, Li J, Guo WJ. Oncogenic role of the chromobox protein CBX7 in gastric cancer. J Exp Clin Cancer Res. 2010;29:114. doi: 10.1186/1756-9966-29-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Wang C, Zhang P, Gao K, Wang D, Yu H, Zhang T, Jiang S, Hexige S, Hong Z, et al. Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis. J Biol Chem. 2013;288:529–539. doi: 10.1074/jbc.M111.338996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 31.Coradini D, Boracchi P, Oriana S, Biganzoli E, Ambrogi F. Differential expression of genes involved in the epigenetic regulation of cell identity in normal human mammary cell commitment and differentiation. Chin J Cancer. 2014;33:501–510. doi: 10.5732/cjc.014.10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crea F, Di Paolo A, Liu HH, Polillo M, Clermont PL, Guerrini F, Ciabatti E, Ricci F, Baratè C, Fontanelli G, et al. Polycomb genes are associated with response to imatinib in chronic myeloid leukemia. Epigenomics. 2015;7:757–765. doi: 10.2217/epi.15.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF, Liao YJ, Deng HX, Chen YC, Guan XY, et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31:1576–1583. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- 34.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herranz N, Pasini D, Díaz VM, Francí C, Gutierrez A, Dave N, Escrivà M, Hernandez-Muñoz I, Di Croce L, Helin K, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du J, Li L, Ou Z, Kong C, Zhang Y, Dong Z, Zhu S, Jiang H, Shao Z, Huang B, Lu J. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast Cancer Res Treat. 2012;131:65–73. doi: 10.1007/s10549-011-1396-3. [DOI] [PubMed] [Google Scholar]
- 37.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 38.Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- 39.Cho KW, Bae J, Lee SJ, Chun T. Expression pattern and functional role of Phc2 during activation of helper T cells after antigenic stimulation. In Vitro Cell Dev Biol Anim. 2013;49:360–370. doi: 10.1007/s11626-013-9618-0. [DOI] [PubMed] [Google Scholar]